Genome-Wide Identification and Characterization of the YTH Domain-Containing RNA-Binding Protein Family in Liriodendron chinense
Abstract
:1. Introduction
2. Results
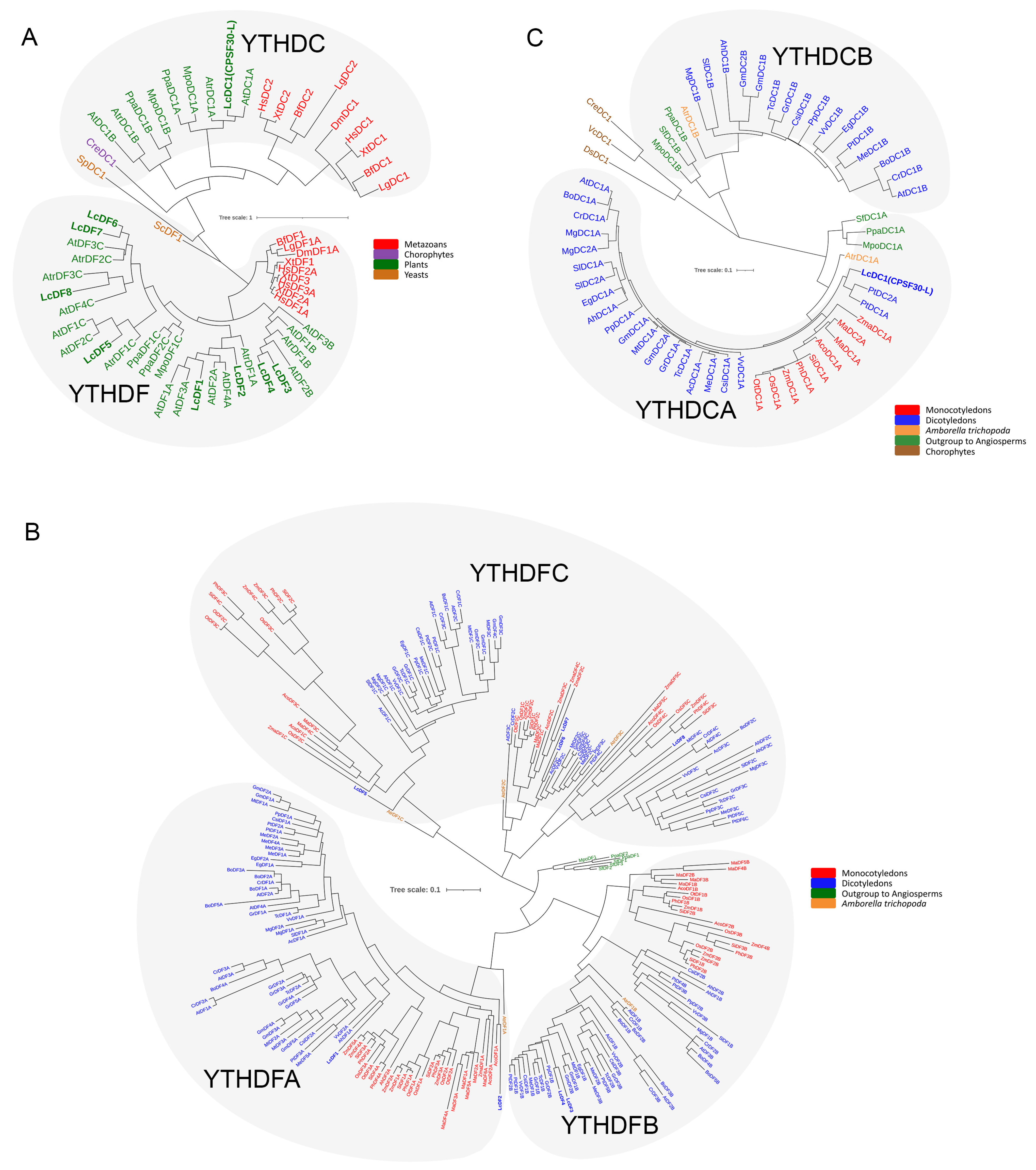
2.1. Genome-Wide Identification and Phylogenetic Analysis of L. chinense YTH Genes
2.2. Sequence Analysis of L. chinense YTH Genes
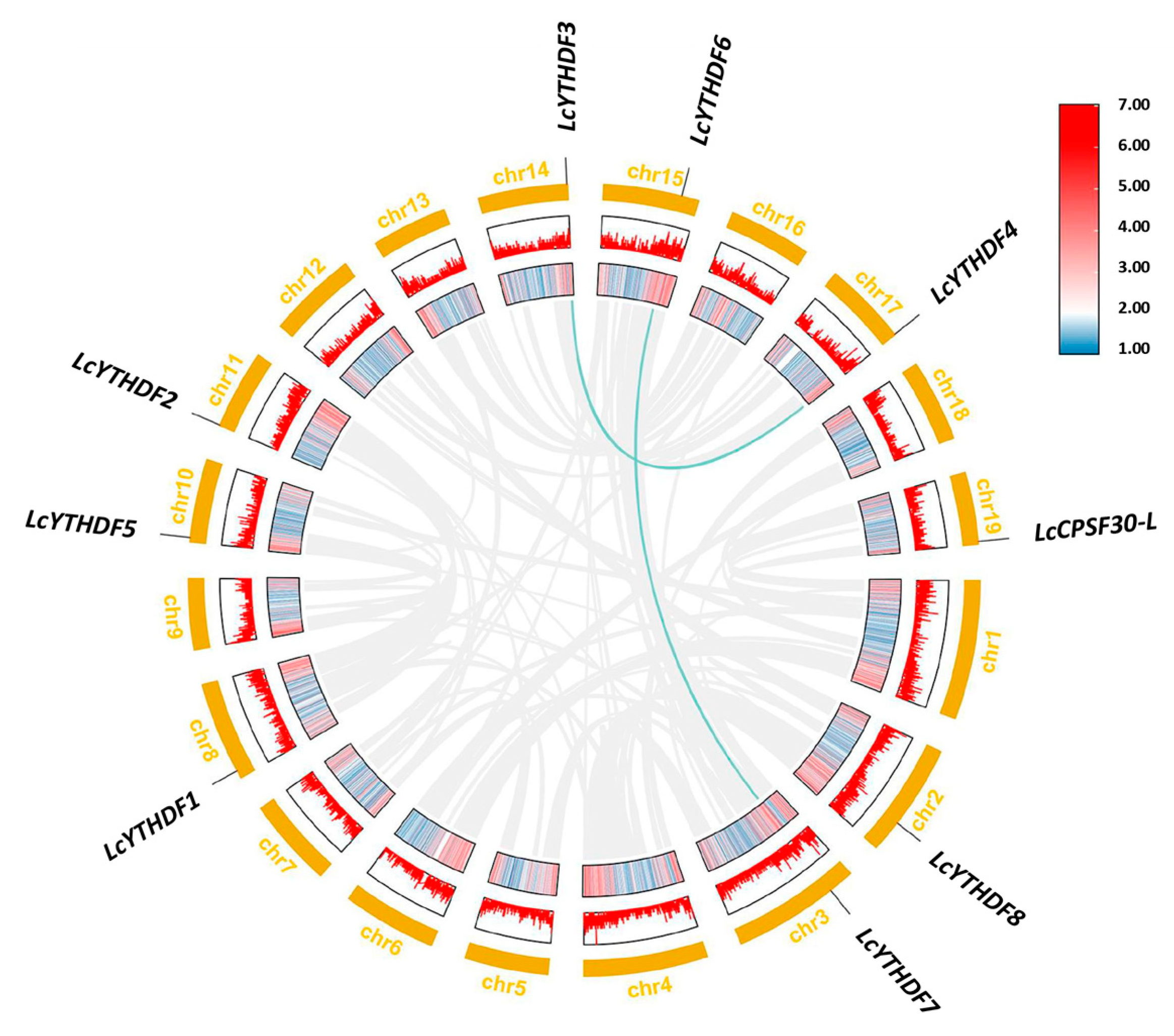
2.3. Duplication Events of LcYTH Genes
2.4. Gene Structure, Conserved Domain, and Motif Analysis of LcYTH Genes
2.5. LcYTHDF1, LcYTHDF2, LcYTHDF4, LcYTHDF5, LcYTHDF7, and LcCPSF30-L May Participate in the Process of Phase Separation
2.6. Expression of LcYTHs in Different Tissues
2.7. Analysis of Cis-Acting Elements in LcYTH Promoters
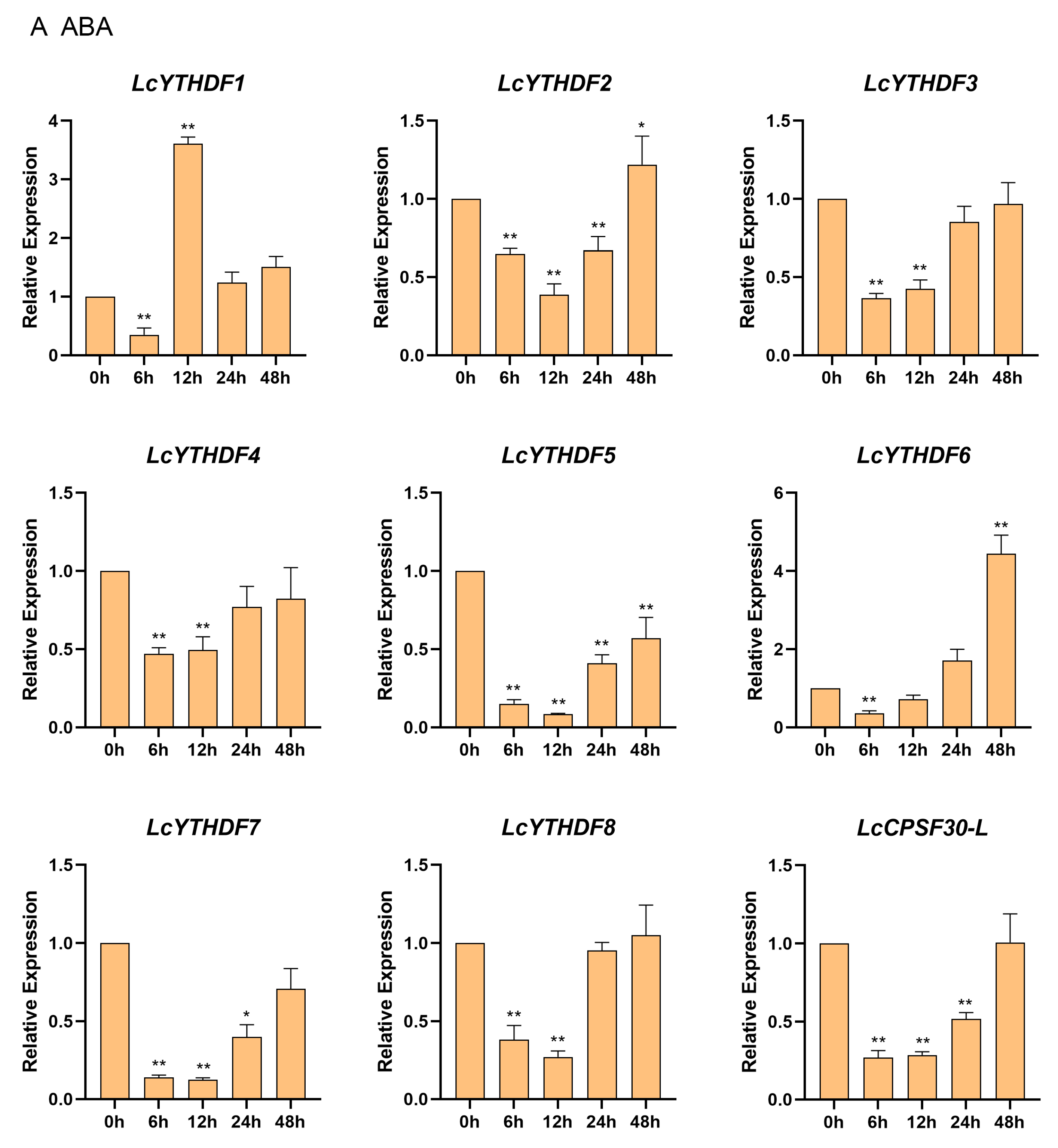
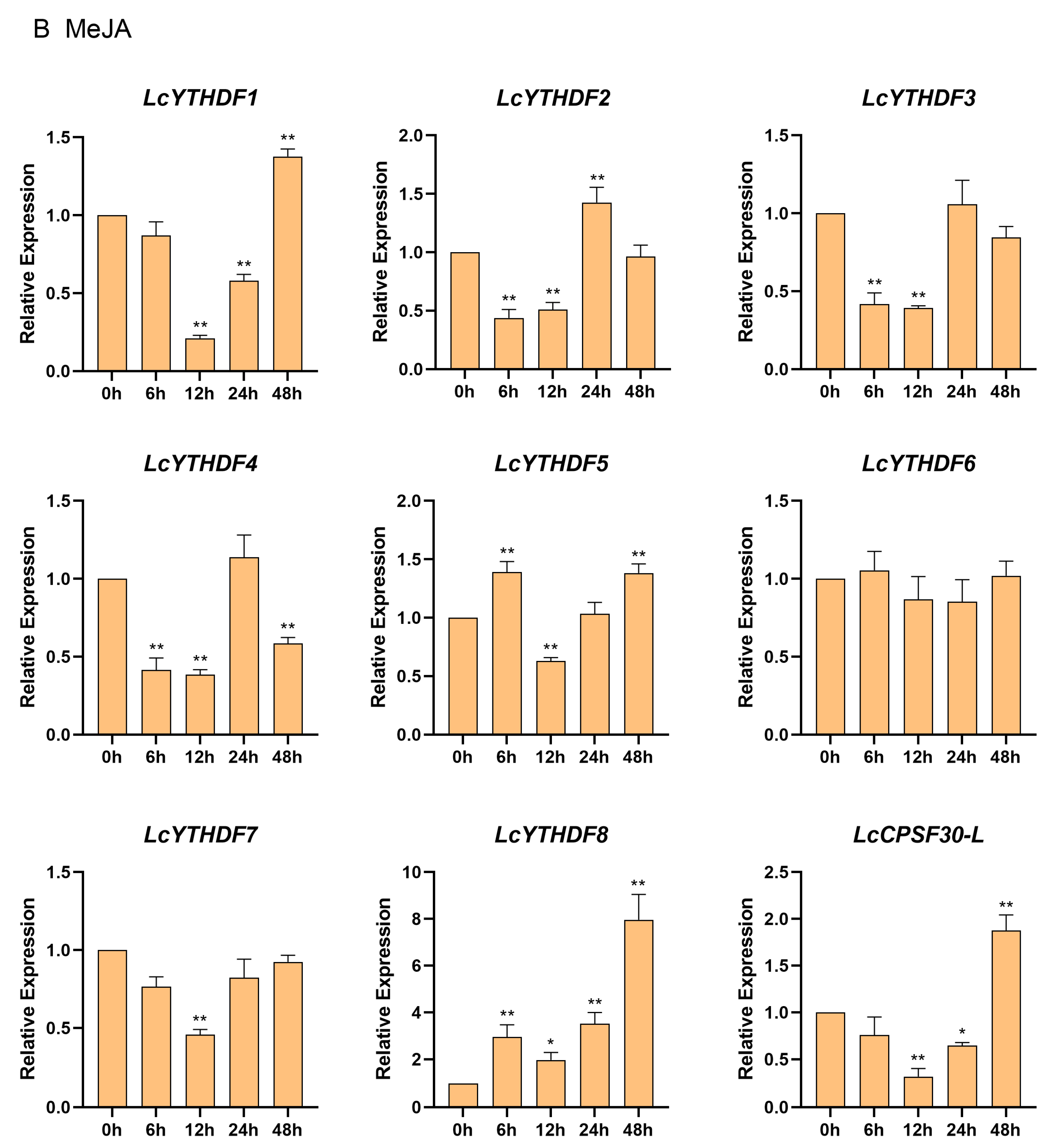
2.8. Differential Expression Profiling of LcYTH Genes Treated by ABA and MeJA
3. Discussion
3.1. The Function of YTH Protein in L. chinense May Be More Complex or Redundant Than That in Animals
3.2. The LcYTHDC Proteins May Have a Similar but Unique Molecular Mechanism to the YTHDC1 Protein in Animals
3.3. LcYTH Proteins May Participate in LLPS Process through PrLDs Domain
3.4. LcYTHDF2 Has an Obvious Expression Advantage in the LcYTH Gene Family
3.5. The LcYTH Genes May Play Important Roles in ABA and MeJA Pathways through Different Forms
4. Materials and Methods
4.1. Identification of YTH Genes in the L. chinense Genome
4.2. Amino Acid Sequence Analysis, Multiple Sequence Alignments, and Phylogenetic Analysis
4.3. Chromosomal Distribution of YTH Genes
4.4. Gene Structure, Conserved Motifs Analysis, and Cis-Regulatory Elements
4.5. LLPS Prediction
4.6. Plant Materials and Treatments
4.7. RNA Extraction and qRT-PCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willyard, C. An epigenetics gold rush: New controls for gene expression. Nature 2017, 542, 406–408. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; He, C.; Kaye, E.G.; Li, J.; Mu, M.; Nelson, G.M.; Dong, L.; Wang, J.; Wu, F.; Shi, Y.G.; et al. Dynamic control of chromatin-associated m6A methylation regulates nascent RNA synthesis. Mol. Cell 2022, 82, 1156–1168. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef] [PubMed]
- Arribas-Hernández, L.; Brodersen, P. Occurrence and functions of m6A and other covalent modifications in plant mRNA. Plant Physiol. 2020, 182, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Reichel, M.; Köster, T.; Staiger, D. Marking RNA: m6A writers, readers, and functions in Arabidopsis. J. Mol. Cell Biol. 2019, 11, 899–910. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. M6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 2017, 6, e31311. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Zaccara, S.; Jaffrey, S.R. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 2020, 181, 1582–1595. [Google Scholar] [CrossRef] [PubMed]
- Chokkalla, A.K.; Mehta, S.L.; Vemuganti, R. Epitranscriptomic regulation by m6A RNA methylation in brain development and diseases. J. Cereb. Blood Flow Metab. 2020, 40, 2331–2349. [Google Scholar] [CrossRef]
- Song, P.; Yang, J.; Wang, C.; Lu, Q.; Shi, L.; Tayier, S.; Jia, G. Arabidopsis N6-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies. Mol. Plant 2021, 14, 571–587. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967. [Google Scholar] [CrossRef]
- Scutenaire, J.; Deragon, J.-M.; Jean, V.; Benhamed, M.; Raynaud, C.; Favory, J.-J.; Merret, R.; Bousquet-Antonelli, C. The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell 2018, 30, 986–1005. [Google Scholar] [CrossRef]
- Tang, J.; Chen, S.; Jia, G. Detection, regulation, and functions of RNA N6-methyladenosine modification in plants. Plant Commun. 2023, 4, 100546. [Google Scholar] [CrossRef]
- Xu, C.; Liu, K.; Ahmed, H.; Loppnau, P.; Schapira, M.; Min, J. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Res. 2015, 290, 24902–24913. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Li, X.; Zhang, Y.; Ouyang, Z.; Song, F. Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain containing RNA-binding protein family in Arabidopsis and rice. Plant Mol. Biol. Rep. 2014, 32, 1169–1186. [Google Scholar] [CrossRef]
- Wei, L.-H.; Song, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.-C.; Jia, G. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 2018, 30, 968–985. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Wei, L.; Chen, Z.; Cai, Z.; Lu, Q.; Wang, C.; Tian, E.; Jia, G. m6A readers ECT2/ECT3/ECT4 enhance mRNA stability through direct recruitment of the poly(A) binding proteins in Arabidopsis. Genome Biol. 2023, 24, 103. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Simonini, S.; Hansen, M.H.; Paredes, E.B.; Bressendorff, S.; Dong, Y.; Østergaard, L.; Brodersen, P. Recurrent requirement for the m6A-ECT2/ECT3/ECT4 axis in the control of cell proliferation during plant organogenesis. Development 2020, 147, dev189134. [Google Scholar] [CrossRef]
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading m6A in the transcriptome: m6A binding proteins. Trends Cell Biol. 2018, 28, 113–127. [Google Scholar] [CrossRef]
- Timcheva, K.; Dufour, S.; Touat-Todeschini, L.; Burnard, C.; Carpentier, M.-C.; Chuffart, F.; Merret, R.; Helsmoortel, M.; Ferré, S.; Grézy, A.; et al. Chromatin-associated YTHDC1 coordinates heat-induced reprogramming of gene expression. Cell Rep. 2022, 41, 111784. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, W.; Pickering, B.F.; Chu, K.L.; Savino, A.M.; Yang, X.; Luo, K.; Nguyen, D.T.T.; Mo, S.; Barin, E.; et al. N6-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemia differentiation. Cancer Cell 2021, 39, 958–972. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, B.-F.; Xiao, W.; Yang, X.; Sun, H.-Y.; Zhao, Y.-L.; Yang, Y.-G. Dynamic m6A modification and its emerging regulatory role in mRNA splicing. Sci. Bull. 2015, 60, 21–32. [Google Scholar] [CrossRef]
- Zhu, D.Z.; Zhao, X.F.; Liu, C.Z.; Ma, F.F.; Wang, F.; Gao, X.Q.; Zhang, X.S. Interaction between RNA helicase ROOT INITIATION DEFECTIVE 1 and GAMETOPHYTIC FACTOR 1 is involved in female gametophyte development in Arabidopsis. J. Exp. Bot. 2016, 67, 5757–5768. [Google Scholar] [CrossRef]
- Tanabe, A.; Tanikawa, K.; Tsunetomi, M.; Takai, K.; Ikeda, H.; Konno, J.; Torigoe, T.; Maeda, H.; Kutomi, G.; Okita, K.; et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016, 376, 34–42. [Google Scholar] [CrossRef] [PubMed]
- March, Z.M.; King, O.D.; Shorter, J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 2016, 1647, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Franzmann, T.M.; Jahnel, M.; Pozniakovsky, A.; Mahamid, J.; Holehouse, A.S.; Nüske, E.; Richter, D.; Baumeister, W.; Grill, S.W.; Pappu, R.V.; et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 2018, 359, eaao5654. [Google Scholar] [CrossRef]
- Fu, Y.; Zhuang, X. m6A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 2020, 16, 955–963. [Google Scholar] [CrossRef]
- Ries, R.J.; Zaccara, S.; Klein, P.; Olarerin-George, A.; Namkoong, S.; Pickering, B.F.; Patil, D.P.; Kwak, H.; Lee, J.H.; Jaffrey, S.R. m6A enhances the phase separation potential of mRNA. Nature 2019, 571, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Pei, G.; Li, D.; Li, R.; Shao, Y.; Zhang, Q.C.; Li, P. Multivalent m6A motifs promote phase separation of YTHDF proteins. Cell Res. 2019, 29, 767–769. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, L.; Ishikawa, R.; Li, Y.; Fiedler, M.; Liu, F.; Calder, G.; Rowan, B.; Weigel, D.; Li, P.; et al. Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 2019, 569, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yang, J.; Lu, Q.; Tang, Q.; Chen, S.; Jia, G. The RNA N6-methyladenosine demethylase ALKBH9B modulates ABA responses in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 2361–2373. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.-C.; Wei, L.-H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B is an RNA N6-methyladenosine demethylase affecting arabidopsis floral transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 2019, 5, 328. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]





| Group | Gene ID | Locus ID | Locus | Protein Length (aa) | MW (kDa) | pI | Homolog in A. thaliana |
|---|---|---|---|---|---|---|---|
| DFA | LcYTHDF1 | Lchi30795 | Chr08 | 723 | 78.3 | 6.04 | ECT1/2/3/4 |
| DFA | LcYTHDF2 | Lchi30011 | Chr11 | 744 | 80.4 | 6.56 | ECT1/2/3/4 |
| DFB | LcYTHDF3 | Lchi10804 | Chr14 | 618 | 67.2 | 5.43 | ECT5/9/10 |
| DFB | LcYTHDF4 | Lchi05236 | Chr17 | 679 | 73.3 | 5.31 | ECT5/9/10 |
| DFC | LcYTHDF5 | Lchi06362 | Chr10 | 588 | 64.4 | 6.25 | ECT6/7 |
| DFC | LcYTHDF6 | Lchi04058 | Chr15 | 427 | 48.2 | 5.97 | ECT8 |
| DFC | LcYTHDF7 | Lchi07049 | Chr03 | 637 | 70.1 | 6.07 | ECT8 |
| DFC | LcYTHDF8 | Lchi01573 | Chr04 | 579 | 61.9 | 6.44 | ECT11 |
| DCA | LcCPSF30-L (LcDC1) | Lchi11130 | Chr19 | 654 | 71.8 | 5.81 | CPSF30-L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, S.; Zhang, J.; Cheng, X.; Wang, D.; Yu, W.; Ji, K.; Yu, Q. Genome-Wide Identification and Characterization of the YTH Domain-Containing RNA-Binding Protein Family in Liriodendron chinense. Int. J. Mol. Sci. 2023, 24, 15189. https://doi.org/10.3390/ijms242015189
Yao S, Zhang J, Cheng X, Wang D, Yu W, Ji K, Yu Q. Genome-Wide Identification and Characterization of the YTH Domain-Containing RNA-Binding Protein Family in Liriodendron chinense. International Journal of Molecular Sciences. 2023; 24(20):15189. https://doi.org/10.3390/ijms242015189
Chicago/Turabian StyleYao, Sheng, Jingjing Zhang, Xiang Cheng, Dengbao Wang, Wenya Yu, Kongshu Ji, and Qiong Yu. 2023. "Genome-Wide Identification and Characterization of the YTH Domain-Containing RNA-Binding Protein Family in Liriodendron chinense" International Journal of Molecular Sciences 24, no. 20: 15189. https://doi.org/10.3390/ijms242015189





