Cloning of PmMYB6 in Pinus massoniana and an Analysis of Its Function
Abstract
:1. Introduction
2. Results
2.1. Isolation and Characterization of PmMYB6
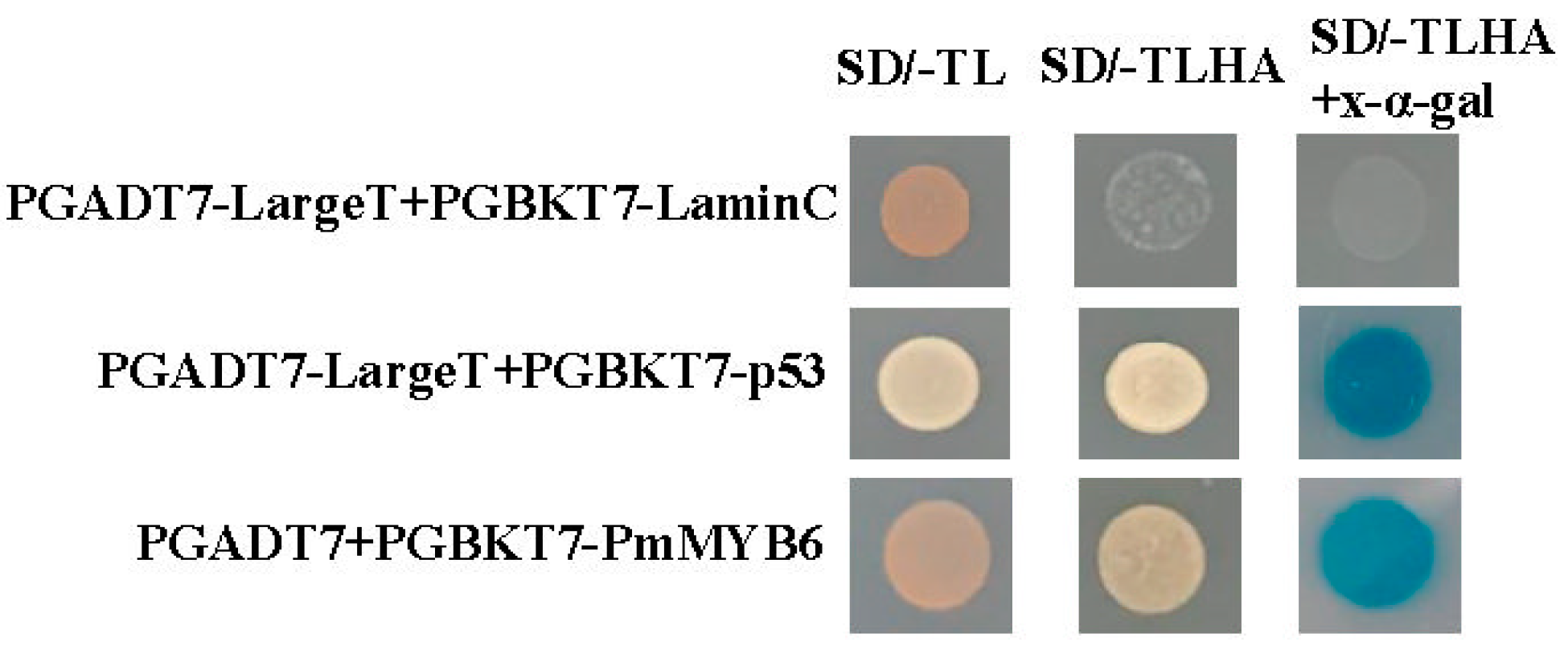
2.2. Transcriptional Activation of PmMYB6
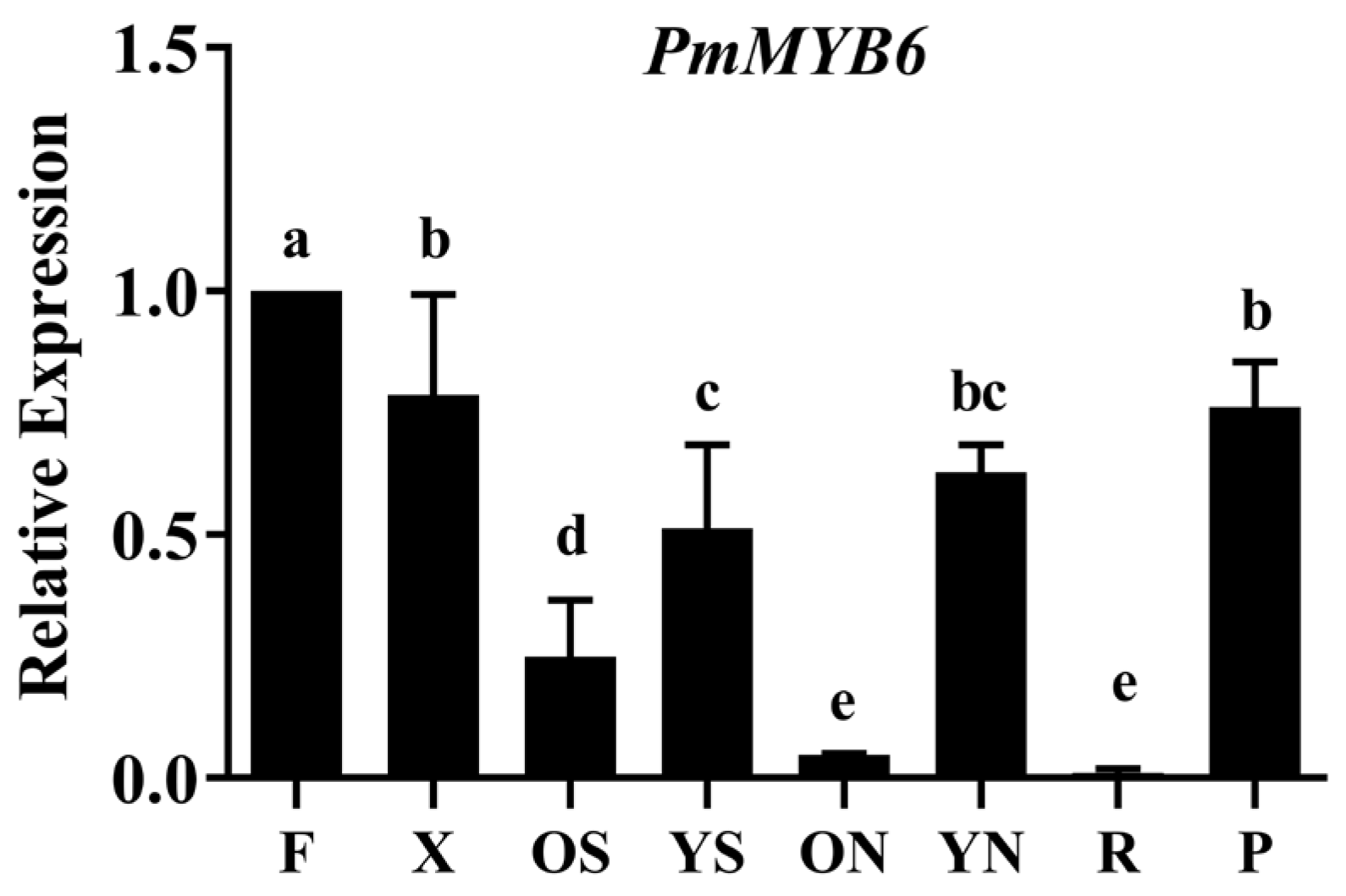
2.3. The Expression Patterns of PmMYB6 in P. massoniana
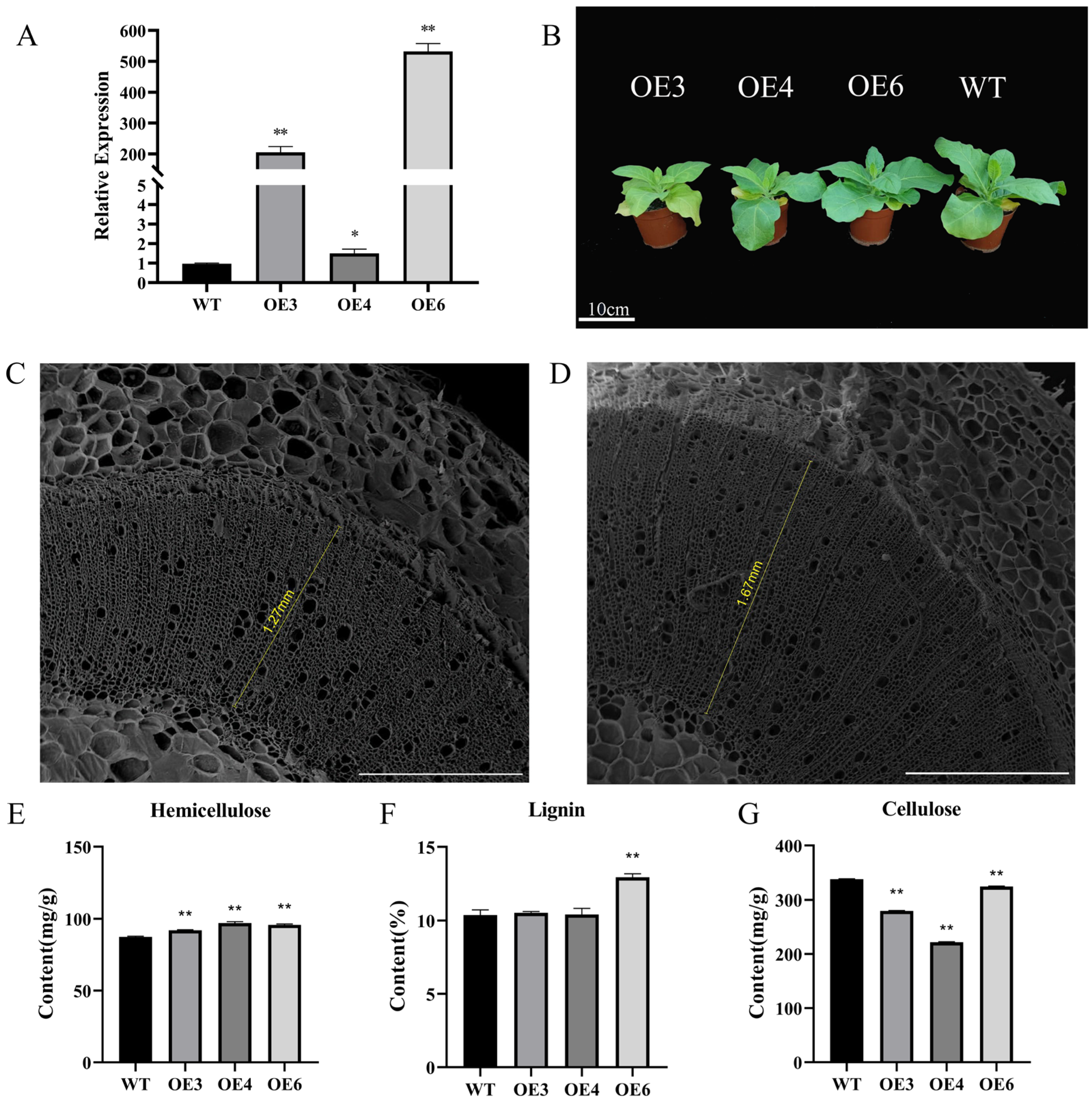
2.4. Overexpression of PmMYB6 in Tobacco Promotes the Accumulation of LIGNIN Content
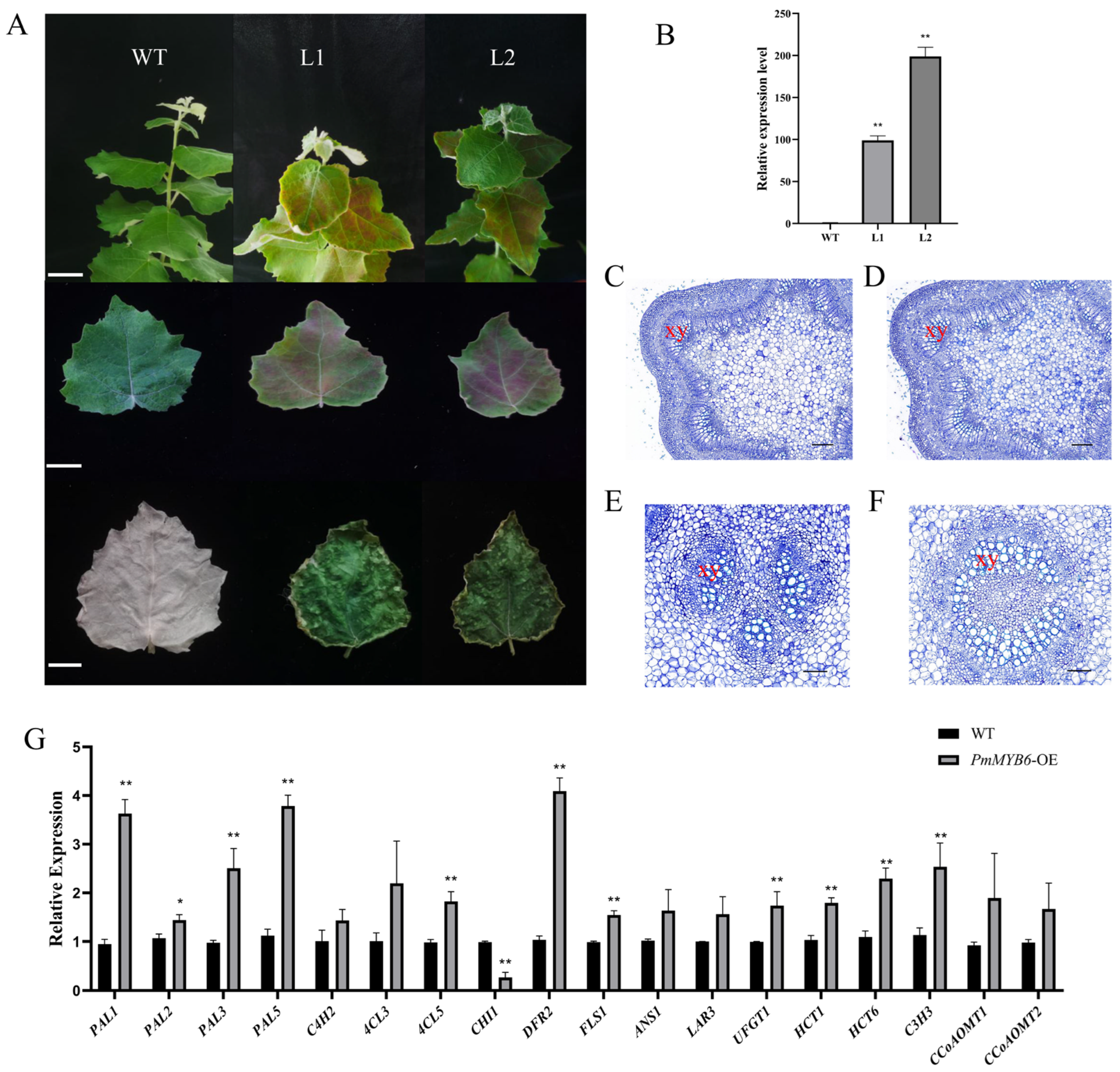
2.5. Overexpression of PmMYB6 in Poplar Promotes the Accumulation of Flavonoid Content
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Isolation and First-Strand cDNA Synthesis
4.3. RT-qPCR
4.4. Agrobacterium Transformation
4.5. Scanning Electron Microscope
4.6. Histological Sectioning
4.7. Determination of Secondary Wall Fraction Content
4.8. DMACA Staining of Poplar Leaves
4.9. Analysis of Flavonoid Composition
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shahivand, M.; Drikvand, R.M.; Gomarian, M.; Samiei, K. Key genes in the phenylpropanoids biosynthesis pathway have different expression patterns under various abiotic stresses in the Iranian red and green cultivars of sweet basil (Ocimum basilicum L.). Plant Biotechnol. Rep. 2021, 15, 585–594. [Google Scholar] [CrossRef]
- Khare, S.; Singh, N.B.; Singh, A.; Hussain, I.; Niharika, K.; Yadav, V.; Bano, C.; Yadav, R.K.; Amist, N. Plant secondary metabolites synthesis and their regulations under biotic and abiotic constraints. J. Plant Biol. 2020, 63, 203–216. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Zhuang, W.-B.; Li, Y.-H.; Shu, X.-C.; Pu, Y.-T.; Wang, X.-J.; Wang, T.; Wang, Z. The classification, molecular structure and biological biosynthesis of flavonoids, and their roles in biotic and abiotic stresses. Molecules 2023, 28, 3599. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Lan, H.-N.; Liu, R.-Y.; Liu, Z.-H.; Li, X.; Li, B.-Z.; Yuan, Y.-J. Biological valorization of lignin to flavonoids. Biotechnol. Adv. 2023, 64, 108107. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Zhu, Y.; Bao, Y. Genome-wide mining of MYB transcription factors in the anthocyanin biosynthesis pathway of Gossypium Hirsutum. Biochem. Genet. 2021, 59, 678–696. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, S.; Li, H.; Wang, L.; Liu, Y.; Niu, L.; Yang, Q.; Meng, D.; Fu, Y. Genome-wide analysis and characterization of R2R3-MYB family in pigeon pea (Cajanus cajan) and their functional identification in phenylpropanoids biosynthesis. Planta 2021, 64, 254. [Google Scholar] [CrossRef]
- Cavallini, E.; Zenoni, S.; Finezzo, L.; Guzzo, F.; Zamboni, A.; Avesani, L.; Tornielli, G.B. Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol. 2014, 55, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Deluc, L.; Barrieu, F.; Marchive, C.; Lauvergeat, V.; Decendit, A.; Richard, T.; Carde, J.-P.; Mérillon, J.-M.; Hamdi, S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006, 140, 499–511. [Google Scholar] [CrossRef]
- Bhargava, A.; Mansfield, S.D.; Hall, H.C.; Douglas, C.J.; Ellis, B.E. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 2010, 154, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Shan, H.; Chen, S.; Jiang, J.; Gu, C.; Zhou, G.; Chen, Y.; Song, A.; Chen, F. The heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB1 alters lignin composition and represses flavonoid synthesis in Arabidopsis thaliana. PLoS ONE 2013, 8, e65680. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Guan, Y.; Zhang, Z.; Song, A.; Chen, S.; Jiang, J.; Chen, F. CmMYB8 encodes an R2R3 MYB transcription factor which represses lignin and flavonoid synthesis in Chrysanthemum. Plant Physiol. Biochem. 2020, 149, 217–224. [Google Scholar] [CrossRef]
- Sonbol, F.-M.; Fornalé, S.; Capellades, M.; Encina, A.; Touriño, S.; Torres, J.-L.; Rovira, P.; Ruel, K.; Puigdomènech, P.; Rigau, J.; et al. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 2009, 70, 283–296. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
- Ni, Z.; Han, X.; Yang, Z.; Xu, M.; Feng, Y.; Chen, Y.; Xu, L.-A. Integrative analysis of wood biomass and developing xylem transcriptome provide insights into mechanisms of lignin biosynthesis in wood formation of Pinus massoniana. Int. J. Biol. Macromol. 2020, 163, 1926–1937. [Google Scholar] [CrossRef]
- Chen, P.; Li, R.; Zhu, L.; Hao, Q.; Yao, S.; Liu, J.; Ji, K. Characterization and interaction analysis of the secondary cell wall synthesis-related transcription factor PmMYB7 in Pinus massoniana Lamb. Int. J. Mol. Sci. 2022, 23, 2079. [Google Scholar] [CrossRef]
- Yao, S.; Chen, P.; Yu, Y.; Zhang, M.; Wang, D.; Liu, J.; Hao, Q.; Ji, K. PmMYB4, a transcriptional activator from Pinus massoniana, regulates secondary cell wall formation and lignin biosynthesis. Forests 2021, 12, 1618. [Google Scholar] [CrossRef]
- Lou, X.; Yao, S.; Chen, P.; Wang, D.; Agassin, R.H.; Hou, Y.; Zhang, C.; Ji, K. Transcriptome identification of R2R3-MYB gene family members in Pinus massoniana and PmMYB4 response to drought stress. Forests 2023, 14, 410. [Google Scholar] [CrossRef]
- He, Y.; Zhang, X.; Li, L.; Sun, Z.; Li, J.; Chen, X.; Hong, G. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorusstatus-dependent anthocyanin biosynthesis. New Phytol. 2021, 230, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Biała, W.; Jasiński, M. The phenylpropanoid case—It is transport that matters. Front. Plant Sci. 2018, 9, 1610. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, Z.; Maximova, S.; Payne, M.J.; Guiltinan, M.J. Proanthocyanidin synthesis in Theobroma cacao: Genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol. 2013, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Weng, J.; Chapple, C. The origin and evolution of lignin biosynthesis. New Phytol. 2010, 187, 273–285. [Google Scholar] [CrossRef]
- Bomal, C.; Bedon, F.; Caron, S.; Mansfield, S.D.; Levasseur, C.; Cooke, J.E.K.; Blais, S.; Tremblay, L.; Morency, M.-J.; Pavy, N.; et al. Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: A comparative in planta analysis. J. Exp. Bot. 2008, 59, 3925–3939. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Mellway, R.D.; Tran, L.T.; Prouse, M.B.; Campbell, M.M.; Constabel, C.P. The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol. 2009, 150, 924–941. [Google Scholar] [CrossRef]
- James, A.M.; Ma, D.; Mellway, R.; Gesell, A.; Yoshida, K.; Walker, V.; Tran, L.; Stewart, D.; Reichelt, M.; Suvanto, J.; et al. Poplar MYB115 and MYB134 transcription factors regulate proanthocyanidin synthesis and structure. Plant Physiol. 2017, 174, 154–171. [Google Scholar] [CrossRef]
- Fraser, C.M.; Chapple, C. The phenylpropanoid pathway in Arabidopsis. Arab. Book 2011, 9, e0152. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 1999, 11, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Andrea, M.; Francesco, E.F.; Sergio, I.; Alessandra, G.; Maria, A.M.; Cinzia, C.; Lorenzo, B.; Arianna, M.; Cecilia, C.; Patrizia, R.; et al. Identification of a new R3 MYB type repressor and functional characterization of the members of the MBW transcriptional complex involved in anthocyanin biosynthesis in eggplant (S. melongena L.). PLoS ONE 2020, 15, e0232986. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhao, Y.Q.; Zhao, B.; Ren, S.; Guo, Y.D. Influencing factors and structural characterization of hyperhydricity of in vitro regeneration in Brassica oleracea var. italica. Can. J. Plant Sci. 2011, 91, 159–165. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Ran, L.; Tian, Q.; Fan, D.; Luo, K. PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa. Plant Cell Physiol. 2015, 56, 2436–2446. [Google Scholar] [CrossRef]
- Klason, P. On the lignin content of spruce wood. Sven. Papperstidn 1923, 26, 319–321. [Google Scholar]
- Alves, A.; Cisneros, E.F.; Balmelli, G.; Poltri, S.N.M.; Rodrigues, J. Assessment of eucalypts wood lignin content by analytical pyrolysis, comparison with Klason and total lignin contents. J. Wood Chem. Technol. 2021, 41, 229–235. [Google Scholar] [CrossRef]
- Hossain, Z.; McGarvey, B.; Amyot, L.; Gruber, M.; Jung, J.; Hannoufa, A. DIMINUTO 1 affects the lignin profile and secondary cell wall formation in Arabidopsis. Planta 2012, 235, 485–498. [Google Scholar] [CrossRef]
- Li, Y.-G.; Tanner, G.; Larkin, P. The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in Forage legumes. J. Sci. Food Agric. 1996, 70, 89–101. [Google Scholar] [CrossRef]
- Feucht, W.; Treutter, D. Flavan-3-ols in trichomes, pistils and phelloderm of some tree species. Ann. Bot. 1990, 65, 225–230. [Google Scholar] [CrossRef]
- Yao, L.; Jiang, Y.; Datta, N.; Singanusong, R.; Liu, X.; Duan, J.; Raymont, K.; Lisle, A.; Xu, Y. HPLC analyses of flavanols and phenolic acids in the fresh young shoots of tea (Camellia sinensis) grown in Australia. Food Chem. 2004, 84, 253–263. [Google Scholar] [CrossRef]
- Yari, A.; Rashnoo, S. Optimization of a new method for extraction of cyanidin chloride and pelargonidin chloride anthocyanins with magnetic solid phase extraction and determination in fruit samples by HPLC with central composite design. J. Chromatogr. B 2017, 1067, 38–44. [Google Scholar] [CrossRef] [PubMed]

| WT (ng/g) | PmMYB6-OE (ng/g) | |
|---|---|---|
| 13-Gallic acid | 120.177 ± 2.483 | 157.650 ± 2.730 ** |
| 22-Catechin | 5465.054 ± 54.855 | 34,525.780 ± 111.787 ** |
| 21-Epicatechin | 375.651 ± 6.638 | 1862.982 ± 41.593 ** |
| 78-Epicatechin Gallate | 1048.251 ± 1.191 | 700.262 ± 13.386 ** |
| WT (ng/g) | PmMYB6-OE (ng/g) | |
|---|---|---|
| Pelargonidin | 0.031 ± 0.005 | 0.685 ± 0.087 ** |
| Cyanidin | 16.170 ± 0.200 | 67.260 ± 7.330 ** |
| Delphinidin | 3.272 ± 0.045 | 4.023 ± 0.258 * |
| Petunidin | 0.349 ± 0.003 | 0.364 ± 0.034 |
| Malvidin | 0.085 ± 0.004 | 0.162 ± 0.005 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Hao, Q.; Chen, P.; Qin, Y.; Peng, M.; Yao, S.; He, X.; Yu, Q.; Agassin, R.H.; Ji, K. Cloning of PmMYB6 in Pinus massoniana and an Analysis of Its Function. Int. J. Mol. Sci. 2023, 24, 13766. https://doi.org/10.3390/ijms241813766
He Y, Hao Q, Chen P, Qin Y, Peng M, Yao S, He X, Yu Q, Agassin RH, Ji K. Cloning of PmMYB6 in Pinus massoniana and an Analysis of Its Function. International Journal of Molecular Sciences. 2023; 24(18):13766. https://doi.org/10.3390/ijms241813766
Chicago/Turabian StyleHe, Yuan, Qingqing Hao, Peizhen Chen, Yiyun Qin, Manqing Peng, Sheng Yao, Xin He, Qiong Yu, Romaric Hippolyte Agassin, and Kongshu Ji. 2023. "Cloning of PmMYB6 in Pinus massoniana and an Analysis of Its Function" International Journal of Molecular Sciences 24, no. 18: 13766. https://doi.org/10.3390/ijms241813766





