Metabolomic Analysis of Pediatric Patients with Idiosyncratic Drug-Induced Liver Injury According to the Updated RUCAM
Abstract
:1. Introduction
2. Results
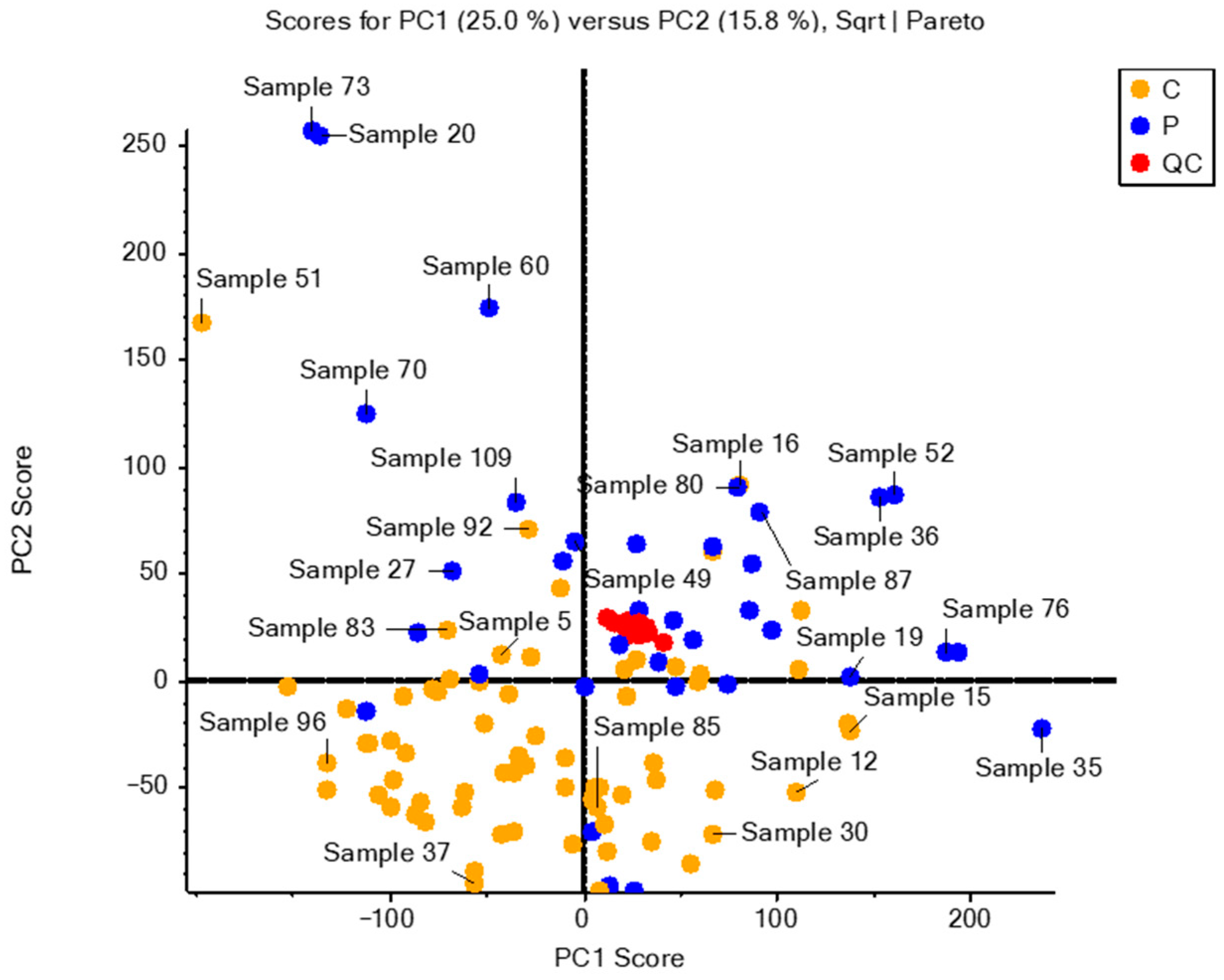
2.1. Data Matrix and Analytical Validation
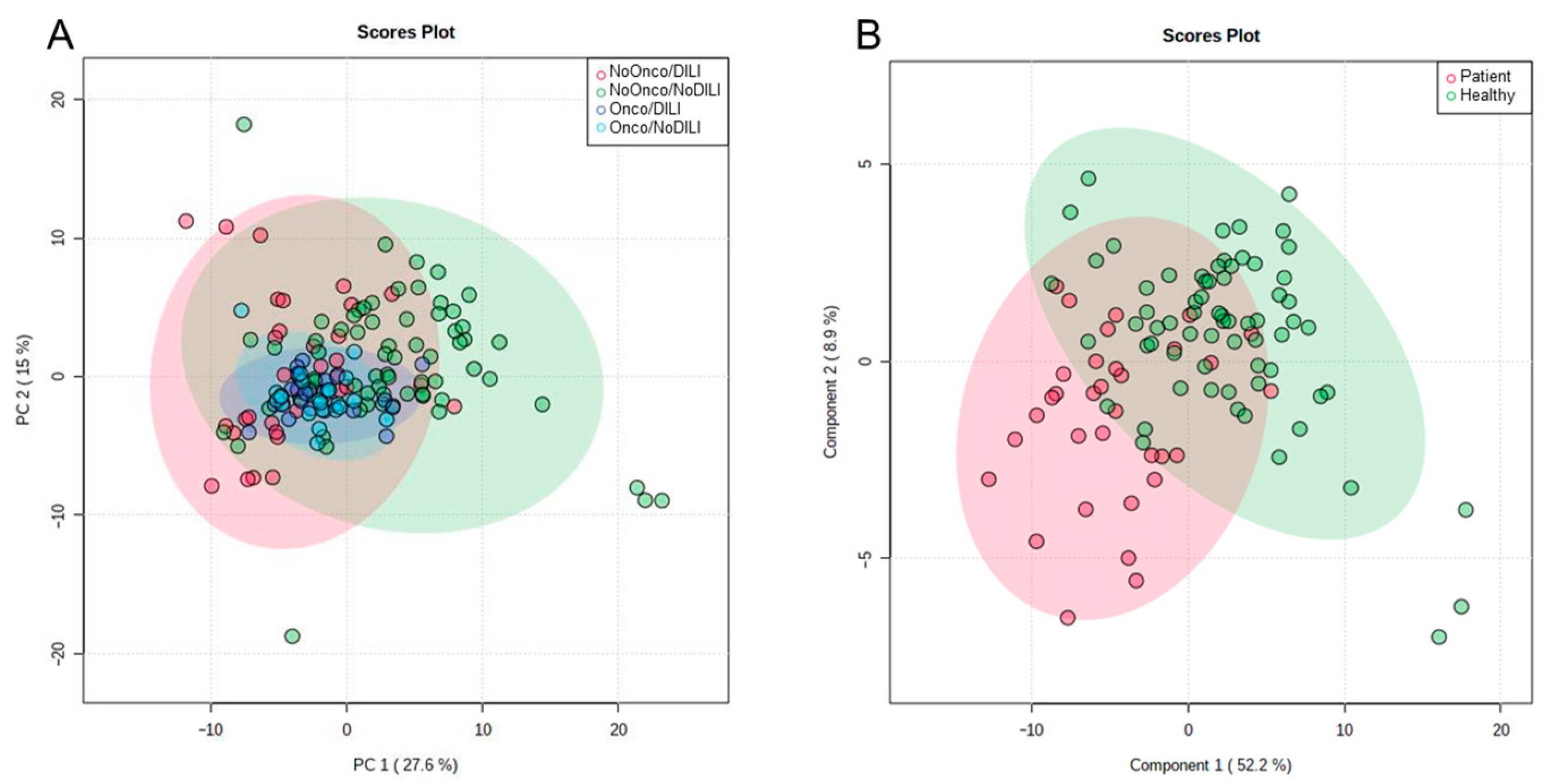
2.2. Statistical Analysis
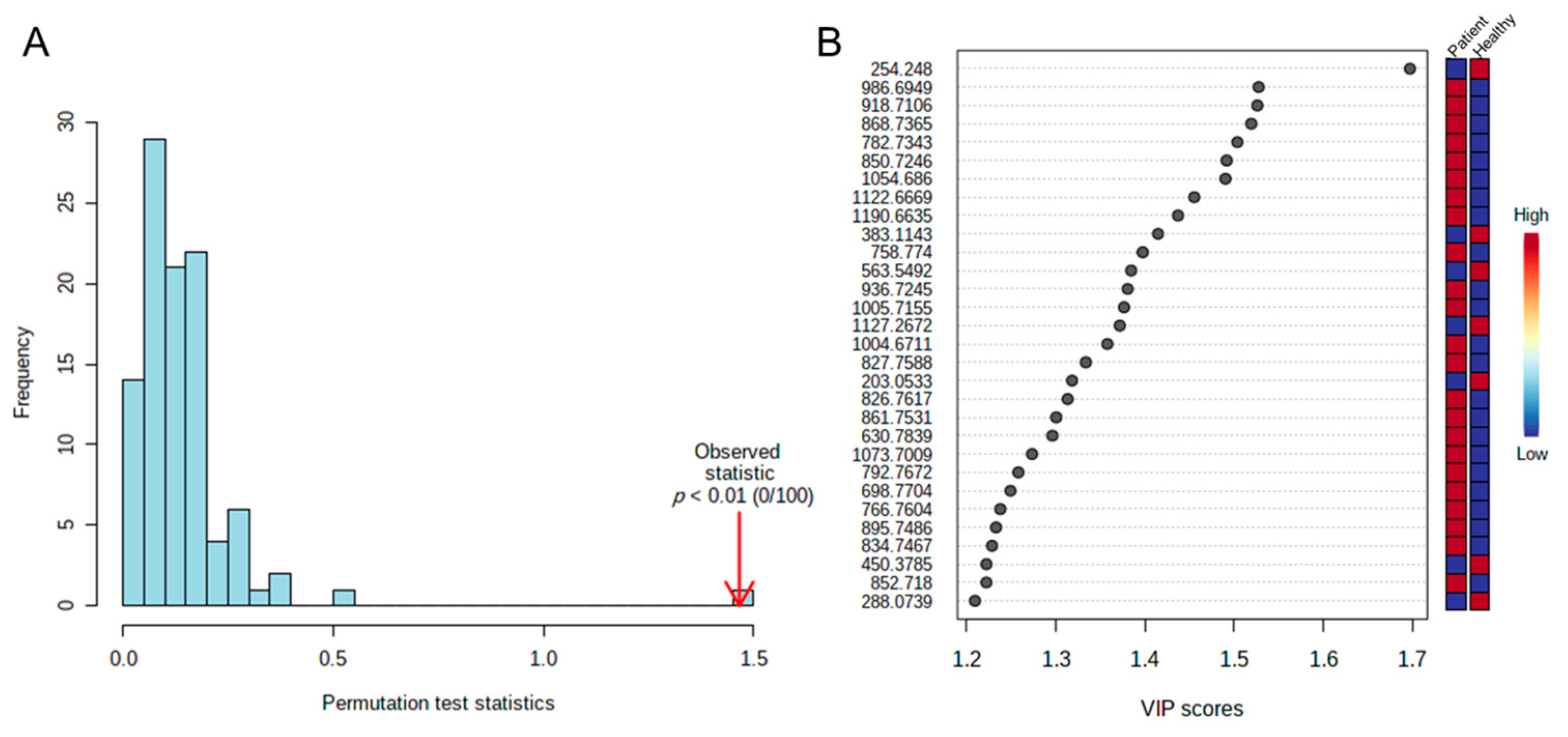
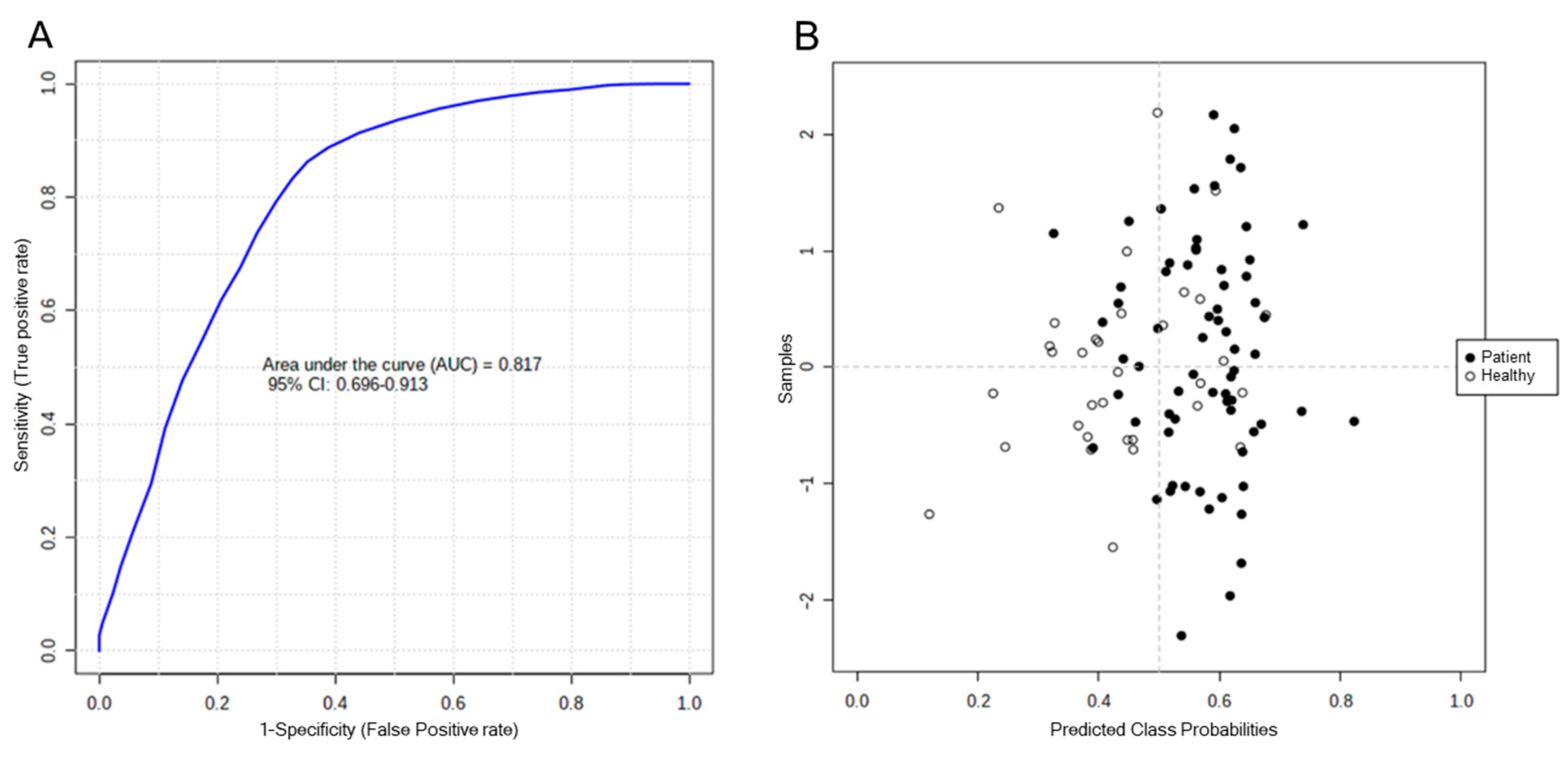
2.3. Identifying Significant Variables
2.4. Analysis of Biological Pathways
2.5. Analysis of Biomarkers
3. Discussion
4. Materials and Methods
4.1. Study Population
- (a)
- The criteria for diagnosing DILI among the patients at the time of their inclusion in the study include a chronological relationship between drug intake and the onset of hepatitis, and the presence of any of the following conditions: (1) level of alanine aminotransferase (ALT) more than five times the upper limit of normality (ULN); (2) level of alkaline phosphatase (ALP) more than double the ULN; (3) level of ALT more than three times the ULN, with concomitant elevation of bilirubin levels to more than double the ULN. The pattern of liver injury can be assessed by the R value, where R = (ALT/ULN)/(ALP/ULN); R ≥ 5 reflects a hepatocellular pattern, 2 < R < 5 reflects a mixed pattern, and R ≤ 2 reflects a cholestatic pattern [61].
- (b)
- If one of the above red flags is detected, cases are reported using a structured protocol to exclude possible alternative causes. A detailed medical history is obtained from all patients regarding biliary or liver disease, as well as information on risk factors associated with liver disease. Serological markers of acute viral hepatitis are determined in all patients prior to diagnosis of DILI, together with serum ceruloplasmin and a battery of autoantibodies related to autoimmune liver disease.
- (c)
- Causality is determined by the RUCAM evaluation method [61].
- (d)
- In every case, a serum sample is extracted for the metabolomic study, at a time selected during the follow-up to coincide with the scheduled clinical control visits.
4.2. Ethics Approval and Consent to Participate
4.3. Preparation of the Plasma Samples
4.4. Analysis by Liquid Chromatography Coupled with High-Resolution Mass Spectrometry (HPLC–MS/MS)
4.5. Data Matrix for Statistical Analysis and Analytical Validation
4.6. Statistical Analysis
4.7. Identifying Significant Variables
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teschke, R. Idiosyncratic DILI: Analysis of 46,266 Cases Assessed for Causality by RUCAM and Published From 2014 to Early 2019. Front. Pharmacol. 2019, 10, 730. [Google Scholar] [CrossRef] [PubMed]
- Teschke, R.; Uetrecht, J. Mechanism of Idiosyncratic Drug Induced Liver Injury (DILI): Unresolved Basic Issues. Ann. Transl. Med. 2021, 9, 730. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Yang, X.; Greenhaw, J.J.; Salminen, A.T.; Russotti, G.M.; Salminen, W.F. Drug-Induced Liver Injury in Children: Clinical Observations, Animal Models, and Regulatory Status. Int. J. Toxicol. 2017, 36, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.D.; Harpavat, S.; Leung, D.H. Drug-Induced Liver Injury in Children. Curr. Opin. Pediatr. 2015, 27, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Feng, Z.; Huang, L.; Guo, C.; Wu, X.; He, L.; Tan, W.; Wang, Y.; Wu, X.; Hu, B.; et al. Causality Evaluation of Drug-Induced Liver Injury in Newborns and Children in the Intensive Care Unit Using the Updated Roussel Uclaf Causality Assessment Method. Front. Pharmacol. 2021, 12, 790108. [Google Scholar] [CrossRef]
- Kuna, L.; Bozic, I.; Kizivat, T.; Bojanic, K.; Mrso, M.; Kralj, E.; Smolic, R.; Wu, G.Y.; Smolic, M. Models of Drug Induced Liver Injury (DILI)—Current Issues and Future Perspectives. Curr. Drug Metab. 2018, 19, 830–838. [Google Scholar] [CrossRef]
- Andrade, R.J.; Aithal, G.P.; Björnsson, E.S.; Kaplowitz, N.; Kullak-Ublick, G.A.; Larrey, D.; Karlsen, T.H. EASL Clinical Practice Guidelines: Drug-Induced Liver Injury. J. Hepatol. 2019, 70, 1222–1261. [Google Scholar] [CrossRef]
- DiPaola, F.; Molleston, J.P.; Gu, J.; Cirulli, E.T.; Chalasani, N.; Barnhart, H.; Kleiner, D.E.; Hoofnagle, J.H.; Fontana, R.J. US Drug Induced Liver Injury Network Antimicrobials and Antiepileptics Are the Leading Causes of Idiosyncratic Drug-Induced Liver Injury in American Children. J. Pediatr. Gastroenterol. Nutr. 2019, 69, 152–159. [Google Scholar] [CrossRef]
- Ocete Hita, E.; Martín García, J.A.; Giménez Sánchez, F.; Flores González, J.C.; Abril Molina, A.; Salmerón Escobar, J.; Ruiz Extremera, A. Hepatotoxicity due to drugs or natural products in children. An. Pediatr. (Barc) 2013, 78, 248–259. [Google Scholar] [CrossRef]
- Hey-Hadavi, J.; Seekins, D.; Palmer, M.; Coffey, D.; Caminis, J.; Abdullaev, S.; Patwardhan, M.; Tyler, H.; Raheja, R.; Stanley, A.M.; et al. Overview of Causality Assessment for Drug-Induced Liver Injury (DILI) in Clinical Trials. Drug Saf. 2021, 44, 619–634. [Google Scholar] [CrossRef]
- Hosack, T.; Damry, D.; Biswas, S. Drug-Induced Liver Injury: A Comprehensive Review. Ther. Adv. Gastroenterol. 2023, 16, 17562848231163410. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Mao, X.; Zhou, Z.; Huai, C.; Li, Z. Research Progress of Pharmacogenomics in Drug-Induced Liver Injury. Front. Pharmacol. 2021, 12, 735260. [Google Scholar] [CrossRef]
- Mattes, W.; Davis, K.; Fabian, E.; Greenhaw, J.; Herold, M.; Looser, R.; Mellert, W.; Groeters, S.; Marxfeld, H.; Moeller, N.; et al. Detection of Hepatotoxicity Potential with Metabolite Profiling (Metabolomics) of Rat Plasma. Toxicol. Lett. 2014, 230, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Robles-Díaz, M.; Medina-Caliz, I.; Stephens, C.; Andrade, R.J.; Lucena, M.I. Biomarkers in DILI: One More Step Forward. Front. Pharmacol. 2016, 7, 267. [Google Scholar] [CrossRef]
- Araújo, A.M.; Carvalho, M.; Carvalho, F.; de Bastos, M.L.; Guedes de Pinho, P. Metabolomic Approaches in the Discovery of Potential Urinary Biomarkers of Drug-Induced Liver Injury (DILI). Crit. Rev. Toxicol. 2017, 47, 638–654. [Google Scholar] [CrossRef]
- García-Cañaveras, J.C.; Jiménez, N.; Gómez-Lechón, M.J.; Castell, J.V.; Donato, M.T.; Lahoz, A. LC-MS Untargeted Metabolomic Analysis of Drug-Induced Hepatotoxicity in HepG2 Cells. Electrophoresis 2015, 36, 2294–2302. [Google Scholar] [CrossRef]
- Iruzubieta, P.; Arias-Loste, M.T.; Barbier-Torres, L.; Martinez-Chantar, M.L.; Crespo, J. The Need for Biomarkers in Diagnosis and Prognosis of Drug-Induced Liver Disease: Does Metabolomics Have Any Role? BioMed Res. Int. 2015, 2015, e386186. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Xu, Q. Chapter 11: Metabolomics investigations of drug-induced hepatotoxicity. In Metabolic Profiling: Disease and Xenobiotics; Issues in Toxicology 21; Grootveld, M., Ed.; The Chemical Society: London, UK, 2015; pp. 323–356. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Teschke, R. Top-Ranking Drugs out of 3312 Drug-Induced Liver Injury Cases Evaluated by the Roussel Uclaf Causality Assessment Method. Expert Opin. Drug Metab. Toxicol. 2018, 14, 1169–1187. [Google Scholar] [CrossRef]
- Teschke, R.; Danan, G. Idiosyncratic Drug-Induced Liver Injury (DILI) and Herb-Induced Liver Injury (HILI): Diagnostic Algorithm Based on the Quantitative Roussel Uclaf Causality Assessment Method (RUCAM). Diagnostics 2021, 11, 458. [Google Scholar] [CrossRef]
- Gonzalez, E.; van Liempd, S.; Conde-Vancells, J.; Gutierrez-de Juan, V.; Perez-Cormenzana, M.; Mayo, R.; Berisa, A.; Alonso, C.; Marquez, C.A.; Barr, J.; et al. Serum UPLC-MS/MS Metabolic Profiling in an Experimental Model for Acute-Liver Injury Reveals Potential Biomarkers for Hepatotoxicity. Metabolomics 2012, 8, 997–1011. [Google Scholar] [CrossRef]
- Kumar, B.S.; Chung, B.C.; Kwon, O.-S.; Jung, B.H. Discovery of Common Urinary Biomarkers for Hepatotoxicity Induced by Carbon Tetrachloride, Acetaminophen and Methotrexate by Mass Spectrometry-Based Metabolomics. J. Appl. Toxicol. JAT 2012, 32, 505–520. [Google Scholar] [CrossRef]
- Li, Z.; Li, Q.; Geng, L.; Chen, X.; Bi, K. Use of the Local False Discovery Rate for Identification of Metabolic Biomarkers in Rat Urine Following Genkwa Flos-Induced Hepatotoxicity. PLoS ONE 2013, 8, e67451. [Google Scholar] [CrossRef]
- Lu, C.; Wang, Y.; Sheng, Z.; Liu, G.; Fu, Z.; Zhao, J.; Zhao, J.; Yan, X.; Zhu, B.; Peng, S. NMR-Based Metabonomic Analysis of the Hepatotoxicity Induced by Combined Exposure to PCBs and TCDD in Rats. Toxicol. Appl. Pharmacol. 2010, 248, 178–184. [Google Scholar] [CrossRef]
- Buness, A.; Roth, A.; Herrmann, A.; Schmitz, O.; Kamp, H.; Busch, K.; Suter, L. Identification of Metabolites, Clinical Chemistry Markers and Transcripts Associated with Hepatotoxicity. PLoS ONE 2014, 9, e97249. [Google Scholar] [CrossRef]
- Conotte, R.; Colet, J.-M. A Metabonomic Evaluation of the Monocrotaline-Induced Sinusoidal Obstruction Syndrome (SOS) in Rats. Toxicol. Appl. Pharmacol. 2014, 276, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Cai, S.; Wang, L.; Zhao, N.; Zhang, T.; Chen, Z.; Meng, F. A Metabonomic Analysis of Serum from Rats Treated with Ricinine Using Ultra Performance Liquid Chromatography Coupled with Mass Spectrometry. PLoS ONE 2014, 9, e90416. [Google Scholar] [CrossRef]
- Rodríguez-Suárez, E.; Gonzalez, E.; Hughes, C.; Conde-Vancells, J.; Rudella, A.; Royo, F.; Palomo, L.; Elortza, F.; Lu, S.C.; Mato, J.M.; et al. Quantitative Proteomic Analysis of Hepatocyte-Secreted Extracellular Vesicles Reveals Candidate Markers for Liver Toxicity. J. Proteom. 2014, 103, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Slavov, S.; Schnackenberg, L.K.; Ando, Y.; Greenhaw, J.; Yang, X.; Salminen, W.; Mendrick, D.L.; Beger, R. Identification of a Metabolic Biomarker Panel in Rats for Prediction of Acute and Idiosyncratic Hepatotoxicity. Comput. Struct. Biotechnol. J. 2014, 10, 78–89. [Google Scholar] [CrossRef]
- Van den Hof, W.F.P.M.; Ruiz-Aracama, A.; Van Summeren, A.; Jennen, D.G.J.; Gaj, S.; Coonen, M.L.J.; Brauers, K.; Wodzig, W.K.W.H.; van Delft, J.H.M.; Kleinjans, J.C.S. Integrating Multiple Omics to Unravel Mechanisms of Cyclosporin A Induced Hepatotoxicity in Vitro. Toxicol. Vitr. 2015, 29, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Jeong, E.S.; Kim, G.; Shin, H.J.; Park, S.-M.; Oh, J.-H.; Kim, Y.-B.; Moon, K.-S.; Choi, H.-K.; Jeong, J.; Shin, J.-G.; et al. Increased Serum Bile Acid Concentration Following Low-Dose Chronic Administration of Thioacetamide in Rats, as Evidenced by Metabolomic Analysis. Toxicol. Appl. Pharmacol. 2015, 288, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Mast, C.; Lyan, B.; Joly, C.; Centeno, D.; Giacomoni, F.; Martin, J.-F.; Mosoni, L.; Dardevet, D.; Pujos-Guillot, E.; Papet, I. Assessment of Protein Modifications in Liver of Rats under Chronic Treatment with Paracetamol (Acetaminophen) Using Two Complementary Mass Spectrometry-Based Metabolomic Approaches. J. Proteom. 2015, 120, 194–203. [Google Scholar] [CrossRef]
- Van den Hof, W.F.P.M.; Van Summeren, A.; Lommen, A.; Coonen, M.L.J.; Brauers, K.; van Herwijnen, M.; Wodzig, W.K.W.H.; Kleinjans, J.C.S. Integrative Cross-Omics Analysis in Primary Mouse Hepatocytes Unravels Mechanisms of Cyclosporin A-Induced Hepatotoxicity. Toxicology 2014, 324, 18–26. [Google Scholar] [CrossRef]
- Xiong, Y.-H.; Xu, Y.; Yang, L.; Wang, Z.-T. Gas Chromatography–Mass Spectrometry-Based Profiling of Serum Fatty Acids in Acetaminophen-Induced Liver Injured Rats. J. Appl. Toxicol. 2014, 34, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Yan, K.; Pence, L.; Simpson, P.M.; Gill, P.; Letzig, L.G.; Beger, R.D.; Sullivan, J.E.; Kearns, G.L.; Reed, M.D.; et al. Targeted Liquid Chromatography-Mass Spectrometry Analysis of Serum Acylcarnitines in Acetaminophen Toxicity in Children. Biomark. Med. 2014, 8, 147–159. [Google Scholar] [CrossRef]
- Andrew Clayton, T.; Lindon, J.C.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.-P.; Le Net, J.-L.; Baker, D.; Walley, R.J.; et al. Pharmaco-Metabonomic Phenotyping and Personalized Drug Treatment. Nature 2006, 440, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Shao, L.; Gong, Y.; Mao, Y.; Liu, C.; Qu, H.; Cheng, Y. A Metabonomic Characterization of CCl4-Induced Acute Liver Failure Using Partial Least Square Regression Based on the GC/MS Metabolic Profiles of Plasma in Mice. J. Chromatogr. B 2008, 870, 178–185. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, R.; Liu, L.; Peng, J.; Xiao, B.; Yang, J.; Miao, Z.; Huang, H. Metabonomics Study of Urine from Sprague–Dawley Rats Exposed to Huang-Yao-Zi Using 1H NMR Spectroscopy. J. Pharm. Biomed. Anal. 2010, 52, 136–141. [Google Scholar] [CrossRef]
- Lu, X.; Hu, B.; Shao, L.; Tian, Y.; Jin, T.; Jin, Y.; Ji, S.; Fan, X. Integrated Analysis of Transcriptomics and Metabonomics Profiles in Aflatoxin B1-Induced Hepatotoxicity in Rat. Food Chem. Toxicol. 2013, 55, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Shima, N.; Miyawaki, I.; Bando, K.; Horie, H.; Zaitsu, K.; Katagi, M.; Bamba, T.; Tsuchihashi, H.; Fukusaki, E. Influences of Methamphetamine-Induced Acute Intoxication on Urinary and Plasma Metabolic Profiles in the Rat. Toxicology 2011, 287, 29–37. [Google Scholar] [CrossRef]
- Winnike, J.H.; Li, Z.; Wright, F.A.; Macdonald, J.M.; O’Connell, T.M.; Watkins, P.B. Use of Pharmaco-Metabonomics for Early Prediction of Acetaminophen-Induced Hepatotoxicity in Humans. Clin. Pharmacol. Ther. 2010, 88, 45–51. [Google Scholar] [CrossRef]
- Ma, H.; Patti, M.E. Bile Acids, Obesity, and the Metabolic Syndrome. Best Pract. Res. Clin. Gastroenterol. 2014, 28, 573–583. [Google Scholar] [CrossRef]
- Manco, M. Insulin Resistance and NAFLD: A Dangerous Liaison beyond the Genetics. Children 2017, 4, 74. [Google Scholar] [CrossRef]
- Dasarathy, S.; Yang, Y.; McCullough, A.J.; Marczewski, S.; Bennett, C.; Kalhan, S.C. Elevated Hepatic Fatty Acid Oxidation, High Plasma Fibroblast Growth Factor 21, and Fasting Bile Acids in Nonalcoholic Steatohepatitis. Eur. J. Gastroenterol. Hepatol. 2011, 23, 382–388. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma Metabolomic Profile in Nonalcoholic Fatty Liver Disease. Metabolism 2011, 60, 404–413. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Ratziu, V.; Loomba, R.; Rinella, M.; Anstee, Q.M.; Goodman, Z.; Bedossa, P.; Geier, A.; Beckebaum, S.; Newsome, P.N.; et al. Obeticholic Acid for the Treatment of Non-Alcoholic Steatohepatitis: Interim Analysis from a Multicentre, Randomised, Placebo-Controlled Phase 3 Trial. Lancet 2019, 394, 2184–2196. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, E.; Ouyang, X.; Xu, X.; Ma, S.; Ji, F.; Wu, D.; Zhang, S.; Zhao, Y.; Li, L. Metabolomics and Cytokine Analysis for Identification of Severe Drug-Induced Liver Injury. J. Proteome Res. 2019, 18, 2514–2524. [Google Scholar] [CrossRef]
- He, X.; Zhou, M.-X.; Cheng, C.; Li, S.-S.; Gao, Y.; Ma, Z.-T.; Song, X.-H.; Bai, Z.-F.; Zou, Z.-S.; Xiao, X.-H.; et al. Metabolomic Profiling for Histologically Fibrotic Stage in Chronic Drug-Induced Liver Injury. Front. Pharmacol. 2022, 13, 896198. [Google Scholar] [CrossRef]
- Zhao, S.; Fu, H.; Zhou, T.; Cai, M.; Huang, Y.; Gan, Q.; Zhang, C.; Qian, C.; Wang, J.; Zhang, Z.; et al. Alteration of Bile Acids and Omega-6 PUFAs Are Correlated with the Progression and Prognosis of Drug-Induced Liver Injury. Front. Immunol. 2022, 13, 772368. [Google Scholar] [CrossRef]
- Wang, M.-G.; Wu, S.-Q.; Zhang, M.-M.; He, J.-Q. Urine Metabolomics and Microbiome Analyses Reveal the Mechanism of Anti-Tuberculosis Drug-Induced Liver Injury, as Assessed for Causality Using the Updated RUCAM: A Prospective Study. Front. Immunol. 2022, 13, 1002126. [Google Scholar] [CrossRef]
- Shen, R.; Zhou, Y.; Zhang, L.; Yang, S. The Value of Bile Acid Spectrum in the Evaluation of Hepatic Injury in Children with Infectious Mononucleosis Caused by Epstein Barr Virus Infection. Front. Pediatr. 2023, 11, 1109762. [Google Scholar] [CrossRef]
- Alotaibi, S.A.; Alanazi, A.; Bakheet, S.A.; Alharbi, N.O.; Nagi, M.N. Prophylactic and Therapeutic Potential of Acetyl-L-Carnitine against Acetaminophen-Induced Hepatotoxicity in Mice. J. Biochem. Mol. Toxicol. 2016, 30, 5–11. [Google Scholar] [CrossRef]
- Bodaghi-Namileh, V.; Sepand, M.R.; Omidi, A.; Aghsami, M.; Seyednejad, S.A.; Kasirzadeh, S.; Sabzevari, O. Acetyl-l-Carnitine Attenuates Arsenic-Induced Liver Injury by Abrogation of Mitochondrial Dysfunction, Inflammation, and Apoptosis in Rats. Environ. Toxicol. Pharmacol. 2018, 58, 11–20. [Google Scholar] [CrossRef]
- Lheureux, P.E.R.; Penaloza, A.; Zahir, S.; Gris, M. Science Review: Carnitine in the Treatment of Valproic Acid-Induced Toxicity—What Is the Evidence? Crit. Care 2005, 9, 431–440. [Google Scholar] [CrossRef]
- Malaguarnera, M.; Gargante, M.P.; Russo, C.; Antic, T.; Vacante, M.; Malaguarnera, M.; Avitabile, T.; Li Volti, G.; Galvano, F. L-Carnitine Supplementation to Diet: A New Tool in Treatment of Nonalcoholic Steatohepatitis—A Randomized and Controlled Clinical Trial. Am. J. Gastroenterol. 2010, 105, 1338–1345. [Google Scholar] [CrossRef]
- Hamza, R.T.; Elkabbany, Z.A.; Shedid, A.M.; Hamed, A.I.; Ebrahim, A.O. Serum Chemerin in Obese Children and Adolescents Before and After L-Carnitine Therapy: Relation to Nonalcoholic Fatty Liver Disease and Other Features of Metabolic Syndrome. Arch. Med. Res. 2016, 47, 541–549. [Google Scholar] [CrossRef]
- Hatamkhani, S.; Khalili, H.; Karimzadeh, I.; Dashti-Khavidaki, S.; Abdollahi, A.; Jafari, S. Carnitine for Prevention of Antituberculosis Drug-Induced Hepatotoxicity: A Randomized, Clinical Trial. J. Gastroenterol. Hepatol. 2014, 29, 997–1004. [Google Scholar] [CrossRef]
- Zhang, J.; He, K.; Cai, L.; Chen, Y.-C.; Yang, Y.; Shi, Q.; Woolf, T.F.; Ge, W.; Guo, L.; Borlak, J.; et al. Inhibition of Bile Salt Transport by Drugs Associated with Liver Injury in Primary Hepatocytes from Human, Monkey, Dog, Rat, and Mouse. Chem. Biol. Interact. 2016, 255, 45–54. [Google Scholar] [CrossRef]
- Moreno-Torres, M.; Quintás, G.; Castell, J.V. The Potential Role of Metabolomics in Drug-Induced Liver Injury (DILI) Assessment. Metabolites 2022, 12, 564. [Google Scholar] [CrossRef]
- Danan, G.; Teschke, R. RUCAM in Drug and Herb Induced Liver Injury: The Update. Int. J. Mol. Sci. 2015, 17, 14. [Google Scholar] [CrossRef]
- Díaz, C.; González-Olmedo, C. Untargeted Metabolomics by Liquid Chromatography-Mass Spectrometry in Biomedical Research. In Mass Spectrometry for Metabolomics; Methods in Molecular Biology; Humana: New York, NY, USA, 2023; Volume 2571, pp. 57–69. [Google Scholar] [CrossRef]


| Clinical Characteristics | n | % | Days (Median ± SD) |
|---|---|---|---|
| Type of injury | |||
| Hepatocellular injury | 29 | 90.63 | |
| Cholestatic and mixed injury | 3 | 9.38 | |
| Clinical presentation | |||
| Fulminant course (liver transplant, death) | 2 | 6.25 | |
| Severe injury | 5 | 15.63 | |
| Chronicity | 1 | 3.13 | |
| Clinical signs and symptoms | |||
| Abdominal pain | 25 | 78.13 | |
| Nausea, vomiting | 22 | 68.75 | |
| Anorexia, asthenia | 19 | 59.38 | |
| Days from drug start to onset of symptoms | 8 ± 3 | ||
| Total days of drug use | 5 ± 6 |
| Mass/ Charge | p-Value | FDR * | Retention Time | Fold Change | Log (Fold Change) | Molecular Formula | Δppm | Structure | Adduct |
|---|---|---|---|---|---|---|---|---|---|
| 152.0695 | 2.6 × 10−3 | 6.5 × 10−3 | 3.08 | 13.243 | 1.122 | C8H9NO2 | −4.0 | Acetaminophen | H |
| 203.0533 | 1.2 × 10−6 | 1.5 × 10−5 | 1.33 | 1.554 | 0.191 | C6H12O6 | 1.4 | Glucose | Na |
| 383.1143 | 2.3 × 10−7 | 7.5 × 10−6 | 1.32 | 1.643 | 0.216 | C6H12O6 | 2M + Na | ||
| 205.1227 | 1.9 × 10−3 | 4.9 × 10−3 | 6.59 | 4.187 | 0.622 | C13H16O2 | 0.5 | Hydroxy-ibuprofen | H-H20 |
| 218.1282 | 1.3 × 10−2 | 2.6 × 10−2 | 2.26 | 0.127 | −0.897 | C12H15N3O | 1.0 | Noramidopyrine | H |
| 245.0742 | 1.6 × 10−6 | 1.7 × 10−5 | 1.36 | 1.608 | 0.206 | C9H12N2O6 | 7.8 | Uridine | H |
| 254.248 | 1.8 × 10−8 | 1.6 × 10−6 | 13.14 | 2.009 | 0.303 | C16H31NO | 0.6 | Palmitoleamide | H |
| 265.2514 | 1.6 × 10−8 | 1.6 × 10−6 | 15.27 | 1.764 | 0.247 | C18H32O | −4.2 | 9-Octadecenamide | H-NH3 |
| 282.2787 | 1.5 × 10−8 | 1.6 × 10−6 | 15.27 | 1.824 | 0.261 | C18H35NO | 0.6 | H | |
| 563.5492 | 1.1 × 10−7 | 5.8 × 10−6 | 15.3 | 2.269 | 0.356 | C36H70N2O2 | −1.8 | 2M + H | |
| 280.2641 | 4.5 × 10−7 | 9.5 × 10−6 | 13.63 | 1.744 | 0.242 | C18H33NO | 0.4 | Linoleic acid conjugate | H |
| 288.2169 | 5.2 × 10−3 | 1.2 × 10−2 | 5.81 | 1.515 | 0.180 | C15H29NO4 | 0.2 | O-octanoylcarnitine | H |
| 305.2464 | 1.4 × 10−3 | 3.9 × 10−3 | 15.03 | 1.449 | 0.161 | C20H32O2 | 1.6 | cis-5,8,11,14-eicosatetraenoic acid | H |
| 317.0541 | 9.3 × 10−3 | 1.9 × 10−2 | 1.41 | 4.143 | 0.617 | C10H12N4O6 | 2.7 | Xanthosine | |
| 320.256 | 7.0 × 10−5 | 2.9 × 10−4 | 10.71 | 1.364 | 0.135 | C17H36O5 | −0.7 | Tetraethylene glycol | Na |
| 320.2575 | 2.8 × 10−5 | 1.5 × 10−4 | 11.01 | 1.377 | 0.139 | C18H35NO2 | 1.6 | 3-ketosphingosine | H |
| 328.1383 | 3.9 × 10−4 | 1.3 × 10−3 | 1.79 | 1.479 | 0.170 | C15H21NO7 | 0.4 | N-(1-Deoxy−1-fructosyl)phenylalanine | H |
| 329.2467 | 1.4 × 10−3 | 3.8 × 10−3 | 14.56 | 1.342 | 0.128 | C22H32O2 | 0.0 | Docosapentaenoic acid (22n-6) | H |
| 332.2425 | 1.8 × 10−4 | 7.0 × 10−4 | 5.81 | 1.333 | 0.125 | C17H33NO5 | −0.8 | Hydroxidecanoylcarnitine | H |
| 363.2164 | 8.2 × 10−4 | 2.6 × 10−3 | 6.58 | 1.441 | 0.159 | C21H30O5 | 0.2 | Cortisol | H |
| 412.283 | 5.0 × 10−3 | 1.1 × 10−2 | 7.37 | 0.119 | −0.924 | C26H37NO3 | 0.4 | Glycocholic acid | H-3H20 |
| 430.2934 | 5.5 × 10−3 | 1.2 × 10−2 | 7.37 | 0.124 | −0.908 | C26H39NO4 | −0.2 | H-2H20 | |
| 448.3064 | 5.2 × 10−3 | 1.2 × 10−2 | 7.37 | 0.118 | −0.928 | C26H41NO5 | 0.3 | H-H20 | |
| 466.3168 | 8.1 × 10−3 | 1.7 × 10−2 | 7.37 | 0.113 | −0.946 | C26H43NO6 | 0.2 | H | |
| 488.2989 | 4.2 × 10−3 | 9.5 × 10−3 | 7.37 | 0.181 | −0.743 | C26H43NO6 | −1.1 | Na | |
| 494.3248 | 7.7 × 10−3 | 1.6 × 10−2 | 9.96 | 0.485 | −0.314 | C24H48NO7P | 0.8 | Phosphatidylcholine (16:1/0:0) | H |
| 516.2975 | 4.1 × 10−3 | 9.3 × 10−3 | 7.9 | 0.107 | −0.971 | C26H45NO7S | −0.1 | Taurocholic acid | H |
| 533.3235 | 4.0 × 10−3 | 9.3 × 10−3 | 7.94 | 0.127 | −0.895 | C26H45NO7S | −0.4 | NH4 |
| Pathway Name | Total Metabolites in the Pathway | Common | p-Value | Impact on the Pathway |
|---|---|---|---|---|
| Biosynthesis of primary bile acids | 46 | 2 | 0.0271 | 0.0161 |
| Metabolism of taurine and hypotaurine | 8 | 1 | 0.04562 | 0.0000 |
| Metabolism of glycerophospholipids | 36 | 1 | 0.19108 | 0.0174 |
| Metabolism of pyridines | 39 | 1 | 0.20543 | 0.0158 |
| Metabolism of purines | 65 | 1 | 0.32062 | 0.0000 |
| Biosynthesis of steroid hormones | 85 | 1 | 0.39887 | 0.0273 |
| Time (min) | Flow (μL/min) | % Mobile Phase A | % Mobile Phase B |
|---|---|---|---|
| 0.0 | 300 | 99 | 1 |
| 0.5 | 300 | 99 | 1 |
| 11.0 | 300 | 1 | 99 |
| 15.5 | 300 | 1 | 99 |
| 15.6 | 300 | 99 | 1 |
| 20.0 | 300 | 99 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andújar-Vera, F.; Alés-Palmer, M.L.; Muñoz-de-Rueda, P.; Iglesias-Baena, I.; Ocete-Hita, E. Metabolomic Analysis of Pediatric Patients with Idiosyncratic Drug-Induced Liver Injury According to the Updated RUCAM. Int. J. Mol. Sci. 2023, 24, 13562. https://doi.org/10.3390/ijms241713562
Andújar-Vera F, Alés-Palmer ML, Muñoz-de-Rueda P, Iglesias-Baena I, Ocete-Hita E. Metabolomic Analysis of Pediatric Patients with Idiosyncratic Drug-Induced Liver Injury According to the Updated RUCAM. International Journal of Molecular Sciences. 2023; 24(17):13562. https://doi.org/10.3390/ijms241713562
Chicago/Turabian StyleAndújar-Vera, Francisco, María Luisa Alés-Palmer, Paloma Muñoz-de-Rueda, Iván Iglesias-Baena, and Esther Ocete-Hita. 2023. "Metabolomic Analysis of Pediatric Patients with Idiosyncratic Drug-Induced Liver Injury According to the Updated RUCAM" International Journal of Molecular Sciences 24, no. 17: 13562. https://doi.org/10.3390/ijms241713562





