Blue Light Regulates Cell Wall Structure and Carbohydrate Metabolism of Soybean Hypocotyl
Abstract
:1. Introduction
2. Results
2.1. Effects of Blue Light on the Anatomical Structure of Soybean Hypocotyls
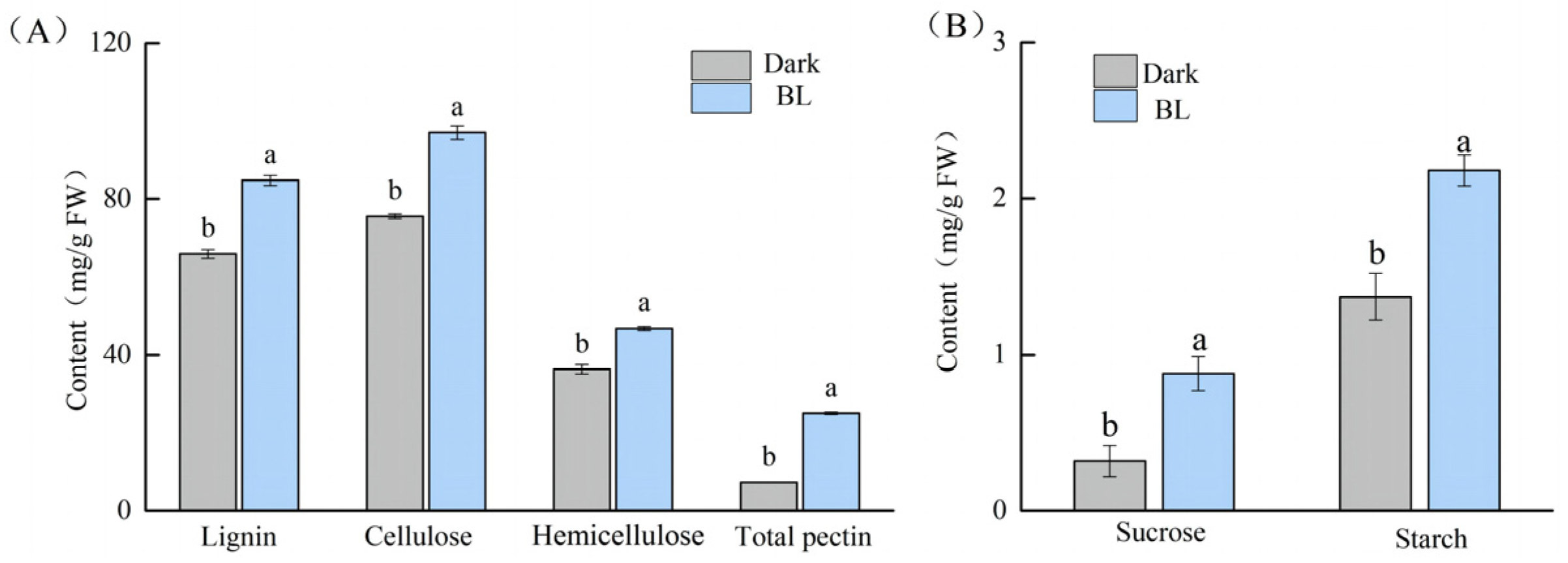
2.2. Effects of Blue Light on the Cell Wall Composition and Carbohydrate Contents of Soybean Hypocotyls
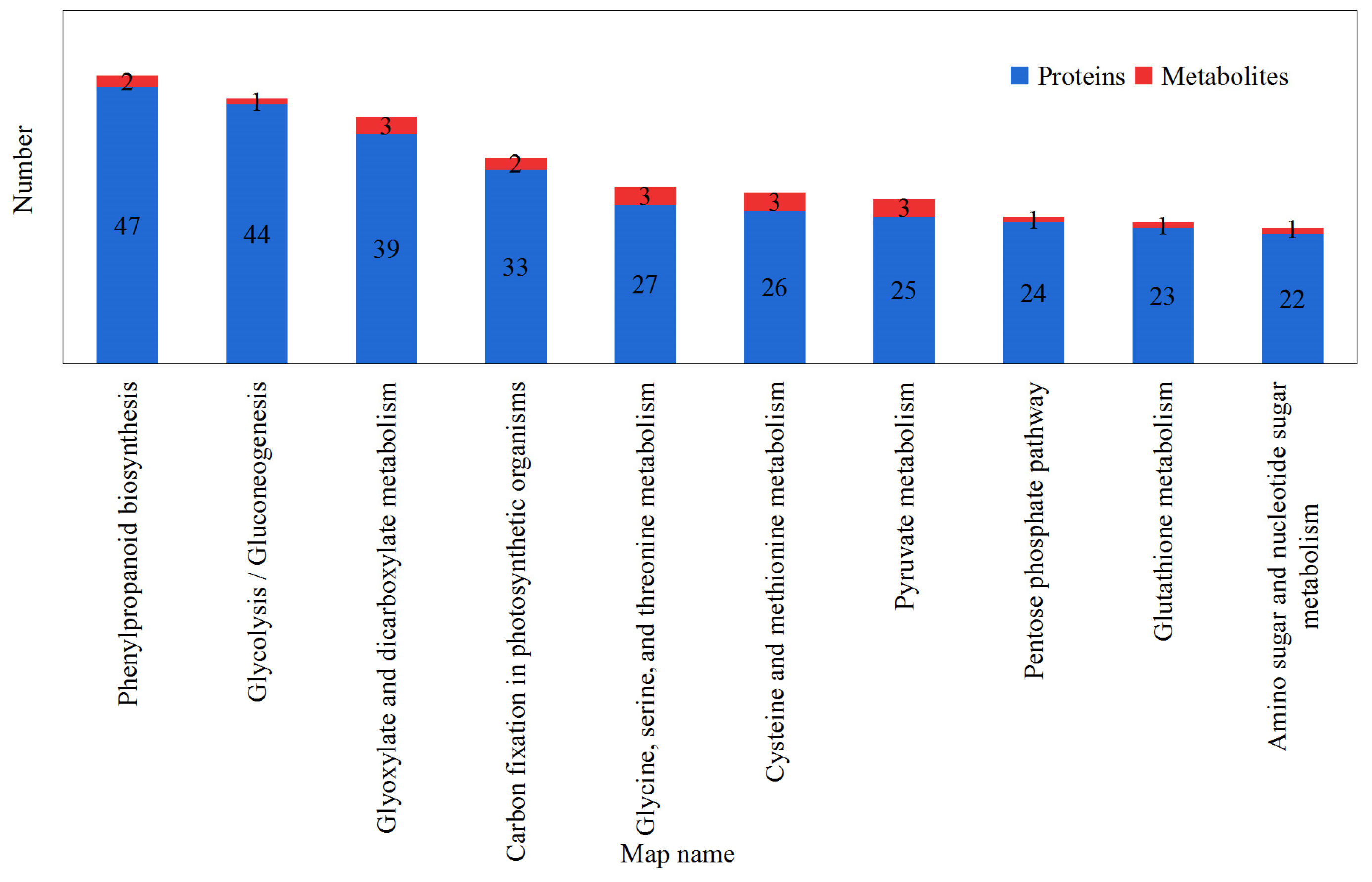
2.3. Proteomic Analysis
2.4. Metabolome Analysis
2.5. Integrated Proteome and Metabolome Analysis
3. Discussion
3.1. Effects of Blue Light on Structural Carbohydrate Metabolism
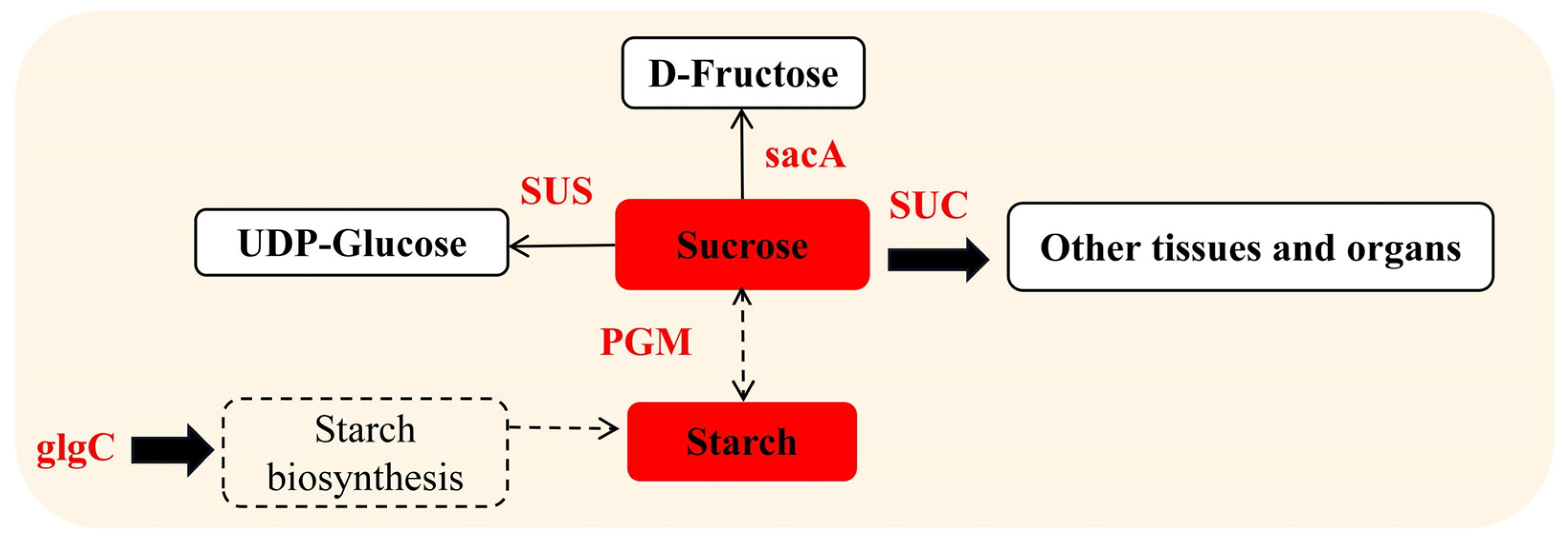
3.2. Effects of Blue Light on Nonstructural Carbohydrate Metabolism
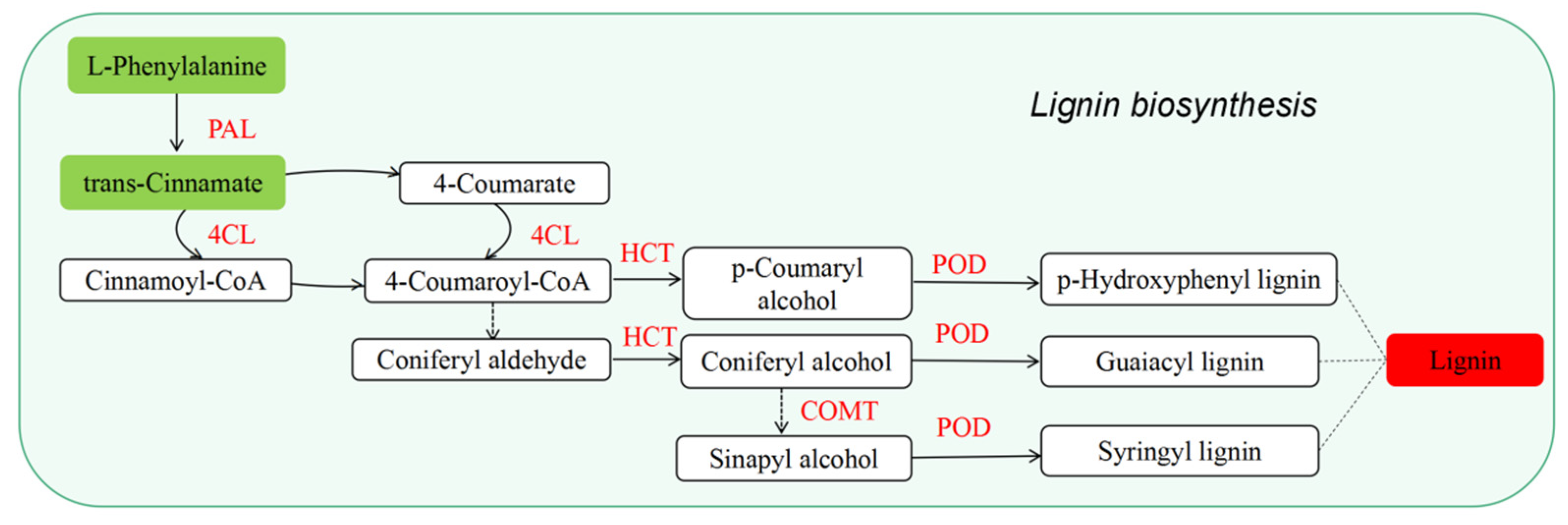
3.3. Effects of Blue Light on Lignin Synthesis
4. Materials and Methods
4.1. Experimental Design
4.2. Anatomical Structure Sampling and Observation
4.3. Determination of Cell Wall Composition and Carbohydrate Contents
4.4. Proteomic Analysis
4.5. Metabolomic Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muhammad, A.; Hao, H.H.; Xue, Y.L.; Alam, A.; Bai, S.M.; Hu, W.C.; Sajid, M.; Hu, Z.; Samad, R.A.; Li, Z.H.; et al. Survey of wheat straw stem characteristics for enhanced resistance to lodging. Cellulose 2020, 27, 2469–2484. [Google Scholar] [CrossRef]
- Mulsanti, I.W.; Yamamoto, T.; Ueda, T.; Samadi, A.F.; Kamahora, E.; Rumanti, I.A.; Thanh, V.C.; Adachi, S.; Suzuki, S.; Kanekatsu, M.; et al. Finding the superior allele of japonica-type for increasing stem lodging resistance in indica rice varieties using chromosome segment substitution lines. Rice 2018, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, R.; Hou, Z.F.; Yan, C.; Xia, X.; Ma, C.M.; Dong, S.K.; Gong, Z.P. Mechanical properties of soybean plants under various plant densities. Crop Pasture Sci. 2020, 71, 249. [Google Scholar] [CrossRef]
- Hussain, S.; Iqbal, N.; Pang, T.; Naeem Khan, M.; Liu, W.G.; Yang, W.Y. Weak stem under shade reveals the lignin reduction behavior. J. Integr. Agric. 2018, 17, 496–505. [Google Scholar] [CrossRef]
- Hussain, S.; Liu, T.; Iqbal, N.; Brestic, M.; Pang, T.; Mumtaz, M.; Shafiq, I.; Li, S.X.; Wang, L.; Gao, Y.; et al. Effects of lignin, cellulose, hemicellulose, sucrose and monosaccharide carbohydrates on soybean physical stem strength and yield in intercropping. Photochem. Photobiol. Sci. 2020, 19, 462–472. [Google Scholar] [CrossRef]
- Huang, X.M.; Wang, H.C.; Gao, F.F.; Huang, H.B. A comparative study of the pericarp of litchi cultivars susceptible and resistant to fruit cracking. J. Hortic. Sci. Biotechnol. 1999, 74, 351–354. [Google Scholar] [CrossRef]
- Sirisomboon, P.; Tanaka, M.; Fujita, S.; Kojima, T. Relationship between the texture and pectin constituents of japanese pear. J. Texture Stud. 2010, 31, 679–690. [Google Scholar] [CrossRef]
- Zheng, M.J.; Chen, J.; Shi, Y.H.; Li, Y.X.; Yin, Y.P.; Yang, D.Q.; Luo, Y.L.; Pang, D.W.; Xu, X.; Li, W.Q.; et al. Manipulation of lignin metabolism by plant densities and its relationship with lodging resistance in wheat. Sci. Rep. 2017, 7, 41805. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Ruan, R.W.; Yuan, X.H.; Hu, D.; Yang, H.; Li, Y.; Yi, Z.L. Effects of nitrogen fertilizer and planting density on the lignin synthesis in the culm in relation to lodging resistance of buckwheat. Plant Prod. Sci. 2015, 18, 218–227. [Google Scholar] [CrossRef]
- Xue, J.; Zhao, Y.S.; Gou, L.; Shi, Z.G.; Yao, M.N.; Zhang, W.F. How high plant density of maize affects basal internode development and strength formation. Crop Sci. 2016, 56, 3295–3306. [Google Scholar] [CrossRef]
- Liu, W.G.; Deng, Y.C.; Hussain, S.; Zou, J.L.; Yuan, J.; Luo, L.; Yang, C.Y.; Yuan, X.Q.; Yang, W.Y. Relationship between cellulose accumulation and lodging resistance in the stem of relay intercropped soybean [ Glycine max (L.) Merr.]. Field Crops Res. 2016, 196, 261–267. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- McCartney, L.; Ormerod, A.P.; Gidley, M.J.; Knox, J.P. Temporal and spatial regulation of pectic (1→4)-β-D-galactan in cell walls of developing pea cotyledons: Implications for mechanical properties. Plant J. 2000, 22, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Barnes, W.J.; Anderson, C.T. Cytosolic invertases contribute to cellulose biosynthesis and influence carbon partitioning in seedlings of Arabidopsis thaliana. Plant J. 2018, 94, 956–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.L.; Song, X.L.; Wang, Z.L.; Sun, X.Z. Sucrose metabolism for cellulose biosynthesis in colored cotton fibers. Pak. J. Agric. Sci. 2017, 54, 51–56. [Google Scholar]
- Tanaka, K.; Murata, K.; Yamazaki, M.; Onosato, K.; Miyao, A.; Hirochika, H. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 2003, 133, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Ishimaru, K.; Togawa, E.; Ookawa, T.; Kashiwagi, T.; Madoka, Y.; Hirotsu, N. New target for rice lodging resistance and its effect in a typhoon. Planta 2008, 227, 601–609. [Google Scholar] [CrossRef]
- Hussain, S.; Mumtaz, M.; Manzoor, S.; Li, S.X.; Ahmed, I.; Skalicky, M.; Brestic, M.; Rastogi, A.; Ulhassan, Z.; Shafiq, I.; et al. Foliar application of silicon improves growth of soybean by enhancing carbon metabolism under shading conditions. Plant Physiol. Biochem. 2021, 159, 43–52. [Google Scholar] [CrossRef]
- Hussain, S.; Iqbal, N.; Rahman, T.; Liu, T.; Brestic, M.; Safdar, M.E.; Asghar, M.A.; Farooq, M.U.; Shafiq, I.; Ali, A.; et al. Shade effect on carbohydrates dynamics and stem strength of soybean genotypes. Environ. Exp. Bot. 2019, 162, 374–382. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, H.; Sun, X.; Muhammad, A.A.; Liu, J.; Liu, W.; Shu, K.; Shang, J.; Yang, F.; Wu, X.; et al. Quantitative proteomic analyses identified multiple sugar metabolic proteins in soybean under shade stress. J. Biochem. 2019, 165, 277–288. [Google Scholar] [CrossRef]
- Hernández, R.; Kubota, C. Physiological responses of cucumber seedlings under different blue and red photon flux ratios using LEDs. Environ. Exp. Bot. 2016, 121, 66–74. [Google Scholar] [CrossRef]
- Piszczek, P.; Głowacka, B. Effect of the colour of light on cucumber (Cucumis sativus L.) seedlings. Veg. Crops Res. Bull. 2008, 68, 71–80. [Google Scholar] [CrossRef]
- Barro, F.; De La Haba, P.; Maldonado, J.M.; Fontes, A.G. Effect of light quality on growth, contents of carbohydrates, protein and pigments, and nitrate reductase activity in soybean plants. J. Plant Physiol. 1989, 134, 586–591. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, C.; Zhang, R.; Ma, C.M.; Gong, Z.P. The relationship between internode elongation of soybean stems and spectral distribution of light in the canopy under different plant densities. Plant Prod. Sci. 2020, 24, 326–338. [Google Scholar] [CrossRef]
- Laskowski, M.J.; Briggs, W.R. Regulation of pea epicotyl elongation by blue light 1. Plant Physiol. 1989, 89, 293–298. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, D.J. Mechanism of rapid suppression of cell expansion in cucumber hypocotyls after blue-light irradiation. Planta 1988, 176, 109–116. [Google Scholar] [CrossRef]
- Cosgrove, D.J.; Green, P.B. Rapid suppression of growth by blue light. Plant Physiol. 1981, 68, 1447–1453. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Xie, Z.; Zhang, R.; Xu, P.; Liu, H.T.; Yang, H.Q.; Doblin, M.S.; Bacic, A.; Li, L.G. Blue light regulates secondary cell wall thickening via MYC2/MYC4 activation of the NST1-Directed transcriptional network in Arabidopsis. Plant Cell 2018, 30, 2512–2528. [Google Scholar] [CrossRef] [Green Version]
- OuYang, F.Q.; Mao, J.F.; Wang, J.H.; Zhang, S.G.; Li, Y. Transcriptome analysis reveals that red and blue light regulate growth and phytohormone metabolism in Norway Spruce [Picea abies (L.) Karst]. PLoS ONE 2015, 10, e0127896. [Google Scholar] [CrossRef] [Green Version]
- Li, H.M.; Tang, C.M.; Xu, Z.G. The effects of different light qualities on rapeseed (Brassica napus L.) plantlet growth and morphogenesis in vitro. Sci. Hortic. 2013, 150, 117–124. [Google Scholar] [CrossRef]
- Li, H.M.; Xu, Z.G.; Tang, C.M. Effect of light-emitting diodes on growth and morphogenesis of upland cotton (Gossypium hirsutum L.) plantlets in vitro. Plant Cell Tissue Organ Cult. 2010, 103, 155–163. [Google Scholar] [CrossRef]
- He, W.; Pu, M.; Li, J.; Xu, Z.G.; Gan, L. Potato tuber growth and yield under red and blue LEDs in plant factories. J. Plant Growth Regul. 2021, 41, 40–51. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Perez-Llorca, M.; Munne-Bosch, S.; Gibin, M.S.; Sato, F.; Pelozo, A.; Pattaro, M.C.; Giacomelli, M.E.; Ruggeberg, M.; et al. Increased gibberellins and light levels promotes cell wall thickness and enhance lignin deposition in xylem fibers. Front. Plant Sci. 2018, 203, 111745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato-Noguchi, H.; Kasai, T. Recovery of growth of soybean hypocotyls under continuous red light. J. Plant Physiol. 1991, 139, 252–254. [Google Scholar] [CrossRef]
- Samimy, C. Effect of light on ethylene production and hypocotyl growth of soybean seedlings. Plant Physiol. 1978, 61, 772–774. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.G.; Wen, B.X.; Zhou, T.; Wang, L.; Gao, Y.; Li, S.X.; Qin, S.S.; Liu, J.; Yang, W.Y. iTRAQ protein profile analysis of soybean stems reveals new aspects critical for lodging in intercropping systems. J. Integr. Agric. 2019, 18, 2029–2040. [Google Scholar] [CrossRef]
- Ryan, D.; Robards, K. Metabolomics: The greatest omics of them all? Anal. Chem. 2006, 78, 7954–7958. [Google Scholar] [CrossRef]
- Yates, J.R.; Ruse, C.I.; Nakorchevsky, A. Proteomics by mass spectrometry: Approaches, advances, and applications. Annu. Rev. Biomed. Eng. 2009, 11, 49–79. [Google Scholar] [CrossRef] [Green Version]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef]
- Sánchez-Rodríguez, C.; Rubio-Somoza, I.; Sibout, R.; Persson, S. Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 2010, 15, 291–301. [Google Scholar] [CrossRef]
- Suzuki, S.; Li, L.G.; Sun, Y.H.; Chiang, V.L. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 2006, 142, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Beeckman, T.; Przemeck, G.K.; Stamatiou, G.; Lau, R.; Terryn, N.; De Rycke, R.; Inze, D.; Berleth, T. Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol. 2002, 130, 1883–1893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, T. Xyloglucans in the primary cell wall. Annu. Rev. Plant Physiol. Plant Mol. Bioi. 1989, 40, 139–168. [Google Scholar] [CrossRef]
- Shani, Z.; Dekel, M.; Roiz, L.; Horowitz, M.; Kolosovski, N.; Lapidot, S.; Alkan, S.; Koltai, H.; Tsabary, G.; Goren, R.; et al. Expression of endo-1,4-beta-glucanase (cel1) in Arabidopsis thaliana is associated with plant growth, xylem development and cell wall thickening. Plant Cell Rep. 2006, 25, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Shimuzu, Y.; Aotsuka, S.; Hasegawa, O.; Kawada, T.; Sakuno, T.; Sakai, F.; Hayashi, T. Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant Cell Physiol. 1997, 38, 375–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Sandt, V.S.; Suslov, D.; Verbelen, J.P.; Vissenberg, K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 2007, 100, 1467–1473. [Google Scholar] [CrossRef] [Green Version]
- Sasidharan, R.; Chinnappa, C.C.; Staal, M.; Elzenga, J.T.; Yokoyama, R.; Nishitani, K.; Voesenek, L.A.; Pierik, R. Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 2010, 154, 978–990. [Google Scholar] [CrossRef] [Green Version]
- Pérez, J.; Muñoz-Dorado, J.; de la Rubia, T.; Martínez, J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: An overview. Int. Microbiol. 2002, 5, 53–63. [Google Scholar] [CrossRef]
- Moreira, L.R.; Filho, E.X. An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 2008, 79, 165–178. [Google Scholar] [CrossRef]
- Pauly, M.; Gille, S.; Liu, L.F.; Mansoori, N.; de Souza, A.; Schultink, A.; Xiong, G.Y. Hemicellulose biosynthesis. Planta 2013, 238, 627–642. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Braybrook, S.; Höfte, H. Cell wall mechanics and growth control in plants: The role of pectins revisited. Front. Plant Sci. 2012, 3, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mertens, J.A.; Burdick, R.C.; Rooney, A.P. Identification, biochemical characterization, and evolution of the Rhizopus oryzae 99-880 polygalacturonase gene family. Fungal. Genet. Biol. 2008, 45, 1616–1624. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.D.; Bo, W.; West, G.; Lycett, G.W.; Tucker, G.A. Silencing of the major salt-dependent isoform of pectinesterase in tomato alters fruit softening. Plant Physiol. 2007, 144, 1960–1967. [Google Scholar] [CrossRef] [Green Version]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kolli, V.S.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar] [CrossRef] [Green Version]
- Hadfield, K.A.; Rose, J.K.C.; Yaver, D.S.; Berka, R.M.; Bennett, A.B. Polygalacturonase gene expression in ripe melon fruit supports a role for polygalacturonase in ripening-associated pectin disassembly. Plant Physiol. 1998, 117, 363–373. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Qiu, H.; Chen, Y.; Xu, Y.; Shan, F.X.; Li, H.Y.; Yan, C.; Ma, C.M. Red light regulates metabolic pathways of soybean hypocotyl elongation and thickening. Environ. Exp. Bot. 2022, 199, 104890. [Google Scholar] [CrossRef]
- Ruan, Y.L.; Llewellyn, D.J.; Furbank, R.T. Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 2003, 15, 952–964. [Google Scholar] [CrossRef] [Green Version]
- Delmer, D.P.; Amor, Y. Cellulose Biosynthesis. Plant Cell 1995, 7, 987–1000. [Google Scholar]
- Sturm, A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999, 121, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Gottwald, J.R.; Krysan, P.J.; Young, J.C.; Evert, R.F.; Sussman, M.R. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 2000, 97, 13979–13984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuncel, A.; Okita, T.W. Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: Expectations and unanticipated outcomes. Plant Sci. 2013, 211, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Tauberger, E.; Lytovchenko, A.; Roessner, U.; Willmitzer, L.; Trethewey, R.N. Antisense repression of cytosolic phosphoglucomutase in potato (Solanum tuberosum) results in severe growth retardation, reduction in tuber number and altered carbon metabolism. Planta 2002, 214, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gu, M.; Cui, J.X.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Effects of light quality on CO2 assimilation, chlorophyll-fluorescence quenching, expression of Calvin cycle genes and carbohydrate accumulation in Cucumis sativus. J. Photochem. Photobiol. B 2009, 96, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.G.; Hussain, S.; Liu, T.; Zou, J.L.; Ren, M.L.; Zhou, T.; Liu, J.; Yang, F.; Yang, W.Y. Shade stress decreases stem strength of soybean through restraining lignin biosynthesis. J. Integr. Agric. 2018, 17, 43–53. [Google Scholar] [CrossRef]
- Fan, W.; Ge, G.T.; Liu, Y.H.; Wang, W.; Liu, L.Y.; Jia, Y.S. Proteomics integrated with metabolomics: Analysis of the internal causes of nutrient changes in alfalfa at different growth stages. BMC Plant Biol. 2018, 18, 78. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Rohde, A.; Morreel, K.; Ralph, J.; Goeminne, G.; Hostyn, V.; De Rycke, R.; Kushnir, S.; Van Doorsselaere, J.; Joseleau, J.P.; Vuylsteke, M.; et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 2004, 16, 2749–2771. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Ruan, R.W.; Yuan, X.H.; Hu, D.; Yang, H.; Li, Y.; Yi, Z.L. Relationship between lignin metabolism and lodging resistance of culm in buckwheat. J. Agric. Sci. 2014, 6, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.N.; Son, S.; Jordan, M.C.; Levin, D.B.; Ayele, B.T. Lignin biosynthesis in wheat (Triticum aestivum L.): Its response to waterlogging and association with hormonal levels. BMC Plant Biol. 2016, 16, 28. [Google Scholar] [CrossRef]
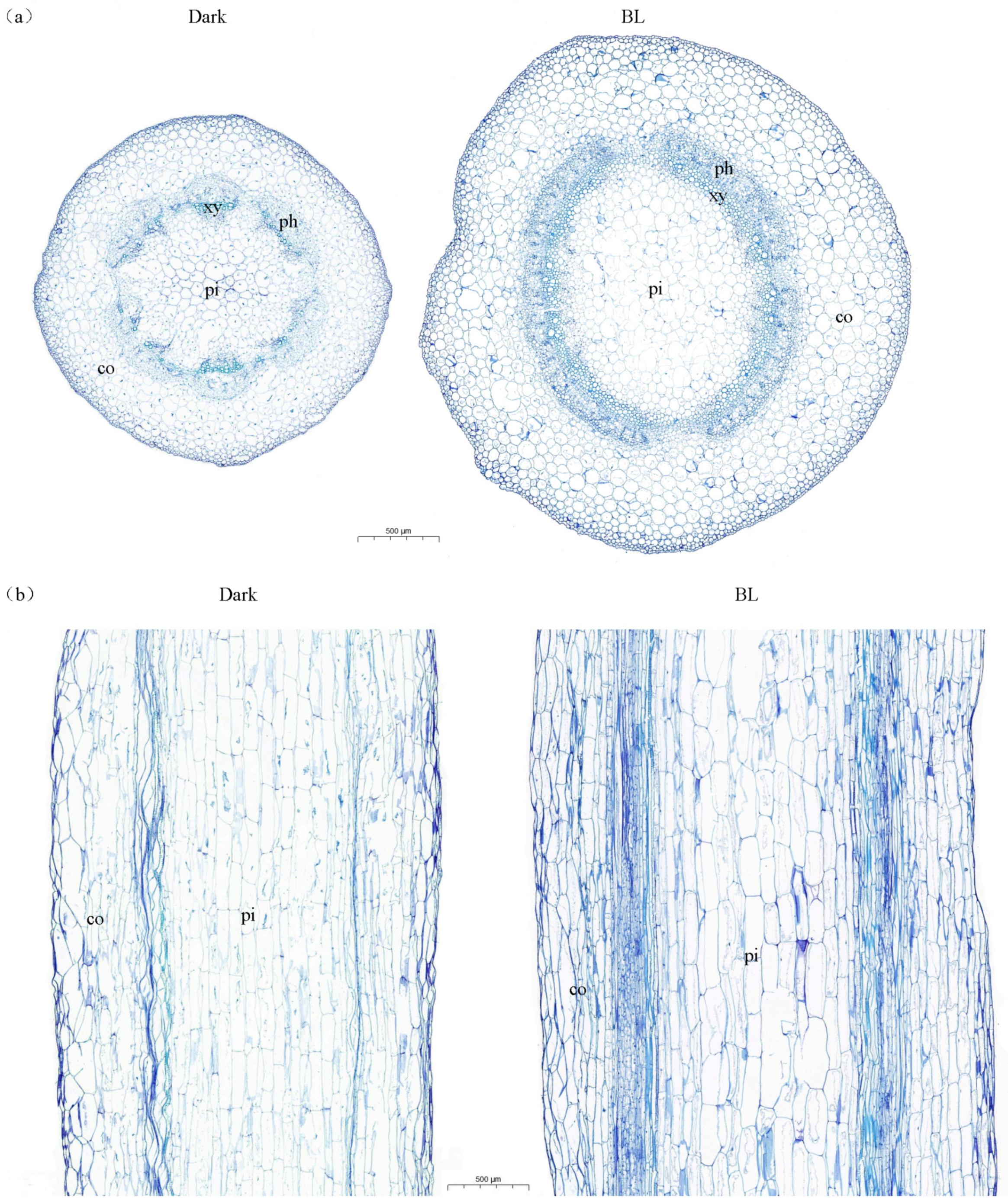




| Treatments | Length (cm) | Diameter (mm) |
|---|---|---|
| Dark | 16.25 ± 0.66a | 2.67 ± 0.01b |
| BL | 6.00 ± 0.00b | 3.15 ± 0.12a |
| Treatments | Length of Longitudinal Pith Cells (mm) | Area of Pith Cell (mm2) | Cross-Sectional Diameter (mm) | Cross-Sectional Area (mm2) | Ratio of Pith Cell Area to Cross-Sectional Area (%) |
|---|---|---|---|---|---|
| Dark | 0.37 ± 0.01a | 0.98 ± 0.05b | 2.12 ± 0.01b | 3.55 ± 0.08b | 27.36 ± 1.13a |
| BL | 0.34 ± 0.01b | 1.53 ± 0.11a | 3.09 ± 0.03a | 7.32 ± 0.23a | 20.71 ± 0.84b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Chen, Y.; Cui, C.; Shan, F.; Zhang, R.; Lyu, X.; Lyu, L.; Chang, H.; Yan, C.; Ma, C. Blue Light Regulates Cell Wall Structure and Carbohydrate Metabolism of Soybean Hypocotyl. Int. J. Mol. Sci. 2023, 24, 1017. https://doi.org/10.3390/ijms24021017
Wang C, Chen Y, Cui C, Shan F, Zhang R, Lyu X, Lyu L, Chang H, Yan C, Ma C. Blue Light Regulates Cell Wall Structure and Carbohydrate Metabolism of Soybean Hypocotyl. International Journal of Molecular Sciences. 2023; 24(2):1017. https://doi.org/10.3390/ijms24021017
Chicago/Turabian StyleWang, Chang, Yu Chen, Can Cui, Fuxin Shan, Rui Zhang, Xiaochen Lyu, Lin Lyu, Hanwen Chang, Chao Yan, and Chunmei Ma. 2023. "Blue Light Regulates Cell Wall Structure and Carbohydrate Metabolism of Soybean Hypocotyl" International Journal of Molecular Sciences 24, no. 2: 1017. https://doi.org/10.3390/ijms24021017






