Long Non-Coding RNAs and Their “Discrete” Contribution to IBD and Johne’s Disease—What Stands out in the Current Picture? A Comprehensive Review
Abstract
:1. Introduction
1.1. Introduction to Intestine-Related Pathologies
1.2. LncRNAs as Aides in the Disease Research
2. Morphology and Biological Characteristics of Mycobacterium avium
3. MAP, Crohn’s Disease, and Relative Pathologies
4. Long Non-Coding RNAs (LncRNAs) and Their Footprint in Gene Regulation
4.1. LncRNAs in the Disease State
4.2. Principles of Classification for Long Non-Coding RNAs
5. LncRNAs’ Involvement in Immune Dysregulation in IBD and JD
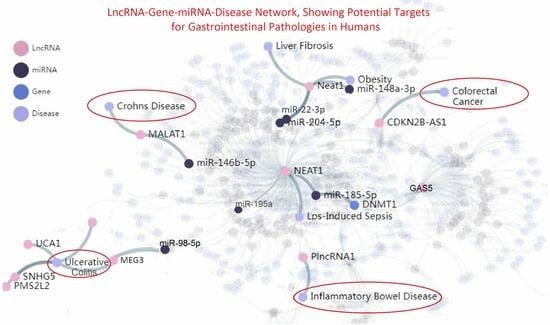
5.1. LncRNA Evidence in IBD-Related Pathologies
5.2. LncRNA Evidence from JD-Related Pathologies
5.3. Differences in LncRNA Profiles of CD and JD
5.4. Expression Profiling in Crohn’s Disease vs. Healthy Controls
6. The Epigenetic Role of the LncRNAs Involved in Human IBD-Related Pathologies and Mycobacterial Infections of the Host
7. The Triarchy of Infection, lncRNA Intervention, and Regulation (IIR)
7.1. The Infection Stage
7.2. Signaling Event Pathways and lncRNA Intervention
7.3. Coding Genes and LncRNAs Regulate the Pathological Phenotype Variables
8. The Prospect of LncRNAs in Contemporary Therapies of GI
- Post-transcriptional RNA degradation pathways aimed at the knockdown of pathogenic RNAs could be an option, either by using siRNAs that elicit a DICER- and ARGONAUTE (AGO)-dependent cleavage pathway or by targeting the RNA of interest for degradation through an RNase H-dependent mechanism employing antisense oligonucleotides (ASOs).
- The modulation of lncRNA genes is also feasible by exploiting steric hindrances of the promoter under study using genome-editing techniques, e.g., CRISPR/Cas9 etc., and
- Loss-of-function, gain-of-function, and dominant-negative mutations have profoundly different effects on the protein structure, therefore providing great insight into the molecular mechanisms underlying genetic diseases. Thus, loss of function by steric inhibition of RNA–protein interactions, e.g., studying HuR binding or the disruption of its secondary structure using the RNA binding of small molecules or the aforementioned ASOs is another attractive approach.
9. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Reis, A.C.; Ramos, B.; Pereira, A.C.; Cunha, M.V. Global trends of epidemiological research in livestock tuberculosis for the last four decades. Transbound. Emerg. Dis. 2021, 68, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Gopalaswamy, R.; Shanmugam, S.; Mondal, R.; Subbian, S. Of tuberculosis and non-tuberculous mycobacterial infections—A comparative analysis of epidemiology, diagnosis and treatment. J. Biomed. Sci. 2020, 27, 74. [Google Scholar] [CrossRef] [PubMed]
- McAloon, C.G.; Roche, S.; Ritter, C.; Barkema, H.W.; Whyte, P.; More, S.J.; O’Grady, L.; Green, M.J.; Doherty, M.L. A review of paratuberculosis in dairy herds—Part 2: On-farm control. Vet. J. 2019, 246, 54–58. [Google Scholar] [CrossRef]
- McAloon, C.G.; Roche, S.; Ritter, C.; Barkema, H.W.; Whyte, P.; More, S.J.; O’Grady, L.; Green, M.J.; Doherty, M.L. A review of paratuberculosis in dairy herds—Part 1: Epidemiology. Vet. J. 2019, 246, 59–65. [Google Scholar] [CrossRef] [PubMed]
- More, S.; Botner, A.; Butterworth, A.; Calistri, P.; Depner, K.; Edwards, S.; Garin-Bastuji, B.; Good, M.; Gortazar Schmidt, C.; Michel, V.; et al. Assessment of listing and categorisation of animal diseases within the framework of the Animal Health Law (Regulation (EU) No 2016/429): Paratuberculosis. EFSA J. Eur. Food Saf. Auth. 2017, 15, e04960. [Google Scholar] [CrossRef]
- Barratt, A.S.; Arnoult, M.H.; Ahmadi, B.V.; Rich, K.M.; Gunn, G.J.; Stott, A.W. A framework for estimating society’s economic welfare following the introduction of an animal disease: The case of Johne’s disease. PLoS ONE 2018, 13, e0198436. [Google Scholar] [CrossRef]
- Taylor, A.W. The experimental infection of cattle with varieties of Mycobacterium johnei isolated from sheep. J. Comp. Pathol. 1953, 63, 368–373. [Google Scholar] [CrossRef]
- Taylor, A.W. Experimental Johne’s disease in cattle. J. Comp. Pathol. 1953, 63, 355–367. [Google Scholar] [CrossRef]
- Alexejeff-Goloff, N.A. Zur Frage der Pathogenese und Bazillenausscheidung bei Rinderparatuberkulose. Krankenhaus-Hygiene + Infektionsverhütung 1929, 36, 313–317. [Google Scholar]
- Sweeney, R.W.; Whitlock, R.H.; Rosenberger, A.E. Mycobacterium paratuberculosis cultured from milk and supramammary lymph nodes of infected asymptomatic cows. J. Clin. Microbiol. 1992, 30, 166–171. [Google Scholar] [CrossRef]
- Streeter, R.N.; Hoffsis, G.F.; Bech-Nielsen, S.; Shulaw, W.P.; Rings, D.M. Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am. J. Vet. Res. 1995, 56, 1322–1324. [Google Scholar]
- Ponnusamy, D.; Periasamy, S.; Tripathi, B.N.; Pal, A. Mycobacterium avium subsp. paratuberculosis invades through M cells and enterocytes across ileal and jejunal mucosa of lambs. Res. Vet. Sci. 2013, 94, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Mataragka, A.; Leousi, E.; Liandris, E.; Ntafis, V.; Leontides, L.; Aggelidou, E.; Bossis, I.; Triantaphyllopoulos, K.A.; Theodoropoulou, I.; Ikonomopoulos, J. Faecal shedding of Mycobacterium avium subspecies paratuberculosis reduces before parturition in sheep? Small Rumin. Res. 2017, 147, 32–36. [Google Scholar] [CrossRef]
- Mataragka, A.; Sotirakoglou, K.; Gazouli, M.; Triantaphyllopoulos, K.A.; Ikonomopoulos, J. Parturition affects test-positivity in sheep with subclinical paratuberculosis; investigation following a preliminary analysis. J. King Saud Univ. Sci. 2019, 31, 1399–1403. [Google Scholar] [CrossRef]
- Liandris, E.; Gazouli, M.; Taka, S.; Andreadou, M.; Vaiopoulou, A.; Tzimotoudis, N.; Kasampalidis, I.; Mpaseas, D.; Fyliousis, G.; Poltronieri, P.; et al. Evaluation of the microbial safety of child food of animal origin in Greece. J. Food Sci. 2014, 79, M362–M368. [Google Scholar] [CrossRef]
- Stefanova, E.P.; Quesada-Canales, O.; Paz-Sanchez, Y.; Caballero, M.J.; Quintana-Montesdeoca, M.D.P.; Espinosa de Los Monteros, A.; Rivero, M.A.; Castro, A.; Perez, V.; Andrada, M. Morphological Assessment of Concomitant Lesions Detected in Goat Herds Naturally Infected with Paratuberculosis (Johne’s Disease). Animals 2023, 13, 1693. [Google Scholar] [CrossRef]
- Rathnaiah, G.; Zinniel, D.K.; Bannantine, J.P.; Stabel, J.R.; Grohn, Y.T.; Collins, M.T.; Barletta, R.G. Pathogenesis, Molecular Genetics, and Genomics of Mycobacterium avium subsp. paratuberculosis, the Etiologic Agent of Johne’s Disease. Front. Vet. Sci. 2017, 4, 187. [Google Scholar] [CrossRef]
- Arsenault, R.J.; Li, Y.; Bell, K.; Doig, K.; Potter, A.; Griebel, P.J.; Kusalik, A.; Napper, S. Mycobacterium avium subsp. paratuberculosis inhibits gamma interferon-induced signaling in bovine monocytes: Insights into the cellular mechanisms of Johne’s disease. Infect. Immun. 2012, 80, 3039–3048. [Google Scholar] [CrossRef] [PubMed]
- Ariel, O.; Gendron, D.; Dudemaine, P.L.; Gevry, N.; Ibeagha-Awemu, E.M.; Bissonnette, N. Transcriptome Profiling of Bovine Macrophages Infected by Mycobacterium avium spp. paratuberculosis Depicts Foam Cell and Innate Immune Tolerance Phenotypes. Front. Immunol. 2019, 10, 2874. [Google Scholar] [CrossRef]
- Field, N.L.; McAloon, C.G.; Gavey, L.; Mee, J.F. Mycobacterium avium subspecies paratuberculosis infection in cattle—A review in the context of seasonal pasture-based dairy herds. Ir. Vet. J. 2022, 75, 12. [Google Scholar] [CrossRef]
- Windsor, P.A. Paratuberculosis in sheep and goats. Vet. Microbiol. 2015, 181, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Badia-Bringue, G.; Canive, M.; Alonso-Hearn, M. Control of Mycobacterium avium subsp. paratuberculosis load within infected bovine monocyte-derived macrophages is associated with host genetics. Front. Immunol. 2023, 14, 1042638. [Google Scholar] [CrossRef] [PubMed]
- Pickrodt, C.; Donat, K.; Moog, U.; Kohler, H. Mycobacterium avium subsp. Paratuberculosis in Different Environmental Samples from a Dairy Goat Barn-Implications for Sampling Strategies for Paratuberculosis Diagnostic and Prevention. Animals 2023, 13, 1688. [Google Scholar] [CrossRef] [PubMed]
- Davis, W.C. On deaf ears, Mycobacterium avium paratuberculosis in pathogenesis Crohn’s and other diseases. World J. Gastroenterol. 2015, 21, 13411–13417. [Google Scholar] [CrossRef] [PubMed]
- Eslami, M.; Shafiei, M.; Ghasemian, A.; Valizadeh, S.; Al-Marzoqi, A.H.; Shokouhi Mostafavi, S.K.; Nojoomi, F.; Mirforughi, S.A. Mycobacterium avium paratuberculosis and Mycobacterium avium complex and related subspecies as causative agents of zoonotic and occupational diseases. J. Cell. Physiol. 2019, 234, 12415–12421. [Google Scholar] [CrossRef]
- McNees, A.L.; Markesich, D.; Zayyani, N.R.; Graham, D.Y. Mycobacterium paratuberculosis as a cause of Crohn’s disease. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 1523–1534. [Google Scholar] [CrossRef]
- Sechi, L.A.; Dow, C.T. Mycobacterium avium ss. paratuberculosis Zoonosis—The Hundred Year War—Beyond Crohn’s Disease. Front. Immunol. 2015, 6, 96. [Google Scholar] [CrossRef]
- Timms, V.J.; Daskalopoulos, G.; Mitchell, H.M.; Neilan, B.A. The Association of Mycobacterium avium subsp. paratuberculosis with Inflammatory Bowel Disease. PLoS ONE 2016, 11, e0148731. [Google Scholar] [CrossRef]
- van der Sloot, K.W.J.; Voskuil, M.D.; Blokzijl, T.; Dinkla, A.; Ravesloot, L.; Visschedijk, M.C.; van Dullemen, H.M.; Festen, E.A.M.; Alizadeh, B.Z.; van Leer-Buter, C.; et al. Isotype-specific Antibody Responses to Mycobacterium avium paratuberculosis Antigens Are Associated with the Use of Biologic Therapy in Inflammatory Bowel Disease. J. Crohn’s Colitis 2021, 15, 1253–1263. [Google Scholar] [CrossRef]
- Proietti, E.; Fuhler, G.M.; Peppelenbosch, M.P. Mycobacterium avium Subspecies Paratuberculosis Infection and Biological Treatment of IBD: Cause or Consequence? J. Crohn’s Colitis 2021, 15, 1247–1249. [Google Scholar] [CrossRef]
- Zhao, M.; Burisch, J. Impact of Genes and the Environment on the Pathogenesis and Disease Course of Inflammatory Bowel Disease. Dig. Dis. Sci. 2019, 64, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Zarei-Kordshouli, F.; Geramizadeh, B.; Khodakaram-Tafti, A. Prevalence of Mycobacterium avium subspecies paratuberculosis IS 900 DNA in biopsy tissues from patients with Crohn’s disease: Histopathological and molecular comparison with Johne’s disease in Fars province of Iran. BMC Infect. Dis. 2019, 19, 23. [Google Scholar] [CrossRef]
- Cullen, C.M.; Aneja, K.K.; Beyhan, S.; Cho, C.E.; Woloszynek, S.; Convertino, M.; McCoy, S.J.; Zhang, Y.; Anderson, M.Z.; Alvarez-Ponce, D.; et al. Emerging Priorities for Microbiome Research. Front. Microbiol. 2020, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Liu, J.; Li, L.; Guo, Y.; Luo, Q.; Li, J. Long non-coding RNA expression profiling of macrophage line RAW264.7 infected by Mycobacterium tuberculosis. Biotech. Histochem. 2020, 95, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Liu, K.; Jia, Q.; Zhang, H.; Bie, Q.; Zhang, B. The Roles of Host Noncoding RNAs in Mycobacterium tuberculosis Infection. Front. Immunol. 2021, 12, 664787. [Google Scholar] [CrossRef]
- Yang, X.; Yang, J.; Wang, J.; Wen, Q.; Wang, H.; He, J.; Hu, S.; He, W.; Du, X.; Liu, S.; et al. Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci. Rep. 2016, 6, 38963. [Google Scholar] [CrossRef]
- Pekarek, L.; Torres-Carranza, D.; Fraile-Martinez, O.; Garcia-Montero, C.; Pekarek, T.; Saez, M.A.; Rueda-Correa, F.; Pimentel-Martinez, C.; Guijarro, L.G.; Diaz-Pedrero, R.; et al. An Overview of the Role of MicroRNAs on Carcinogenesis: A Focus on Cell Cycle, Angiogenesis and Metastasis. Int. J. Mol. Sci. 2023, 24, 7268. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Chen, X.; Zhang, S. Long non-coding RNAs: From disease code to drug role. Acta Pharm. Sin. B 2021, 11, 340–354. [Google Scholar] [CrossRef]
- Zhang, Z.; Shi, S.; Li, J.; Costa, M. Long Non-Coding RNA MEG3 in Metal Carcinogenesis. Toxics 2023, 11, 157. [Google Scholar] [CrossRef]
- Wang, B.; Wang, M.; Jia, S.; Li, T.; Yang, M.; Ge, F. Systematic Survey of the Regulatory Networks of the Long Noncoding RNA BANCR in Cervical Cancer Cells. J. Proteome Res. 2022, 21, 1137–1152. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Gao, F.; Li, H.; Qin, W.; Chai, C.; Shi, G.; Yang, H. Long noncoding RNA THRIL promotes foam cell formation and inflammation in macrophages. Cell Biol. Int. 2023, 47, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Bates, A.; O’Brien, R.; Liggett, S.; Griffin, F. The effect of sub-clinical infection with Mycobacterium avium subsp. paratuberculosis on milk production in a New Zealand dairy herd. BMC Vet. Res. 2018, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- King, H.C.; Khera-Butler, T.; James, P.; Oakley, B.B.; Erenso, G.; Aseffa, A.; Knight, R.; Wellington, E.M.; Courtenay, O. Environmental reservoirs of pathogenic mycobacteria across the Ethiopian biogeographical landscape. PLoS ONE 2017, 12, e0173811. [Google Scholar] [CrossRef]
- Riojas, M.A.; McGough, K.J.; Rider-Riojas, C.J.; Rastogi, N.; Hazbon, M.H. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycobacterium tuberculosis. Int. J. Syst. Evol. Microbiol. 2018, 68, 324–332. [Google Scholar] [CrossRef]
- Rossini, N.O.; Dias, M.V.B. Mutations and insights into the molecular mechanisms of resistance of Mycobacterium tuberculosis to first-line. Genet. Mol. Biol. 2023, 46, e20220261. [Google Scholar] [CrossRef]
- Fecteau, M.E. Paratuberculosis in Cattle. Vet. Clin. N. Am. Food Anim. Pract. 2018, 34, 209–222. [Google Scholar] [CrossRef]
- Galiero, A.; Fratini, F.; Mataragka, A.; Turchi, B.; Nuvoloni, R.; Ikonomopoulos, J.; Cerri, D. Detection of Mycobacterium avium subsp. paratuberculosis in cheeses from small ruminants in Tuscany. Int. J. Food Microbiol. 2016, 217, 195–199. [Google Scholar] [CrossRef]
- Espeschit, I.F.; Bastos, D.S.S.; Fonseca Junior, A.; Cardoso, S.A.; Ferrari, M.L.A.; Moreira, M.A.S. Mycobacterium avium subsp. paratuberculosis and Crohn’s disease: Characterization of the interaction with different aspects of the disease. Braz. J. Microbiol. 2023, 54, 1239–1249. [Google Scholar] [CrossRef]
- Corbett, C.S.; De Buck, J.; Barkema, H.W. Effects of freezing on ability to detect Mycobacterium avium subsp. paratuberculosis from bovine tissues following culture. J. Vet. Diagn. Investig. 2018, 30, 743–746. [Google Scholar] [CrossRef]
- Tuberquia-Lopez, B.C.; Correa-Valencia, N.M.; Hernandez-Agudelo, M.; Fernandez-Silva, J.A.; Ramirez-Vasquez, N.F. Paratuberculosis control strategies in dairy cattle: A systematic review. Open Vet. J. 2022, 12, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.B.; Shalloo, L. Invited review: The economic impact and control of paratuberculosis in cattle. J. Dairy Sci. 2015, 98, 5019–5039. [Google Scholar] [CrossRef]
- Triantaphyllopoulos, K.A.; Baltoumas, F.A.; Hamodrakas, S.J. Structural characterization and molecular dynamics simulations of the caprine and bovine solute carrier family 11 A1 (SLC11A1). J. Comput.-Aided Mol. Des. 2019, 33, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Thoen, C.; Jarnagin, J.; Richards, W. Isolation and identification of mycobacteria from porcine tissues: A three-year summary. Am. J. Vet. Res. 1975, 36, 1383–1386. [Google Scholar]
- Greig, A.; Stevenson, K.; Perez, V.; Pirie, A.; Grant, J.; Sharp, J. Paratuberculosis in wild rabbits (Oryctolagus cuniculus). Vet. Rec. 1997, 140, 141–143. [Google Scholar] [CrossRef]
- Beard, P.; Daniels, M.; Henderson, D.; Pirie, A.; Rudge, K.; Buxton, D.; Rhind, S.; Greig, A.; Hutchings, M.; McKendrick, I.; et al. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 2001, 39, 1517–1521. [Google Scholar] [CrossRef] [PubMed]
- Fechner, K.; Matz-Rensing, K.; Lampe, K.; Kaup, F.J.; Czerny, C.P.; Schafer, J. Detection of Mycobacterium avium subsp. paratuberculosis in non-human primates. J. Med. Primatol. 2017, 46, 211–217. [Google Scholar] [CrossRef]
- Byrne, A.; Ollier, S.; Tahlan, K.; Biet, F.; Bissonnette, N. Genomic epidemiology of Mycobacterium avium subsp. paratuberculosis isolates from Canadian dairy herds provides evidence for multiple infection events. Front. Genet. 2023, 14, 1043598. [Google Scholar] [CrossRef]
- Elmagzoub, W.A.; Idris, S.M.; Isameldin, M.; Arabi, N.; Abdo, A.; Ibrahim, M.; Khan, M.A.A.; Tanneberger, F.; Bakhiet, S.M.; Okuni, J.B.; et al. Mycobacterium avium subsp. paratuberculosis and microbiome profile of patients in a referral gastrointestinal diseases centre in the Sudan. PLoS ONE 2022, 17, e0266533. [Google Scholar] [CrossRef]
- Dow, C.T. Hermon-Taylor: M. paratuberculosis and Crohn’s Disease-The Book of Revelation According to John. Pathogens 2021, 10, 1469. [Google Scholar] [CrossRef]
- Agrawal, G.; Clancy, A.; Huynh, R.; Borody, T. Profound remission in Crohn’s disease requiring no further treatment for 3-23 years: A case series. Gut Pathog. 2020, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.; Hamblin, H.; Clancy, A.; Borody, T. Anti-Mycobacterial Antibiotic Therapy Induces Remission in Active Paediatric Crohn’s Disease. Microorganisms 2020, 8, 1112. [Google Scholar] [CrossRef]
- Ekundayo, T.C.; Okoh, A.I. Systematic Assessment of Mycobacterium avium Subspecies Paratuberculosis Infections from 1911-2019: A Growth Analysis of Association with Human Autoimmune Diseases. Microorganisms 2020, 8, 1212. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Chaubey, K.K.; Agarwal, P.; Kuenstner, J.T.; Parashar, D.; Singh, S.V. Therapeutic management of Mycobacterium avium subspecies paratuberculosis infection with complete resolution of symptoms and disease in a patient with advanced inflammatory bowel syndrome. Mol. Biol. Rep. 2021, 48, 7013–7020. [Google Scholar] [CrossRef] [PubMed]
- Savarino, E.; Bertani, L.; Ceccarelli, L.; Bodini, G.; Zingone, F.; Buda, A.; Facchin, S.; Lorenzon, G.; Marchi, S.; Marabotto, E.; et al. Antimicrobial treatment with the fixed-dose antibiotic combination RHB-104 for Mycobacterium avium subspecies paratuberculosis in Crohn’s disease: Pharmacological and clinical implications. Expert Opin. Biol. Ther. 2019, 19, 79–88. [Google Scholar] [CrossRef]
- Pierce, E.S. Could Mycobacterium avium subspecies paratuberculosis cause Crohn’s disease, ulcerative colitis… and colorectal cancer? Infect. Agents Cancer 2018, 13, 1. [Google Scholar] [CrossRef]
- Okuni, J.B.; Hansen, S.; Eltom, K.H.; Eltayeb, E.; Amanzada, A.; Omega, J.A.; Czerny, C.P.; Abd El Wahed, A.; Ojok, L. Paratuberculosis: A Potential Zoonosis and a Neglected Disease in Africa. Microorganisms 2020, 8, 1007. [Google Scholar] [CrossRef]
- Chaubey, K.K.; Singh, S.V.; Gupta, S.; Singh, M.; Sohal, J.S.; Kumar, N.; Singh, M.K.; Bhatia, A.K.; Dhama, K. Mycobacterium avium subspecies paratuberculosis—An important food borne pathogen of high public health significance with special reference to India: An update. Vet. Q. 2017, 37, 282–299. [Google Scholar] [CrossRef]
- Cossu, A.; Rosu, V.; Paccagnini, D.; Cossu, D.; Pacifico, A.; Sechi, L.A. MAP3738c and MptD are specific tags of Mycobacterium avium subsp. paratuberculosis infection in type I diabetes mellitus. Clin. Immunol. 2011, 141, 49–57. [Google Scholar] [CrossRef]
- Songini, M.; Mannu, C.; Targhetta, C.; Bruno, G. Type 1 diabetes in Sardinia: Facts and hypotheses in the context of worldwide epidemiological data. Acta Diabetol. 2017, 54, 9–17. [Google Scholar] [CrossRef]
- Bitti, M.L.; Masala, S.; Capasso, F.; Rapini, N.; Piccinini, S.; Angelini, F.; Pierantozzi, A.; Lidano, R.; Pietrosanti, S.; Paccagnini, D.; et al. Mycobacterium avium subsp. paratuberculosis in an Italian cohort of type 1 diabetes pediatric patients. Clin. Dev. Immunol. 2012, 2012, 785262. [Google Scholar] [CrossRef] [PubMed]
- Cossu, A.; Ferrannini, E.; Fallahi, P.; Antonelli, A.; Sechi, L.A. Antibodies recognizing specific Mycobacterium avium subsp. paratuberculosis’s MAP3738c protein in type 1 diabetes mellitus children are associated with serum Th1 (CXCL10) chemokine. Cytokine 2013, 61, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Paccagnini, D.; Sieswerda, L.; Rosu, V.; Masala, S.; Pacifico, A.; Gazouli, M.; Ikonomopoulos, J.; Ahmed, N.; Zanetti, S.; Sechi, L.A. Linking chronic infection and autoimmune diseases: Mycobacterium avium subspecies paratuberculosis, SLC11A1 polymorphisms and type-1 diabetes mellitus. PLoS ONE 2009, 4, e7109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xue, C.; Wang, Y.; Shi, J.; Zhang, X.; Li, W.; Nunez, S.; Foulkes, A.S.; Lin, J.; Hinkle, C.C.; et al. Deep RNA Sequencing Uncovers a Repertoire of Human Macrophage Long Intergenic Noncoding RNAs Modulated by Macrophage Activation and Associated with Cardiometabolic Diseases. J. Am. Heart Assoc. 2017, 6, e007431. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Koonin, E.V. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell 2020, 183, 1151–1161. [Google Scholar] [CrossRef]
- Villa, T.; Porrua, O. Pervasive transcription: A controlled risk. FEBS J. 2022, 290, 3723–3736. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [CrossRef]
- Li, J.; Xu, Q.; Huang, Z.J.; Mao, N.; Lin, Z.T.; Cheng, L.; Sun, B.; Wang, G. CircRNAs: A new target for the diagnosis and treatment of digestive system neoplasms. Cell Death Dis. 2021, 12, 205. [Google Scholar] [CrossRef]
- Wang, F.; Nazarali, A.J.; Ji, S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016, 6, 1167–1176. [Google Scholar]
- Zhou, Z.; Wang, X.; Hu, Q.; Yang, Z. CircZfp609 contributes to cerebral infarction via sponging miR-145a-5p to regulate BACH1. Metab. Brain Dis. 2023, 38, 1971–1981. [Google Scholar] [CrossRef]
- Mowel, W.K.; Kotzin, J.J.; McCright, S.J.; Neal, V.D.; Henao-Mejia, J. Control of Immune Cell Homeostasis and Function by lncRNAs. Trends Immunol. 2018, 39, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.; Bai, H.; Wei, Z.; Xie, C.; Wang, J.; Li, J.; Chen, Q. The mechanism and function of circular RNAs in human diseases. Exp. Cell Res. 2018, 368, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Garnica, O.; Dhandayuthapani, S. Modulation of Host miRNAs by Intracellular Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 79. [Google Scholar] [CrossRef]
- Aguilar, C.; Mano, M.; Eulalio, A. Multifaceted Roles of microRNAs in Host-Bacterial Pathogen Interaction. Microbiol. Spectr. 2019, 7, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Kim, T.S.; Basu, J.; Jo, E.K. MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell. Microbiol. 2017, 19, e12687. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Chedin, F. Nascent Connections: R-Loops and Chromatin Patterning. Trends Genet. TIG 2016, 32, 828–838. [Google Scholar] [CrossRef]
- Lan, Y.; Xiao, X.; He, Z.; Luo, Y.; Wu, C.; Li, L.; Song, X. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018, 46, 5809–5821. [Google Scholar] [CrossRef]
- Qin, W.; Li, X.; Xie, L.; Li, S.; Liu, J.; Jia, L.; Dong, X.; Ren, X.; Xiao, J.; Yang, C.; et al. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res. 2016, 44, 6423–6433. [Google Scholar] [CrossRef]
- Sridhar, B.; Rivas-Astroza, M.; Nguyen, T.C.; Chen, W.; Yan, Z.; Cao, X.; Hebert, L.; Zhong, S. Systematic Mapping of RNA-Chromatin Interactions In Vivo. Curr. Biol. 2017, 27, 610–612. [Google Scholar] [CrossRef]
- Asim, M.N.; Ibrahim, M.A.; Imran Malik, M.; Dengel, A.; Ahmed, S. Advances in Computational Methodologies for Classification and Sub-Cellular Locality Prediction of Non-Coding RNAs. Int. J. Mol. Sci. 2021, 22, 8719. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L. Pseudogenes: Four Decades of Discovery. Methods Mol. Biol. 2021, 2324, 3–18. [Google Scholar] [CrossRef]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Hendrich, B.D.; Rupert, J.L.; Lafreniere, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992, 71, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Sleutels, F.; Barlow, D.P. The origins of genomic imprinting in mammals. Adv. Genet. 2002, 46, 119–163. [Google Scholar] [CrossRef]
- Sleutels, F.; Zwart, R.; Barlow, D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002, 415, 810–813. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Song, Y.; Wang, R.; Li, L.W.; Liu, X.; Wang, Y.F.; Wang, Q.X.; Zhang, Q. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int. J. Oncol. 2019, 54, 77–86. [Google Scholar] [CrossRef]
- Mahpour, A.; Mullen, A.C. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep. Innov. Hepatol. 2021, 3, 100177. [Google Scholar] [CrossRef]
- Bocchetti, M.; Scrima, M.; Melisi, F.; Luce, A.; Sperlongano, R.; Caraglia, M.; Zappavigna, S.; Cossu, A.M. LncRNAs and Immunity: Coding the Immune System with Noncoding Oligonucleotides. Int. J. Mol. Sci. 2021, 22, 1741. [Google Scholar] [CrossRef]
- Talyan, S.; Andrade-Navarro, M.A.; Muro, E.M. Identification of transcribed protein coding sequence remnants within lincRNAs. Nucleic Acids Res. 2018, 46, 8720–8729. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Liu, T.; Fu, H.; Li, W.; Zheng, X. Genome-wide analysis of lncRNA stability in human. PLoS Comput. Biol. 2021, 17, e1008918. [Google Scholar] [CrossRef] [PubMed]
- Laham-Karam, N.; Laitinen, P.; Turunen, T.A.; Yla-Herttuala, S. Activating the Chromatin by Noncoding RNAs. Antioxid. Redox Signal. 2018, 29, 813–831. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yang, Y.; Chen, L.; Zhang, Z.; Liu, L.; Zhang, C.; Mai, Q.; Chen, Y.; Chen, Z.; Lin, T.; et al. The gut microbiota mediates protective immunity against tuberculosis via modulation of lncRNA. Gut Microbes 2022, 14, 2029997. [Google Scholar] [CrossRef]
- Weikard, R.; Demasius, W.; Kuehn, C. Mining long noncoding RNA in livestock. Anim. Genet. 2017, 48, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Koufariotis, L.T.; Chen, Y.P.; Chamberlain, A.; Vander Jagt, C.; Hayes, B.J. A catalogue of novel bovine long noncoding RNA across 18 tissues. PLoS ONE 2015, 10, e0141225. [Google Scholar] [CrossRef]
- Zhou, Z.Y.; Li, A.M.; Adeola, A.C.; Liu, Y.H.; Irwin, D.M.; Xie, H.B.; Zhang, Y.P. Genome-wide identification of long intergenic noncoding RNA genes and their potential association with domestication in pigs. Genome Biol. Evol. 2014, 6, 1387–1392. [Google Scholar] [CrossRef]
- El-Khishin, D.A.; Ageez, A.; Saad, M.E.; Ibrahim, A.; Shokrof, M.; Hassan, L.R.; Abouelhoda, M.I. Sequencing and assembly of the Egyptian buffalo genome. PLoS ONE 2020, 15, e0237087. [Google Scholar] [CrossRef]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Mele, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.J.; Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 2019, 20, 599–614. [Google Scholar] [CrossRef]
- Sun, W.; Yang, Y.; Xu, C.; Guo, J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017, 216–217, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.H.; Chen, L.L. Processing and roles of snoRNA-ended long noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 596–606. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- Kern, C.; Wang, Y.; Chitwood, J.; Korf, I.; Delany, M.; Cheng, H.; Medrano, J.F.; Van Eenennaam, A.L.; Ernst, C.; Ross, P.; et al. Genome-wide identification of tissue-specific long non-coding RNA in three farm animal species. BMC Genom. 2018, 19, 684. [Google Scholar] [CrossRef]
- Kern, C.; Wang, Y.; Xu, X.; Pan, Z.; Halstead, M.; Chanthavixay, G.; Saelao, P.; Waters, S.; Xiang, R.; Chamberlain, A.; et al. Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research. Nat. Commun. 2021, 12, 1821. [Google Scholar] [CrossRef]
- Washietl, S.; Kellis, M.; Garber, M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014, 24, 616–628. [Google Scholar] [CrossRef]
- Ma, S.; Ming, Z.; Gong, A.Y.; Wang, Y.; Chen, X.; Hu, G.; Zhou, R.; Shibata, A.; Swanson, P.C.; Chen, X.M. A long noncoding RNA, lincRNA-Tnfaip3, acts as a coregulator of NF-kappaB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. 2017, 31, 1215–1225. [Google Scholar] [CrossRef]
- Zhao, K.; Tan, J.Y.; Mao, Q.D.; Ren, K.Y.; He, B.G.; Zhang, C.P.; Wei, L.Z. Overexpression of long non-coding RNA TUG1 alleviates TNF-alpha-induced inflammatory injury in interstitial cells of Cajal. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zheng, H.; Chan, M.T.; Wu, W.K. HULC: An oncogenic long non-coding RNA in human cancer. J. Cell. Mol. Med. 2017, 21, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Tang, A.; Wang, X.; Chen, X.; Zhao, L.; Xiao, Z.; Shen, S. Inhibition of lncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome-mediated polarization of macrophages. Int. J. Mol. Med. 2018, 42, 2903–2913. [Google Scholar] [CrossRef]
- Ma, D.; Cao, Y.; Wang, Z.; He, J.; Chen, H.; Xiong, H.; Ren, L.; Shen, C.; Zhang, X.; Yan, Y.; et al. CCAT1 lncRNA Promotes Inflammatory Bowel Disease Malignancy by Destroying Intestinal Barrier via Downregulating miR-185-3p. Inflamm. Bowel Dis. 2019, 25, 862–874. [Google Scholar] [CrossRef]
- Qiao, C.; Yang, L.; Wan, J.; Liu, X.; Pang, C.; You, W.; Zhao, G. Long noncoding RNA ANRIL contributes to the development of ulcerative colitis by miR-323b-5p/TLR4/MyD88/NF-kappaB pathway. Biochem. Biophys. Res. Commun. 2019, 508, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Rankin, C.R.; Lokhandwala, Z.A.; Huang, R.; Pekow, J.; Pothoulakis, C.; Padua, D. Linear and circular CDKN2B-AS1 expression is associated with Inflammatory Bowel Disease and participates in intestinal barrier formation. Life Sci. 2019, 231, 116571. [Google Scholar] [CrossRef]
- Rankin, C.R.; Shao, L.; Elliott, J.; Rowe, L.; Patel, A.; Videlock, E.; Benhammou, J.N.; Sauk, J.S.; Ather, N.; Corson, M.; et al. The IBD-associated long noncoding RNA IFNG-AS1 regulates the balance between inflammatory and anti-inflammatory cytokine production after T-cell stimulation. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G34–G40. [Google Scholar] [CrossRef]
- Chen, S.W.; Wang, P.Y.; Liu, Y.C.; Sun, L.; Zhu, J.; Zuo, S.; Ma, J.; Li, T.Y.; Zhang, J.L.; Chen, G.W.; et al. Effect of Long Noncoding RNA H19 Overexpression on Intestinal Barrier Function and Its Potential Role in the Pathogenesis of Ulcerative Colitis. Inflamm. Bowel Dis. 2016, 22, 2582–2592. [Google Scholar] [CrossRef]
- Chen, D.; Liu, J.; Zhao, H.Y.; Chen, Y.P.; Xiang, Z.; Jin, X. Plasma long noncoding RNA expression profile identified by microarray in patients with Crohn’s disease. World J. Gastroenterol. 2016, 22, 4716–4731. [Google Scholar] [CrossRef]
- Ostrik, A.A.; Azhikina, T.L.; Salina, E.G. Small Noncoding RNAs and Their Role in the Pathogenesis of Mycobacterium tuberculosis Infection. Biochemistry 2021, 86, S109–S119. [Google Scholar] [CrossRef]
- Li, N.; Shi, R. Expression alteration of long non-coding RNAs and their target genes in the intestinal mucosa of patients with Crohn’s disease. Clin. Chim. Acta Int. J. Clin. Chem. 2019, 494, 14–21. [Google Scholar] [CrossRef]
- Qiao, Y.Q.; Huang, M.L.; Xu, A.T.; Zhao, D.; Ran, Z.H.; Shen, J. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease. J. Biomed. Sci. 2013, 20, 87. [Google Scholar] [CrossRef]
- Mirza, A.H.; Berthelsen, C.H.; Seemann, S.E.; Pan, X.; Frederiksen, K.S.; Vilien, M.; Gorodkin, J.; Pociot, F. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 2015, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Huang, Y.; Dong, F.; Kwon, J.H. Ulcerative Colitis-Associated Long Noncoding RNA, BC012900, Regulates Intestinal Epithelial Cell Apoptosis. Inflamm. Bowel Dis. 2016, 22, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tao, X.; Wei, J. Effects of LncRNA MEG3 on immunity and autophagy of non-small cell lung carcinoma through IDO signaling pathway. World J. Surg. Oncol. 2021, 19, 244. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Xie, J. LncRNA MALAT1 Promotes Ulcerative Colitis by Upregulating lncRNA ANRIL. Dig. Dis. Sci. 2020, 65, 3191–3196. [Google Scholar] [CrossRef]
- Geng, H.; Bu, H.F.; Liu, F.; Wu, L.; Pfeifer, K.; Chou, P.M.; Wang, X.; Sun, J.; Lu, L.; Pandey, A.; et al. In Inflamed Intestinal Tissues and Epithelial Cells, Interleukin 22 Signaling Increases Expression of H19 Long Noncoding RNA, Which Promotes Mucosal Regeneration. Gastroenterology 2018, 155, 144–155. [Google Scholar] [CrossRef]
- Zou, T.; Jaladanki, S.K.; Liu, L.; Xiao, L.; Chung, H.K.; Wang, J.Y.; Xu, Y.; Gorospe, M. H19 Long Noncoding RNA Regulates Intestinal Epithelial Barrier Function via MicroRNA 675 by Interacting with RNA-Binding Protein HuR. Mol. Cell. Biol. 2016, 36, 1332–1341. [Google Scholar] [CrossRef]
- Yang, F.; Li, X.F.; Cheng, L.N.; Li, X.L. Long non-coding RNA CRNDE promotes cell apoptosis by suppressing miR-495 in inflammatory bowel disease. Exp. Cell Res. 2019, 382, 111484. [Google Scholar] [CrossRef]
- Xiao, L.; Rao, J.N.; Cao, S.; Liu, L.; Chung, H.K.; Zhang, Y.; Zhang, J.; Liu, Y.; Gorospe, M.; Wang, J.Y. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol. Biol. Cell 2016, 27, 617–626. [Google Scholar] [CrossRef]
- Xiao, L.; Gorospe, M.; Wang, J.Y. Long noncoding RNAs in intestinal epithelium homeostasis. Am. J. Physiol. Cell Physiol. 2019, 317, C93–C100. [Google Scholar] [CrossRef]
- Chen, T.; Xue, H.; Lin, R.; Huang, Z. MiR-34c and PlncRNA1 mediated the function of intestinal epithelial barrier by regulating tight junction proteins in inflammatory bowel disease. Biochem. Biophys. Res. Commun. 2017, 486, 6–13. [Google Scholar] [CrossRef]
- Padua, D.; Mahurkar-Joshi, S.; Law, I.K.; Polytarchou, C.; Vu, J.P.; Pisegna, J.R.; Shih, D.; Iliopoulos, D.; Pothoulakis, C. A long noncoding RNA signature for ulcerative colitis identifies IFNG-AS1 as an enhancer of inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G446–G457. [Google Scholar] [CrossRef]
- Quan, Y.; Song, K.; Zhang, Y.; Zhu, C.; Shen, Z.; Wu, S.; Luo, W.; Tan, B.; Yang, Z.; Wang, X. Roseburia intestinalis-derived flagellin is a negative regulator of intestinal inflammation. Biochem. Biophys. Res. Commun. 2018, 501, 791–799. [Google Scholar] [CrossRef]
- Liu, Z.; Lee, J.; Krummey, S.; Lu, W.; Cai, H.; Lenardo, M.J. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 2011, 12, 1063–1070. [Google Scholar] [CrossRef]
- Mahmoud, R.H.; Fouad, N.A.; Hefzy, E.M.; Shaker, O.G.; Ahmed, T.I.; Hussein, H.A.; Nasr, M.H.; Zaki, O.M.; Abdelghaffar, N.K.; Abdelaleem, O.O. The potential role of serum expression profile of long non coding RNAs, Cox2 and HOTAIR as novel diagnostic biomarkers in systemic lupus erythematosus. PLoS ONE 2022, 17, e0268176. [Google Scholar] [CrossRef]
- Bussieres-Marmen, S.; Vinette, V.; Gungabeesoon, J.; Aubry, I.; Perez-Quintero, L.A.; Tremblay, M.L. Loss of T-cell protein tyrosine phosphatase in the intestinal epithelium promotes local inflammation by increasing colonic stem cell proliferation. Cell. Mol. Immunol. 2018, 15, 367–376. [Google Scholar] [CrossRef]
- Longhi, M.S.; Kokkotou, E. Lnc-ing RNA Expression with Disease Pathogenesis: MALAT1 and ANRIL in Ulcerative Colitis. Dig. Dis. Sci. 2020, 65, 3061–3063. [Google Scholar] [CrossRef]
- Deng, W.; Zhang, Y.; Cai, J.; Zhang, J.; Liu, X.; Yin, J.; Bai, Z.; Yao, H.; Zhang, Z. LncRNA-ANRIL promotes gastric cancer progression by enhancing NF-kB signaling. Exp. Biol. Med. 2019, 244, 953–959. [Google Scholar] [CrossRef]
- Cui, C.; Wang, F.; Zheng, Y.; Wei, H.; Peng, J. From birth to death: The hardworking life of Paneth cell in the small intestine. Front. Immunol. 2023, 14, 1122258. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Eghtedarian, R.; Taheri, M. The crucial role of non-coding RNAs in the pathophysiology of inflammatory bowel disease. Biomed. Pharmacother. 2020, 129, 110507. [Google Scholar] [CrossRef]
- Lin, L.; Zhou, G.; Chen, P.; Wang, Y.; Han, J.; Chen, M.; He, Y.; Zhang, S. Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis. 2020, 11, 456. [Google Scholar] [CrossRef]
- Barshir, R.; Fishilevich, S.; Iny-Stein, T.; Zelig, O.; Mazor, Y.; Guan-Golan, Y.; Safran, M.; Lancet, D. GeneCaRNA: A Comprehensive Gene-centric Database of Human Non-coding RNAs in the GeneCards Suite. J. Mol. Biol. 2021, 433, 166913. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Wang, P.; Li, X.; Gao, Y.; Guo, Q.; Wang, Y.; Fang, Y.; Ma, X.; Zhi, H.; Zhou, D.; Shen, W.; et al. LncACTdb 2.0: An updated database of experimentally supported ceRNA interactions curated from low- and high-throughput experiments. Nucleic Acids Res. 2019, 47, D121–D127. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- MacArthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; McMahon, A.; Milano, A.; Morales, J.; et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef]
- Wang, M.; Xie, F.; Lin, J.; Zhao, Y.; Zhang, Q.; Liao, Z.; Wei, P. Diagnostic and Prognostic Value of Circulating CircRNAs in Cancer. Front. Med. 2021, 8, 649383. [Google Scholar] [CrossRef]
- Guindo-Martinez, M.; Amela, R.; Bonas-Guarch, S.; Puiggros, M.; Salvoro, C.; Miguel-Escalada, I.; Carey, C.E.; Cole, J.B.; Rueger, S.; Atkinson, E.; et al. The impact of non-additive genetic associations on age-related complex diseases. Nat. Commun. 2021, 12, 2436. [Google Scholar] [CrossRef]
- Liu, J.Z.; van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef]
- Barrett, J.C.; Hansoul, S.; Nicolae, D.L.; Cho, J.H.; Duerr, R.H.; Rioux, J.D.; Brant, S.R.; Silverberg, M.S.; Taylor, K.D.; Barmada, M.M.; et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008, 40, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Peter, S.; Jung, M.; Lewin, A.; Hemmrich-Stanisak, G.; Franke, A.; von Kleist, M.; Schutte, C.; Einspanier, R.; Sharbati, S.; et al. Analysis of long non-coding RNA and mRNA expression in bovine macrophages brings up novel aspects of Mycobacterium avium subspecies paratuberculosis infections. Sci. Rep. 2019, 9, 1571. [Google Scholar] [CrossRef]
- Marete, A.; Ariel, O.; Ibeagha-Awemu, E.; Bissonnette, N. Identification of Long Non-coding RNA Isolated From Naturally Infected Macrophages and Associated with Bovine Johne’s Disease in Canadian Holstein Using a Combination of Neural Networks and Logistic Regression. Front. Vet. Sci. 2021, 8, 639053. [Google Scholar] [CrossRef]
- Kim, J.; Abdelmohsen, K.; Yang, X.; De, S.; Grammatikakis, I.; Noh, J.H.; Gorospe, M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016, 44, 2378–2392. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef] [PubMed]
- Wooten, S.; Smith, K.N. Long non-coding RNA OIP5-AS1 (Cyrano): A context-specific regulator of normal and disease processes. Clin. Transl. Med. 2022, 12, e706. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, P.; Tian, R.; Wang, S.; Guo, Q.; Luo, M.; Zhou, W.; Liu, G.; Jiang, H.; Jiang, Q. LncRNA2Target v2.0: A comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019, 47, D140–D144. [Google Scholar] [CrossRef]
- Ghoussaini, M.; Mountjoy, E.; Carmona, M.; Peat, G.; Schmidt, E.M.; Hercules, A.; Fumis, L.; Miranda, A.; Carvalho-Silva, D.; Buniello, A.; et al. Open Targets Genetics: Systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021, 49, D1311–D1320. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Yang, M.; Lu, H.; Liu, J.; Wu, S.; Kim, P.; Zhou, X. lncRNAfunc: A knowledgebase of lncRNA function in human cancer. Nucleic Acids Res. 2022, 50, D1295–D1306. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef]
- Zhou, B.; Ji, B.; Liu, K.; Hu, G.; Wang, F.; Chen, Q.; Yu, R.; Huang, P.; Ren, J.; Guo, C.; et al. EVLncRNAs 2.0: An updated database of manually curated functional long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res. 2021, 49, D86–D91. [Google Scholar] [CrossRef]
- Jacq, A.; Becquet, D.; Guillen, S.; Boyer, B.; Bello-Goutierrez, M.M.; Franc, J.L.; Francois-Bellan, A.M. Direct RNA-RNA interaction between Neat1 and RNA targets, as a mechanism for RNAs paraspeckle retention. RNA Biol. 2021, 18, 2016–2027. [Google Scholar] [CrossRef]
- Houtman, M.; Shchetynsky, K.; Chemin, K.; Hensvold, A.H.; Ramskold, D.; Tandre, K.; Eloranta, M.L.; Ronnblom, L.; Uebe, S.; Catrina, A.I.; et al. T cells are influenced by a long non-coding RNA in the autoimmune associated PTPN2 locus. J. Autoimmun. 2018, 90, 28–38. [Google Scholar] [CrossRef]
- Zhang, Q.; Chao, T.C.; Patil, V.S.; Qin, Y.; Tiwari, S.K.; Chiou, J.; Dobin, A.; Tsai, C.M.; Li, Z.; Dang, J.; et al. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019, 38, e100041. [Google Scholar] [CrossRef]
- Xu, J.; Xu, H.M.; Yang, M.F.; Liang, Y.J.; Peng, Q.Z.; Zhang, Y.; Tian, C.M.; Wang, L.S.; Yao, J.; Nie, Y.Q.; et al. New Insights Into the Epigenetic Regulation of Inflammatory Bowel Disease. Front. Pharmacol. 2022, 13, 813659. [Google Scholar] [CrossRef]
- Yarani, R.; Mirza, A.H.; Kaur, S.; Pociot, F. The emerging role of lncRNAs in inflammatory bowel disease. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Wang, S.; Hou, Y.; Chen, W.; Wang, J.; Xie, W.; Zhang, X.; Zeng, L. KIF9-AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease. Mol. Med. Rep. 2018, 17, 2195–2202. [Google Scholar] [CrossRef]
- Zacharopoulou, E.; Gazouli, M.; Tzouvala, M.; Vezakis, A.; Karamanolis, G. The contribution of long non-coding RNAs in Inflammatory Bowel Diseases. Dig. Liver Dis. 2017, 49, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, H.; Wei, Z.; Gou, X.; Liang, S.; Liu, F. A Novel Prognostic Model Based on Autophagy-Related Long Non-Coding RNAs for Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 711736. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.P.; Nobrega-Pereira, S.; Domingues-Silva, B.; Rebelo, K.; Alves-Vale, C.; Marinho, S.P.; Carvalho, T.; Dias, S.; Bernardes de Jesus, B. An antisense transcript mediates MALAT1 response in human breast cancer. BMC Cancer 2019, 19, 771. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Liu, R.; Wu, X.; Ma, K.; Luo, W.; Nie, K.; Zhang, C.; Meng, X.; Tong, T.; Chen, X.; et al. LncRNA NEAT1 mediates intestinal inflammation by regulating TNFRSF1B. Ann. Transl. Med. 2021, 9, 773. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, L.; Chen, P.; Wang, Y.; Yang, G.; Zhou, G.; Li, L.; Feng, R.; Qiu, Y.; Han, J.; et al. MALAT1 Maintains the Intestinal Mucosal Homeostasis in Crohn’s Disease via the miR-146b-5p-CLDN11/NUMB Pathway. J. Crohn’s Colitis 2021, 15, 1542–1557. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Ke, L.; Yang, D.C.; Wang, Y.; Ding, Y.; Gao, G. AnnoLnc2: The one-stop portal to systematically annotate novel lncRNAs for human and mouse. Nucleic Acids Res. 2020, 48, W230–W238. [Google Scholar] [CrossRef]
- Mudge, J.M.; Jungreis, I.; Hunt, T.; Gonzalez, J.M.; Wright, J.C.; Kay, M.; Davidson, C.; Fitzgerald, S.; Seal, R.; Tweedie, S.; et al. Discovery of high-confidence human protein-coding genes and exons by whole-genome PhyloCSF helps elucidate 118 GWAS loci. Genome Res. 2019, 29, 2073–2087. [Google Scholar] [CrossRef]
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801. [Google Scholar] [CrossRef]
- Triantaphyllopoulos, K.A.; Ikonomopoulos, I.; Bannister, A.J. Epigenetics and inheritance of phenotype variation in livestock. Epigenetics Chromatin 2016, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Bujold, D.; Morais, D.A.L.; Gauthier, C.; Cote, C.; Caron, M.; Kwan, T.; Chen, K.C.; Laperle, J.; Markovits, A.N.; Pastinen, T.; et al. The International Human Epigenome Consortium Data Portal. Cell Syst. 2016, 3, 496–499 e492. [Google Scholar] [CrossRef]
- Hagihara, Y.; Yoshimatsu, Y.; Mikami, Y.; Takada, Y.; Mizuno, S.; Kanai, T. Epigenetic regulation of T helper cells and intestinal pathogenicity. Semin. Immunopathol. 2019, 41, 379–399. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P.; Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Gonzalez-Serna, D.; Villanueva-Martin, G.; Acosta-Herrera, M.; Marquez, A.; Martin, J. Approaching Shared Pathophysiology in Immune-Mediated Diseases through Functional Genomics. Genes 2020, 11, 1482. [Google Scholar] [CrossRef]
- Chou, I.J.; Kuo, C.F.; Huang, Y.S.; Grainge, M.J.; Valdes, A.M.; See, L.C.; Yu, K.H.; Luo, S.F.; Huang, L.S.; Tseng, W.Y.; et al. Familial Aggregation and Heritability of Schizophrenia and Co-aggregation of Psychiatric Illnesses in Affected Families. Schizophr. Bull. 2017, 43, 1070–1078. [Google Scholar] [CrossRef]
- Woo, H.J.; Yu, C.; Kumar, K.; Reifman, J. Large-scale interaction effects reveal missing heritability in schizophrenia, bipolar disorder and posttraumatic stress disorder. Transl. Psychiatry 2017, 7, e1089. [Google Scholar] [CrossRef]
- Rajeev, R.; Dwivedi, A.P.; Sinha, A.; Agarwaal, V.; Dev, R.R.; Kar, A.; Khosla, S. Epigenetic interaction of microbes with their mammalian hosts. J. Biosci. 2021, 46, 94. [Google Scholar] [CrossRef]
- Woo, V.; Alenghat, T. Epigenetic regulation by gut microbiota. Gut Microbes 2022, 14, 2022407. [Google Scholar] [CrossRef]
- Han, X.; Huang, S.; Xue, P.; Fu, J.; Liu, L.; Zhang, C.; Yang, L.; Xia, L.; Sun, L.; Huang, S.K.; et al. LncRNA PTPRE-AS1 modulates M2 macrophage activation and inflammatory diseases by epigenetic promotion of PTPRE. Sci. Adv. 2019, 5, eaax9230. [Google Scholar] [CrossRef] [PubMed]
- Zhi, H.; Li, X.; Wang, P.; Gao, Y.; Gao, B.; Zhou, D.; Zhang, Y.; Guo, M.; Yue, M.; Shen, W.; et al. Lnc2Meth: A manually curated database of regulatory relationships between long non-coding RNAs and DNA methylation associated with human disease. Nucleic Acids Res. 2018, 46, D133–D138. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhang, L.; Fu, J.; Tian, T.; Liu, X.; Liu, Y.; Sun, H.; Li, D.; Zhu, L.; Xu, J.; et al. Development and validation of 3-CpG methylation prognostic signature based on different survival indicators for colorectal cancer. Mol. Carcinog. 2021, 60, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Ramos, G.P.; Papadakis, K.A. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin. Proc. 2019, 94, 155–165. [Google Scholar] [CrossRef]
- Schoultz, I.; Keita, A.V. Cellular and Molecular Therapeutic Targets in Inflammatory Bowel Disease-Focusing on Intestinal Barrier Function. Cells 2019, 8, 193. [Google Scholar] [CrossRef]
- Bharath, M.N.; Gupta, S.; Vashistha, G.; Ahmad, S.; Singh, S.V. Bioprospective Role of Ocimum sanctum and Solanum xanthocarpum against Emerging Pathogen: Mycobacterium avium Subspecies paratuberculosis: A Review. Molecules 2023, 28, 3490. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kimura, S.; Hase, K. M cell-dependent antigen uptake on follicle-associated epithelium for mucosal immune surveillance. Inflamm. Regen. 2018, 38, 15. [Google Scholar] [CrossRef]
- Loganes, C.; Pin, A.; Naviglio, S.; Girardelli, M.; Bianco, A.M.; Martelossi, S.; Tommasini, A.; Piscianz, E. Altered pattern of tumor necrosis factor-alpha production in peripheral blood monocytes from Crohn’s disease. World J. Gastroenterol. 2016, 22, 9117–9126. [Google Scholar] [CrossRef]
- Tsukamoto, H.; Takeuchi, S.; Kubota, K.; Kobayashi, Y.; Kozakai, S.; Ukai, I.; Shichiku, A.; Okubo, M.; Numasaki, M.; Kanemitsu, Y.; et al. Lipopolysaccharide (LPS)-binding protein stimulates CD14-dependent Toll-like receptor 4 internalization and LPS-induced TBK1-IKKϵ-IRF3 axis activation. J. Biol. Chem. 2018, 293, 10186–10201. [Google Scholar] [CrossRef]
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106. [Google Scholar] [CrossRef]
- Dong, C. Cytokine Regulation and Function in T Cells. Annu. Rev. Immunol. 2021, 39, 51–76. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef]
- Yang, E.; Shen, J. The roles and functions of Paneth cells in Crohn’s disease: A critical review. Cell Prolif. 2021, 54, e12958. [Google Scholar] [CrossRef] [PubMed]
- Clavero, E.; Sanchez-Maldonado, J.M.; Macauda, A.; Ter Horst, R.; Sampaio-Marques, B.; Jurczyszyn, A.; Clay-Gilmour, A.; Stein, A.; Hildebrandt, M.A.T.; Weinhold, N.; et al. Polymorphisms within Autophagy-Related Genes as Susceptibility Biomarkers for Multiple Myeloma: A Meta-Analysis of Three Large Cohorts and Functional Characterization. Int. J. Mol. Sci. 2023, 24, 8500. [Google Scholar] [CrossRef] [PubMed]
- Gazouli, M.; Pachoula, I.; Panayotou, I.; Mantzaris, G.; Chrousos, G.; Anagnou, N.P.; Roma-Giannikou, E. NOD2/CARD15, ATG16L1 and IL23R gene polymorphisms and childhood-onset of Crohn’s disease. World J. Gastroenterol. 2010, 16, 1753–1758. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa-Ishimoto, Y.; Shono, Y.; Gomez, L.E.; Hubbard-Lucey, V.M.; Cammer, M.; Neil, J.; Dewan, M.Z.; Lieberman, S.R.; Lazrak, A.; Marinis, J.M.; et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 2017, 214, 3687–3705. [Google Scholar] [CrossRef]
- Gao, P.; Liu, H.; Huang, H.; Sun, Y.; Jia, B.; Hou, B.; Zhou, X.; Strober, W.; Zhang, F. The Crohn Disease-associated ATG16L1(T300A) polymorphism regulates inflammatory responses by modulating TLR- and NLR-mediated signaling. Autophagy 2022, 18, 2561–2575. [Google Scholar] [CrossRef]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef]
- Qu, X.; Zheng, C.; Wang, B.; Wang, F.; Sun, X.; Gao, Y.; Xia, Q.; Kong, X. Comprehensive analysis of circular RNAs from steatotic livers after ischemia and reperfusion injury by next-generation RNA sequencing. FEBS Lett. 2021, 595, 99–109. [Google Scholar] [CrossRef]
- Wehkamp, J.; Stange, E.F. An Update Review on the Paneth Cell as Key to Ileal Crohn’s Disease. Front. Immunol. 2020, 11, 646. [Google Scholar] [CrossRef]
- Ntunzwenimana, J.C.; Boucher, G.; Paquette, J.; Gosselin, H.; Alikashani, A.; Morin, N.; Beauchamp, C.; Thauvette, L.; Rivard, M.E.; Dupuis, F.; et al. Functional screen of inflammatory bowel disease genes reveals key epithelial functions. Genome Med. 2021, 13, 181. [Google Scholar] [CrossRef] [PubMed]
- Baillie, J.K.; Arner, E.; Daub, C.; De Hoon, M.; Itoh, M.; Kawaji, H.; Lassmann, T.; Carninci, P.; Forrest, A.R.; Hayashizaki, Y.; et al. Analysis of the human monocyte-derived macrophage transcriptome and response to lipopolysaccharide provides new insights into genetic aetiology of inflammatory bowel disease. PLoS Genet. 2017, 13, e1006641. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Wang, X.; Li, L.; Wei, H.; Peng, J. Multifaceted involvements of Paneth cells in various diseases within intestine and systemically. Front. Immunol. 2023, 14, 1115552. [Google Scholar] [CrossRef]
- Castellanos-Rubio, A.; Kratchmarov, R.; Sebastian, M.; Garcia-Etxebarria, K.; Garcia, L.; Irastorza, I.; Ghosh, S. Cytoplasmic Form of Carlr lncRNA Facilitates Inflammatory Gene Expression upon NF-kappaB Activation. J. Immunol. 2017, 199, 581–588. [Google Scholar] [CrossRef]
- Zhao, Q.; Pang, G.; Yang, L.; Chen, S.; Xu, R.; Shao, W. Long Noncoding RNAs Regulate the Inflammatory Responses of Macrophages. Cells 2021, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I. Evolution to the rescue: Using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016, 17, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, B.; Peng, W.; Ma, Y.; Huang, Y.; Lan, X.; Lei, C.; Qi, X.; Liu, G.E.; Chen, H. LncRNA-MEG3 promotes bovine myoblast differentiation by sponging miR-135. J. Cell. Physiol. 2019, 234, 18361–18370. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Cui, L.; Li, Y.; Cao, Z.; Wu, X.; Wang, Q.; Zhang, B.; Ma, C.; Cheng, Y. Long Non-coding RNA MEG3 Alleviated Ulcerative Colitis Through Upregulating miR-98-5p-Sponged IL-10. Inflammation 2021, 44, 1049–1059. [Google Scholar] [CrossRef]
- Wang, Y.X.; Lin, C.; Cui, L.J.; Deng, T.Z.; Li, Q.M.; Chen, F.Y.; Miao, X.P. Mechanism of M2 macrophage-derived extracellular vesicles carrying lncRNA MEG3 in inflammatory responses in ulcerative colitis. Bioengineered 2021, 12, 12722–12739. [Google Scholar] [CrossRef]
- Xue, Y.L.; Zhang, S.X.; Zheng, C.F.; Li, Y.F.; Zhang, L.H.; Su, Q.Y.; Hao, Y.F.; Wang, S.; Li, X.W. Long non-coding RNA MEG3 inhibits M2 macrophage polarization by activating TRAF6 via microRNA-223 down-regulation in viral myocarditis. J. Cell. Mol. Med. 2020, 24, 12341–12354. [Google Scholar] [CrossRef]
- Jin, Y.; Yang, Q.; Zhang, M.; Zhang, S.; Cai, H.; Dang, R.; Lei, C.; Chen, H.; Lan, X. Identification of a Novel Polymorphism in Bovine lncRNA ADNCR Gene and Its Association with Growth Traits. Anim. Biotechnol. 2019, 30, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, X.; Cai, H.; Sun, Y.; Plath, M.; Li, C.; Lan, X.; Lei, C.; Lin, F.; Bai, Y.; et al. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim. Biophys. Acta 2016, 1859, 871–882. [Google Scholar] [CrossRef]
- Seva, J.; Sanes, J.M.; Mas, A.; Ramis, G.; Sanchez, J.; Parraga-Ros, E. Prevalence of Mycobacterium avium Subsp. paratuberculosis in Feral Pigeons (Columba livia) Associated with Difficulties Controlling Paratuberculosis in a Bovine Herd (Fighting Bull Breed). Animals 2022, 12, 3314. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.K.; Kim, T.S.; Kim, J.I.; Yim, J.J. Whole genome sequencing of Nontuberculous Mycobacterium (NTM) isolates from sputum specimens of co-habiting patients with NTM pulmonary disease and NTM isolates from their environment. BMC Genom. 2020, 21, 322. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.M.F.; DePaolo, R.W. Infectious Scarring: Setting the Trigger for Intestinal Inflammation. Cell Host Microbe 2018, 23, 154–156. [Google Scholar] [CrossRef]
- Yang, W.H.; Heithoff, D.M.; Aziz, P.V.; Sperandio, M.; Nizet, V.; Mahan, M.J.; Marth, J.D. Recurrent infection progressively disables host protection against intestinal inflammation. Science 2017, 358, eaao5610. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Dong, Y.; Lin, G.; Cao, Y. Long Noncoding RNA Antisense Noncoding RNA in the INK4 Locus Correlates With Risk, Severity, Inflammation and Infliximab Efficacy in Crohn’s Disease. Am. J. Med. Sci. 2019, 357, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Aslam, N.; Lo, S.W.; Sikafi, R.; Barnes, T.; Segal, J.; Smith, P.J.; Limdi, J.K. A review of the therapeutic management of ulcerative colitis. Ther. Adv. Gastroenterol. 2022, 15, 17562848221138160. [Google Scholar] [CrossRef]
- Lucafo, M.; Bravin, V.; Tommasini, A.; Martelossi, S.; Rabach, I.; Ventura, A.; Decorti, G.; De Iudicibus, S. Differential expression of GAS5 in rapamycin-induced reversion of glucocorticoid resistance. Clin. Exp. Pharmacol. Physiol. 2016, 43, 602–605. [Google Scholar] [CrossRef]
- Lucafo, M.; Di Silvestre, A.; Romano, M.; Avian, A.; Antonelli, R.; Martelossi, S.; Naviglio, S.; Tommasini, A.; Stocco, G.; Ventura, A.; et al. Role of the Long Non-Coding RNA Growth Arrest-Specific 5 in Glucocorticoid Response in Children with Inflammatory Bowel Disease. Basic Clin. Pharmacol. Toxicol. 2018, 122, 87–93. [Google Scholar] [CrossRef]
- Lucafo, M.; Pugnetti, L.; Bramuzzo, M.; Curci, D.; Di Silvestre, A.; Marcuzzi, A.; Bergamo, A.; Martelossi, S.; Villanacci, V.; Bozzola, A.; et al. Long Non-Coding RNA GAS5 and Intestinal MMP2 and MMP9 Expression: A Translational Study in Pediatric Patients with IBD. Int. J. Mol. Sci. 2019, 20, 5280. [Google Scholar] [CrossRef]
- Maruyama, R.; Yokota, T. Knocking Down Long Noncoding RNAs Using Antisense Oligonucleotide Gapmers. Methods Mol. Biol. 2020, 2176, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Gong, N.; Teng, X.; Li, J.; Liang, X.J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater. Interfaces 2019, 11, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Yuan, Y.; Yi, T.; Dang, W. The Roles of Antisense Long Noncoding RNAs in Tumorigenesis and Development through Cis-Regulation of Neighbouring Genes. Biomolecules 2023, 13, 684. [Google Scholar] [CrossRef]
- Hamazaki, N.; Nakashima, K.; Hayashi, K.; Imamura, T. Detection of Bidirectional Promoter-Derived lncRNAs from Small-Scale Samples Using Pre-Amplification-Free Directional RNA-seq Method. Methods Mol. Biol. 2017, 1605, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Jadaliha, M.; Gholamalamdari, O.; Tang, W.; Zhang, Y.; Petracovici, A.; Hao, Q.; Tariq, A.; Kim, T.G.; Holton, S.E.; Singh, D.K.; et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018, 14, e1007802. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Lin, A.; Li, C.; Liang, K.; Wang, S.; Liu, Y.; Park, P.K.; Qin, L.; Wei, Y.; Hawke, D.H.; et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014, 159, 1110–1125. [Google Scholar] [CrossRef]
- Tian, L.I.; Huang, Y.; Zhang, B.; Song, Y.I.; Yang, L.; Chen, Q.; Wang, Z.; Wang, Y.; He, Q.; Yang, W.; et al. Targeting LncRNA LLNLR-299G3.1 with antisense oligonucleotide inhibits malignancy of esophageal squamous cell carcinoma cells in vitro and in vivo. Oncol. Res. 2023, 31, 463–479. [Google Scholar] [CrossRef]
- Simner, P.J.; Salzberg, S.L. The Human “Contaminome” and Understanding Infectious Disease. N. Engl. J. Med. 2022, 387, 943–946. [Google Scholar] [CrossRef]


| NONCODE | Human | Mouse | Cow | Pig | Chicken |
|---|---|---|---|---|---|
| Genes | 96,411 | 87,890 | 22,227 | 17,811 | 9527 |
| Non-Coding Transcripts | 173,112 | 131,974 | 23,515 | 29,585 | 12,850 |
| GENECODE, NCBI, and Ensembl | Human | Mouse | Cow | Pig | Chicken |
| Coding Genes | 19,988 | 21,833 | 21,880 | 21,303 | 16,878 |
| Coding Transcripts | 87,814 | 59,138 | 43,984 | 63,041 | 39,288 |
| Disease | Source | Gene Expression Change | Transcript/Gene Name | Gene ID | Cited by, Reference | Location (hg38) | Reported Mechanism |
|---|---|---|---|---|---|---|---|
| DSS-induced colitis | Mice serum and tissues | Upregulated | NEAT1 | NEAT1 | [122] | chr11 | Modulated intestinal epithelial barrier |
| UC & CD | Colonic tissues | Upregulated | CCAT1 | lnc-APPL2-1, lnc-BMP6-106, lnc-FAM84B-15 | [123] | chr12, chr6, chr8 | Increased barrier permeability |
| UC | Colonic tissues | Upregulated | ANRIL | CDKN2B-AS1 | [124] | chr9 | Promoted inflammatory cytokines and chemokines production |
| UC | Colonic tissues | Downregulated | CDKN2B-AS1 | CDKN2B-AS1 | [125] | chr9 | Enhanced the barrier formation |
| UC | Colonic tissues | Upregulated | IFNG-AS1 | IFNG-AS1 | [126] | chr12 | Enhanced inflammation |
| UC | Colonic tissues | Upregulated | H19 | H19 | [127] | chr11 | Disrupted intestinal epithelial barrier function |
| CD | Plasma | Upregulated | ENST00000466668 | GUSBP2 | [128] | chr6 | |
| ENST00000422548 | AL022100.2 | chr1 | |||||
| ENST00000502712 | RP11-68L1.2 | chr3 | |||||
| ENST00000425364 | lnc-REV3L-1 | chr6 | |||||
| LINC01272 | |||||||
| NR_037605 | GAS5-AS1 | chr1 | |||||
| ENST00000562996 | FIGNL2-DT divergent transcript | chr12 | |||||
| NR_038927 | DDX11-AS1 | chr12 | |||||
| TCONS_00014043(*) | - | chr7 | |||||
| TCONS_00012771(*) | - | chr6 | |||||
| ENST00000569039 (novel transcript) | Antisense to C16orf53 | chr16 | |||||
| Downregulated | uc001ody.3 | - | chr11 | ||||
| ENST00000575787 | ALOX12P2 | chr17 | |||||
| ENSGO0000259472 | LOC80154(uc010bmo.1) | chr15 | |||||
| ENST00000509252 | TRIM52-AS1 | chr5 | |||||
| ENST00000413954 | AC064871.3 | chr2 | |||||
| ENST00000431104 | RNF217 | chr6 | |||||
| Ileal Biopsy Terminal Ileum | ENST00000452481.1 | HNF4A-AS1 | [129] | chr20 | |||
| ENSGO0000245060.2 (uc011dhd.3) | LINC00847 | chr5 | |||||
| TCONS_00020749(*) | - | chr12 | |||||
| NR_027074 | LINC00928 | chr15 | |||||
| TCONS_00027621(*) | - | chr19 | |||||
| CD | Upregulated | ENST00000460164.1 | lnc-BRF1-9 | chr14 | |||
| ENST00000532855.1 | MMP12 | chr11 | |||||
| ENST00000326227.5 | MMP12 | chr11 | |||||
| Whole Blood | ENST00000419897.1 ENST00000535913.2 | SLC12A5-AS1 | chr20 | ||||
| ENST00000520185.1 | ENST00000520185 | chr11 | |||||
| ENST00000526690.1 | lnc-ZNF705D-2 | chr8 | |||||
| ENST00000445003.1 | lnc-CEBPB-13 | chr20 | |||||
| ENST00000522970.1 | lnc-ADAM2-1 | chr8 | |||||
| ENST00000524555.1 | lnc-SAA2-SAA4-1 | chr11 | |||||
| ENST00000429315.2 | KIF9-AS1 | chr3 | |||||
| Downregulated | ENST00000432658.1 | DPP10-AS1 | chr2 | ||||
| ENST00000401008.2 | lnc-NBPF11-2 | chr1 | |||||
| ENST00000553575.1 | DIO3OS | chr14 | |||||
| ENST00000554694.1 | DIO3OS | chr14 | |||||
| ENST00000557532.1 | DIO3OS | chr14 | |||||
| ENST00000557109.1 | DIO3OS | chr14 | |||||
| ENST00000422420.1 | CDKN2B-AS1 | chr9 | |||||
| ENST00000428597.1 | CDKN2B-AS1 | chr9 | |||||
| ENST00000554441.1 | DIO3OS | chr14 | |||||
| ENST00000554735.1 | DIO3OS | chr14 | |||||
| CD | Ileal tissues | Upregulated | ENST00000487539.1_1 | MMP12 | [130] | chr11 | Involved in the pathogenesis of CD |
| ENST00000409569.2_1 | MIR4435-1HG | chr2 | |||||
| ENST00000392442.6_1 | RSRC2 | chr12 | |||||
| Downregulated | ENST00000524613.5_1 | ENSG00000254645 | chr11 | ||||
| ENST00000465605.5_1 | ENSG00000234539 | chr6 | |||||
| CD | Blood | Upregulated | DQ786243 | - | [131] | chr1 | Affected CREB and Foxp3 expression and regulated Tregs function |
| UC | Colonic tissues | Upregulated | ENST00000460164.1 | lnc-BRF1-9 | [132] | chr14 | |
| ENST00000532855.1 | MMP12 | chr11 | |||||
| ENST00000326227.5 | MMP12 | chr11 | |||||
| ENST00000419897.1 ENST00000535913.2 | SLC12A5-AS1 | chr20 | An IBD that has material basis in variation in the chromosome region 12q15. | ||||
| ENST00000429315.2 | KIF9-AS1 | chr3 | |||||
| ENST00000526690.1 | lnc-ZNF705D-2 | chr8 | |||||
| ENST00000524555.1 | lnc-SAA2-SAA4-1 | chr11 | |||||
| ENST00000476886.1 | CLRN1-AS1 | chr3 | |||||
| ENST00000517774.1 | TNFRSF10A-DT | chr8 | |||||
| ENST00000578280.1 | lnc-IGFBP4-1 | chr17 | |||||
| Downregulated | ENST00000422420.1 | CDKN2B-AS1 | chr9 | ||||
| ENST00000428597.1 | CDKN2B-AS1 | chr9 | |||||
| ENST00000585267.1 | CDKN2B-AS1 | chr9 | |||||
| ENST00000580576.1 | CDKN2B-AS1 | chr9 | |||||
| ENST00000577551.1 | CDKN2B-AS1 | chr9 | |||||
| ENST00000581051.1 | CDKN2B-AS1 | chr9 | |||||
| ENST00000582072.1 | CDKN2B-AS1 | chr9 | |||||
| ENST00000401008.2 | lnc-NBPF11-2 | chr1 | |||||
| ENST00000432658.1 | DPP10-AS1 | chr2 | |||||
| ENST00000421632.1 | CDKN2B-AS1 | chr2 | |||||
| Upregulated | ENSG00000070190 | DAPP1-001 | chr4 | Gene hypomethylated. May act as a B-cell-associated adapter that regulates B-cell antigen receptor (BCR)-signaling downstream of PI3K | |||
| UC | Colonic tissues | Upregulated | BC012900 | - | [133] | chr8 | Regulated intestinal epithelial cells apoptosis |
| AK001903 | - | chr7 | |||||
| AK023330 | - | chr9 | |||||
| Downregulated | BC029135 | - | chr10 | ||||
| CDKN2B-AS1 | CDKN2B-AS1 | chr9 | |||||
| UC | Colonic tissues | Downregulated | ENST00000647780.1 | MEG3 | [134] | Chr14 | LncRNA MEG3 could improve ulcerative colitis by upregulating miR-98-5p-Sponged IL-10 |
| UC | BC062296- | chr21 | |||||
| UC | colonic epithelial cell | Upregulated | ENST00000619449.2 | MALAT1 | [135] | chr11 | promotes ulcerative colitis by upregulating lncRNA ANRIL |
| UC | Colonic tissues | Upregulated | H19 | H19 | [136] | chr11 | Promoted mucosal regeneration |
| UC | MSICT | Upregulated | H19 | H19 | [137] | chr11 | Regulated intestinal epithelial barrier by interacting with HuR |
| DSS-induced colitis, IBD | Mice colonic tissues | Upregulated | CRNDE | lnc-IRX3-80 | [138] | chr16 | Promoted epithelial cells apoptosis |
| UC | Colonic tissues | Downregulated | SPRY4-IT1 (NR_131221.1) | SPRY4-IT1 | [139] | chr5 | Regulated intestinal epithelial barrier function |
| UC | Mice small intestinal tissues | Upregulated | uc.173 | uc.173 | [140] | - | Stimulated intestinal epithelium renewal & regulation intestinal epithelial barrier function |
| DSS-induced injury | IEBM | Upregulated | PlncRNA1 | PlncRNA1 | [141] | chr21 | Regulated tight junction proteins |
| UC | Colonic tissues | Upregulated | IFNG-AS1 | IFNG-AS1 | [142] | chr12 | Regulated pro-inflammatory cascade |
| UC | Blood and monocytes | Upregulated | MROCKI (uc003pwa.3) | MROCKI (ENSG00000227502.2) | [142] | chr6 | Promoted inflammatory cytokines and chemokines production |
| UC | Mice colonic tissues | Upregulated | HIF1A-AS2 | HIF1A-AS2, lnc-TMEM30B-9 | [143] | chr14 | Negatively regulated intestinal inflammation and exerts oncogenic functions in colorectal cancer |
| EC, IBD | Colonic tissues | Downregulated | NRON | NCRNA00194 | [144] | chr9 | NRON is a repressor of NFAT component LRRK2 in EC. |
| SLE, Lupus nephritis | Innate & Acquired immune system, Blood, Nervous system | Upregulated | ENST00000607592.2 | lincRNA-COX2 | [145] | chr3 | Control of inflammatory response. Induction followed by TLR2 and TLR4 stimulation in NFκB dependent manner. Maintain homeostasis within the lung |
| UC & TNF-α-treated HT-29 cells | Colonic tissues | Downregulated | ENST00000644773.3 | TUG1 | [146] | chr22 | TUG1 played protective role in UC by preventing TNF-α-induced cell injury and inflammation |
| UC, IBD | PBMCs | Downregulated | LINC01882 | lnc-PTPN2-2 | [147] | chr18 | Involved in T cells activation and IL-2 expression |
| DSS-induced colitis | Mice colonic tissues | Downregulated | NEAT1 | NEAT1 | [148] | chr11 | Regulated by 5-ALA and involved in PDT therapy treated colitis |
| Chemicaly induced colitis, UC, inflammed lung | Colonic tissues | Upregulated | ENST00000306042.9 | PTPRE-AS1 | [149] | chr10 | controls macrophage function, exacerbates chemical-induced colitis |
| Celiac, intestinal disease | Colonic tissues | Upregulated | ENST00000518376 | LncRNA-CARL, LINC00990 | [150] | chr8 | In celiac disease patients, increased levels of the Carlr transcript were detected in the cytoplasm, alongside elevated expression of NF-κB pathway genes |
| ID | Name | Chromosome | Disease Name | Interaction | Interaction Target | NCBI Accession | Description (Interaction) | Description (Function) | PMID |
|---|---|---|---|---|---|---|---|---|---|
| EL0129 (--) | ADNCR | 13 | N/A | binding | miR-204 | NR_137293 | LncRNA ADNCR functions by targeting miR-204 to significantly regulate the expression of the target SIRT1 gene in preadipocytes at both the mRNA and protein levels, thereby inhibiting adipogenesis. | Identification of a novel polymorphism in the bovine lncRNA ADNCR gene and its association with growth traits. LncRNA ADNCR suppresses adipogenic differentiation by targeting miR-204. | 29631473 27156885 |
| ENSG00000226950 | DANCR | 4 | N/A | miR-4449 (ENSBTAT00000064646) | NR_131910.1 | miR-4449 is enriched in the serum exosomes of diabetic kidney disease patients. These exosomes regulate the expression of proinflammatory cytokines, ROS levels, and pyroptosis through miR-4449 | 17488528 | ||
| EL1185 (--) | H19 | 29 | bovine mastitis | N/A | N/A | NR_003958 | All observations imply that lncRNA H19 modulates the TGF-β1-induced epithelial to mesenchymal transition in BEC through the PI3K/AKT signaling pathway, suggesting that MEC might be one source of myofibroblasts in vivo in the mammary glands under inflammatory conditions, thereby contributing to mammary gland fibrosis. | The overexpression of lncRNA H19 changes the basic characteristics and affects the immune response of bovine mammary epithelial cells. | 29062612 |
| EL1909 (--) | lnc133b | N/A | N/A | binding | miR-133b | N/A | IGF1R is an important target gene of miR-133b in bovine skeletal muscle satellite cells. lnc133b increases IGF1R expression by the “sponge” miR-133b. | Lnc133b promotes bovine skeletal muscle satellite cell proliferation and represses differentiation by acting as a ceRNA for miR-133b. | 28757453 |
| EL1922 (--) | lnc403 | N/A | N/A | regulation | KRAS, Myf6 | N/A | lnc403 negatively regulates the expression of the adjacent gene Myf6 and positively regulates the expression of the interaction protein KRAS. | A novel lncRNA, lnc403, involved in bovine skeletal muscle myogenesis by mediating KRAS/Myf6. | 32387386 |
| EL2003 (--) | lncKBTBD10 | N/A | N/A | regulation | KBTBD10 | N/A | LncKBTBD10 could induce a decrease in the KBTBD10 protein and further affect bovine skeletal muscle myo genesis. | A novel long non-coding RNA, lncKBTBD10, involved in bovine skeletal muscle myogenesis. | 30465303 |
| EL2170 (--) | lncRNA-TUB | N/A | bovine mastitis | N/A | N/A | N/A | A novel long non-coding RNA regulates the immune response in MAC-T cells and contributes to bovine mastitis. | 30771271 | |
| EL2345 (--) | LRRC75A-AS1 | N/A | bovine mastitis | regulation | LRRC75A | N/A | LRRC75A antisense lncRNA1 knockout attenuates inflammatory responses of bovine mammary epithelial cells. | LRRC75A-AS1 may protect LRRC75A from degradation by binding to its CDS region. | 31929753 |
| EL2406 (--) | MDNCR | N/A | N/A | binding | miR-133a | N/A | MDNCR was observed to directly bind to miR-133a with 32 potential binding sites | MDNCR binding to miR-133a promotes cell differentiation by targeting GosB in cattle primary myoblasts. | 30195797 |
| EL2412 (ENSG00000214548) | MEG3 | 21 | N/A | binding | miR-135B | NR_037684 | LncRNA-MEG3 promotes bovine myoblast differentiation by sponging miR-135. | LncRNA-MEG3 was verified as a miR-135 sponge. Overexpression of miR-135 markedly inhibits wild-type lncRNA-MEG3. Also, it has prognostic value in cervical cancer in humans. | 30887511 28057015 |
| EL2415 (ENSG00000258399, ENSG00000225746) | MEG8 | 21 | N/A | N/A | N/A | NR_146189 | Three SNP sites were identified in these three lncRNAs. They showed monoallelic expression in the analyzed tissues, suggesting that they may be imprinted in cattle. | 27925264 | |
| EL2417 (--) | MEG8-IT3 | N/A | N/A | N/A | N/A | N/A | Similar SNP effect to the previous variant detailed above | 27925264 | |
| EL2451 (--) | MIR221HG | N/A | N/A | N/A | N/A | N/A | MIR221HG Is a novel LncRNA that inhibits bovine adipocyte differentiation. | 31887993 | |
| EL2668 (--) | NONBTAT017009.2 | N/A | N/A | N/A | N/A | N/A | MiR-21-3p centric regulatory network in dairy cow MEC proliferation. | 31532202 | |
| EL3546 (--) | TCONS_00041733 | N/A | N/A | co-expression | EFNA1 | N/A | We found that TCONS_00041733 lncRNA targets the node gene EFNA1 (ephrin A1), which is involved in male reproductive physiology. | Integrated analysis of mRNAs and lncRNAs in the semen from Holstein bulls with high and low sperm motility. | 30765858 |
| ENSBTAG00000010766 | OIP5 | 10 | N/A, cancer (human) | expression and binding | ENST00000507296.1 lncRNA, MIS18 binding protein 1, HuR, mir-424 | The RNA-binding protein HuR, which enhances cell prolifer- ation, associates with OIP5-AS1 and stabilizes it. mir-424 interacts with OIP5-AS1 and competes with HuR for binding to OIP5-AS1. | OIP5-AS1 reduces cell proliferation and serves as a sponge or a competing endogenous (ce)RNA for HuR, restricting its availability to HuR target mRNAs and thereby repressing HuR-elicited proliferative phenotypes. | 26819413 | |
| EL3885 (ENSG00000229807) | XIST | X | glioma | binding | miR-34a, miR- 429 | XR_001495596, XR_001495595, XR_001495594 | XIST serves as a ceRNA for miR-34a through sponging miR-34a, competing with MET for miR-34a binding, and finally, modulating thyroid cancer cell proliferation and tumor growth. The MiR-429 inhibitor restores the metastatic and proangiogenic abilities of gliomas abolished by silencing XIST. | LncRNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. The XIST/miR-34a axis modulates cell proliferation and tumor growth of TC. XIST expression is promoted by the activated NF-κB pathway; in turn, XIST generates (-) feedback to regulate the NF-κB/NLRP3 inflammasome pathway. | 30463570 29187887 30463570 30362186 |
| EL3948 (--) | XLOC_059976 | N/A | N/A | N/A | N/A | N/A | XLOC_059976 could be used as candidate marker for milk protein content prediction. | 30105049 |
| Species | Human and Cattle | Human | Cattle | |||||
|---|---|---|---|---|---|---|---|---|
| Pathology | Immune Regulation/ Deregulation | IBD-Related Pathologies | Additional lncRNAs in the CD (Dataset GSE75459) (§) | Crohn’s Disease/Upregulated | Crohn’s Disease/Downregulated | Ulcerative Colitis/Upregulated | Ulcerative Colitis/Downregulated | Johne’s Disease |
| H19 | CBR3-AS1 | Upregulated | GAS5-AS1 | DPP10-AS1 | H19 | NRON | H19 | |
| DQ786243 | GUSBP2 | GUSBP3, 14, 15, 16 | DQ786243 | ALOX12P2 | ANRIL (†) | DPP10-AS1 | ADNCR | |
| BC012900 | GAS5-AS1 | FIGNL2-DT | GUSBP2 | DIO3OS | KIF9-AS1 | lnc-PTPN2-2 | DANCR | |
| CDKN2B-AS1 | NRON | GAS5-AS1 | TALAM1 | CDKN2B-AS1 | IFNG-AS1 | CDKN2B-AS1 | lnc133b | |
| NEAT1 | BC012900 | Downregulated | DDX11-AS1 | TRIM52-AS1 | NEAT1 (*) | NEAT1 (*) | lnc403 | |
| LncRNA | CCAT1 | LINC01272 | ALOX12P2 var1, 2 | CCAT1 | HNF4A-AS1 (#) | CCAT1 | RNF217 | LRRC75A-AS1 |
| MALAT1 | DIO3OS | ALOX12E | MALAT1 | CDKN2B-AS1 | MALAT1 | SLC12A5-AS1 | MEG3 | |
| lincRNA-Cox2 | DDX11-AS1 | GOLGA2P8 | lnc-MICAL2-1 | FOXD2-AS1 | SPRY4-AS1 | MEG8 | ||
| DANCR | IFNG-AS1 | MALAT1 var1,2,3 | SLC12A5-AS1 | SLC12A5-AS1 | Mirt2 | OIP5-AS1 | ||
| MEG3 | KIF9-AS1 | TRIM52 var1,2,3 | LINC01272 (#) | CRNDE (DSS) | MEG3 | MDNCR | ||
| OIP5-AS1 | CRNDE (DSS) | TALAM1 | lncRNA-CARL | MROCKI | TUG1 | lncKBTBD10 | ||
| uc.173 | uc.173 | MALAT1 | ||||||
| lncRNA-CARL | lncRNA-CARL | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Triantaphyllopoulos, K.A. Long Non-Coding RNAs and Their “Discrete” Contribution to IBD and Johne’s Disease—What Stands out in the Current Picture? A Comprehensive Review. Int. J. Mol. Sci. 2023, 24, 13566. https://doi.org/10.3390/ijms241713566
Triantaphyllopoulos KA. Long Non-Coding RNAs and Their “Discrete” Contribution to IBD and Johne’s Disease—What Stands out in the Current Picture? A Comprehensive Review. International Journal of Molecular Sciences. 2023; 24(17):13566. https://doi.org/10.3390/ijms241713566
Chicago/Turabian StyleTriantaphyllopoulos, Kostas A. 2023. "Long Non-Coding RNAs and Their “Discrete” Contribution to IBD and Johne’s Disease—What Stands out in the Current Picture? A Comprehensive Review" International Journal of Molecular Sciences 24, no. 17: 13566. https://doi.org/10.3390/ijms241713566