DNA Oligonucleotides as Antivirals and Vaccine Constituents against SARS Coronaviruses: A Prospective Tool for Immune System Tuning
Abstract
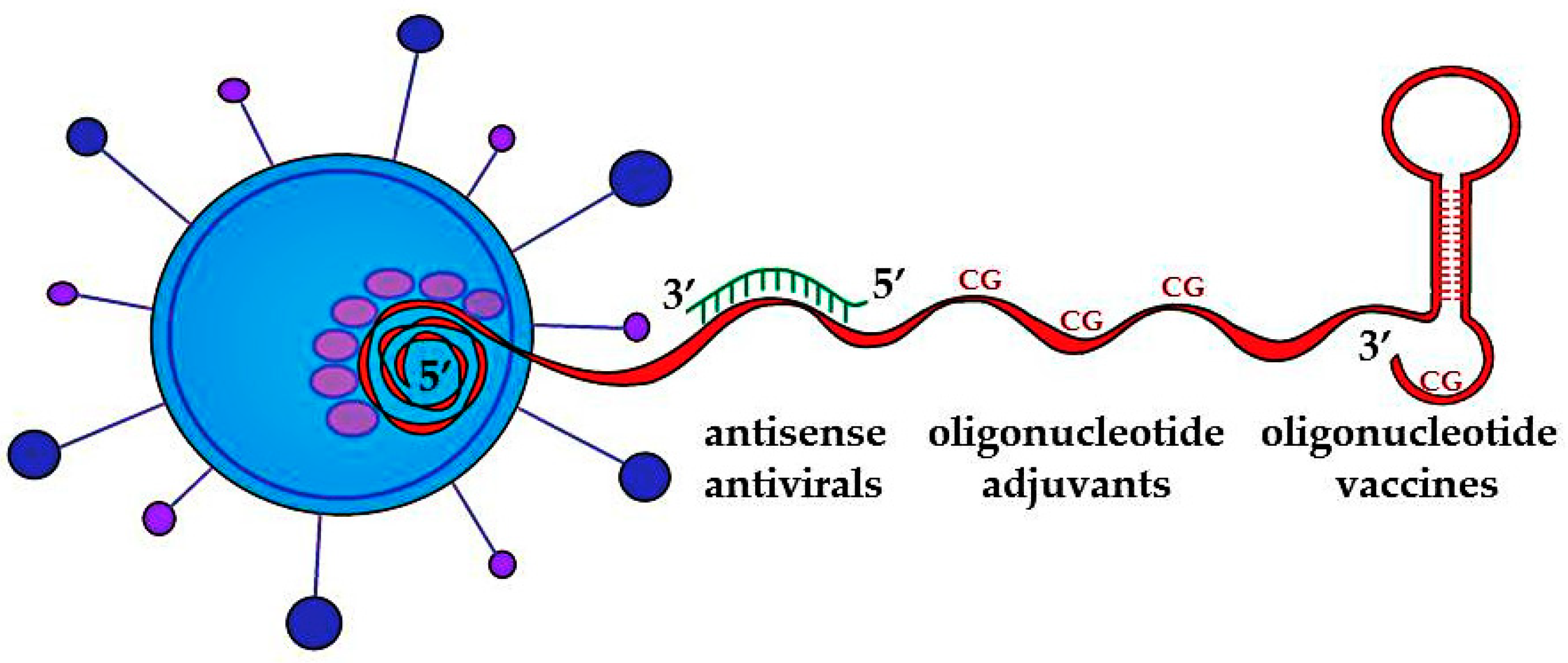
:1. Introduction
2. Antisense Antivirals
3. Oligonucleotide Adjuvants
4. Oligonucleotide Vaccines
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Yu, Q.; Binder, G.K.; Chen, Z.; Slepushkina, T.; Rossi, J.; Dropulic, B. Antisense-mediated inhibition of human immunodeficiency virus (HIV) replication by use of an HIV type 1-based vector results in severely attenuated mutants incapable of developing resistance. J. Virol. 2004, 78, 7079–7088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, S.; Kurreck, J. Oligonucleotide-based antiviral strategies. Handb. Exp. Pharmacol. 2006, 173, 261–287. [Google Scholar] [CrossRef]
- Kurreck, J. Antisense technologies. FEBS J. 2003, 8, 1628–1644. [Google Scholar]
- Soler, M.; McHutchison, J.G.; Kwoh, T.J.; Dorr, F.A.; Pawlotsky, J.-M. Virological Effects of Isis 14803, An Antisense Oligonucleotide Inhibitor of Hepatitis C Virus (HCV) Internal Ribosome Entry Site (IRES), on HCV Ires in Chronic Hepatitis C Patients and Examination of the Potential Role of Primary and Secondary HCV Resistance in the Outcome of Treatment. Antivir. Ther. 2004, 9, 953–968. [Google Scholar] [CrossRef]
- Yokota, T.; Sakamoto, N.; Enomoto, N.; Tanabe, Y.; Miyagishi, M.; Maekawa, S.; Yi, L.; Kurosaki, M.; Taira, K.; Watanabe, M.; et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003, 4, 602–608. [Google Scholar] [CrossRef] [Green Version]
- Stein, C.A.; Castanotto, D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bege, M.; Borbás, A. Rise and fall of fomivirsen, the first approved gene silencing medicine—A historical review. Res. Gate 2022, 92, 38–44. [Google Scholar] [CrossRef]
- Bradley, C.A. First Antisense Drug is Approved with Fleeting Success. 2019. Available online: https://www.nature.com/articles/d42859-019-00080-6 (accessed on 12 December 2022).
- Kim, C.; Hu, C.; Moufawad, E.L.; Achkar, L.E.; Black, J.; Douville, A.; Larson, M.K.; Pendergast, S.F.; Goldkind, E.A.; Lee, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N. Engl. J. Med. 2019, 381, 1644–1652. [Google Scholar] [CrossRef]
- Suzuki, Y.; Ishihara, H. Difference in the lipid nanoparticle technology employed in three approved siRNA (Patisiran) and mRNA (COVID-19 vaccine) drugs. Drug Metab. Pharmacokinet. 2021, 41, 100424. [Google Scholar] [CrossRef]
- Wdowikowska, A.; Janicka, M. Antisense oligonucleotide technology as a research tool in plant biology. Funct. Plant Biol. 2021, 49, 1–12. [Google Scholar] [CrossRef]
- Callaway, E. Fast-spreading COVID variant can elude immune responses. Nature 2021, 589, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Kirby, T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. The Lancet. Respir. Med. 2021, 9, e20–e21. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.R.; Navas-Martin, S. Coronavirus Pathogenesis and the Emerging Pathogen Severe Acute Respiratory Syndrome Coronavirus. Microbiol. Mol. Biol. Rev. 2005, 69, 635–664. [Google Scholar] [CrossRef] [Green Version]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.-H.; et al. Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [Green Version]
- Bermingham, A.; Chand, M.A.; Brown, C.S.; Aarons, E.; Tong, C.; Langrish, C.; Hoschler, K.; Brown, K.; Galiano, M.; Myers, R.; et al. Severe Respiratory Illness Caused by a Novel Coronavirus, in a Patient Transferred to the United Kingdom from the Middle East. Euro. Surveill. 2012, 17, 20290. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- Chen, Y.; Liu, Q.; Guo, D. Emerging Coronaviruses: Genome Structure, Replication, and Pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [Green Version]
- Amoutzias, G.D.; Nikolaidis, M.; Tryfonopoulou, E.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G. The Remarkable Evolutionary Plasticity of Coronaviruses by Mutation and Recombination: Insights for the COVID-19 Pandemic and the Future Evolutionary Paths of SARS-CoV-2. Viruses 2022, 14, 78. [Google Scholar] [CrossRef]
- Shi, Y.; Luo, H.; Jia, J.; Xiong, J.; Yang, D.; Huang, B.; Jin, Y. Antisense downregulation of SARS-CoV gene expression in Vero E6 cells. J. Gene Med. 2005, 7, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; Panariello, F.; Annunziata, P.; Giuliano, T.; Daniele, M.; Pierri, B.; Colantuono, C.; Salvi, M.; Bouché, V.; Manfredi, A.; et al. Improved SARS-CoV-2 sequencing surveillance allows the identification of new variants and signatures in infected patients. Genome Med. 2022, 14, 90. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Li, P.; Ju, X.; Rao, J.; Huang, W.; Ren, L.; Zhang, S.; Xiong, T.; Xu, K.; Zhou, X.; et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell 2021, 184, 1865–1883.e20. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Ragnarsson, L.; Cardoso, F.C.; Lewis, R.J. Transfection Methods for High-Throughput Cellular Assays of Voltage-Gated Calcium and Sodium Channels Involved in Pain. Res. Gate 2021, 16, e0243645. [Google Scholar] [CrossRef]
- Arriaga-Canon, C.; Contreras-Espinosa, L.; Rebollar-Vega, R.; Montiel-Manríquez, R.; Cedro-Tanda, A.; García-Gordillo, J.A.; Álvarez-Gómez, R.M.; Jiménez-Trejo, F.; Castro-Hernández, C.; Herrera, L.A. Transcriptomics and RNA-Based Therapeutics as Potential Approaches to Manage SARS-CoV-2 Infection. Int. J. Mol. Sci. 2022, 23, 11058. [Google Scholar] [CrossRef]
- Nikolaidis, M.; Papakyriakou, A.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G.; Amoutzias, G.D. Comparative Analysis of SARS-CoV-2 Variants of Concern, Including Omicron, Highlights Their Common and Distinctive Amino Acid Substitution Patterns, Especially at the Spike ORF. Viruses 2022, 14, 707. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, M.; Markoulatos, P.; Van de Peer, Y.; Oliver, S.G.; Amoutzias, G.D. The Neighborhood of the Spike Gene Is a Hotspot for Modular Intertypic Homologous and Nonhomologous Recombination in Coronavirus Genomes. Mol. Biol. Evol. 2022, 39, 1. [Google Scholar] [CrossRef] [PubMed]
- Tay, J.; Porter, A.; Wirth, W.; Duchene, S. The Emergence of SARS-CoV-2 Variants of Concern Is Driven by Acceleration of the Substitution Rate. Mol. Biol. Evol. 2022, 39, 2. [Google Scholar] [CrossRef]
- Cameroni, E.; Bowen, J.E.; Rosen, L.E.; Saliba, C.; Zepeda, S.K.; Culap, K.; Pinto, D.; VanBlargan, L.A.; De Marco, A.; di Iulio, J.; et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022, 602, 664–670. [Google Scholar] [CrossRef]
- Su, X.; Ma, W.; Feng, D.; Cheng, B.; Wang, Q.; Guo, Z.; Zhou, D.; Tang, X. Efficient Inhibition of SARS-CoV-2 Using Chimeric Antisense Oligonucleotides through RNase L Activation*. Angew. Chem. Int. Ed. Engl. 2021. [Google Scholar] [CrossRef]
- Pfafenrot, C.; Schneider, T.; Müller, C.; Hung, L.H.; Schreiner, S.; Ziebuhr, J.; Bindereif, A. Inhibition of SARS-CoV-2 coronavirus proliferation by designer antisense-circRNAs. Nucleic Acids Res. 2021, 49, 12502–12516. [Google Scholar] [CrossRef]
- Alavizadeh, S.H.; Doagooyan, M.; Zahedipour, F.; Torghabe, S.Y.; Baharieh, B.; Soleymani, F.; Gheybi, F. Antisense technology as a potential strategy for the treatment of coronaviruses infection: With focus on COVID-19. IET Nanobiotechnol. 2022, 60, 21662–21667. [Google Scholar] [CrossRef] [PubMed]
- Youngren-Ortiz, S.R.; Gandhi, N.S.; España-Serrano; Chougule, M.B. Aerosol delivery of siRNA to the lungs. Part 1: Rationale for gene delivery systems. KONA. Powder. Part. J. Res. Gate 2016, 33, 63–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, T.W.; Yuan, J.; Liu, Z.; Qiu, D.; Sall, A.; Yang, D. Nucleic-acid-based antiviral agents against positive single-stranded RNA viruses. Curr. Opin. Mol. Ther. 2006, 8, 104–107. [Google Scholar]
- Pray, L.A. DNA replication and causes of mutation. Nat. Educ. 2008, 1, 214. [Google Scholar]
- Pulendran, B.; SArunachalam, P.; O’Hagan, D.T. Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 2021, 20, 454–475. [Google Scholar] [CrossRef]
- Gayed, P.M. Toward a modern synthesis of immunity: Charles, A. Janeway Jr. and the immunologist’s dirty little secret. Yale J. Biol. Med. 2011, 84, 131–138. [Google Scholar]
- Bielinska, A.; Shivdasani, R.A.; Zhang, L.Q.; Nabel, G.J. Regulation of gene expression with double stranded phosphorothioate oligonucleotides. Science 1990, 250, 997–1000. [Google Scholar] [CrossRef]
- Krieg, A.M.; Yi, A.K.; Matson, S.; Waldschmidt, T.J.; Bishop, G.A.; Teasdale, R.; Koretzky, G.A.; Klinman, D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 1995, 374, 546–549. [Google Scholar] [CrossRef]
- Bauer, S.; Kirschning, C.J.; Häcker, H.; Redecke, V.; Hausmann, S.; Akira, S.; Wagner, H.; Lipford, G.B. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 2001, 98, 9237–9242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef]
- Chuang, T.H.; Lee, J.; Kline, L.; Mathison, J.C.; Ulevitch, R.J. Toll-like receptor 9 mediates CpG-DNA signaling. J. Leukoc. Biol. 2002, 71, 538–544. [Google Scholar] [CrossRef]
- Hornung, V.; Rothenfusser, S.; Britsch, S.; Krug, A.; Jahrsdörfer, B.; Giese, T.; Endres, S.; Hartmann, G. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002, 168, 4531–4537. [Google Scholar] [CrossRef] [Green Version]
- Jarrossay, D.; Napolitani, G.; Colonna, M.; Sallusto, F.; Lanzavecchia, A. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur. J. Immunol. 2001, 31, 3388–3393. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, N.; Ho, S.; Antonenko, S.; de Waal Malefyt, R.; Kastelein, R.A.; Bazan, F.; Liu, Y.-J. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 2001, 194, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Y.; Yu, G.Y.; Luo, Y.; Xiang, R.; Chuang, T.H. Immunostimulatory Activities of CpG-Oligodeoxynucleotides in Teleosts: Toll-Like Receptors 9 and 21. Front. Immunol. 2019, 10, 179. [Google Scholar] [CrossRef]
- Lin, H.T.; Chen, C.C.; Chiao, D.J.; Chang, T.Y.; Chen, X.A.; Young, J.J.; Kuo, S.C. Nanoparticular CpG-adjuvanted SARS-CoV-2 S1 protein elicits broadly neutralizing and Th1-biased immunoreactivity in mice. Int. J. Biol. Macromol. 2021, 193, 1885–1897. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://covid19.trackvaccines.org/vaccines/approved/#vaccine-list (accessed on 12 December 2022).
- Nanishi, E.; Borriello, F.; O’Meara, T.R.; McGrath, M.E.; Saito, Y.; Haupt, R.E.; Seo, H.S.; van Haren, S.D.; Cavazzoni, C.B.; Brook, B.; et al. An aluminum hydroxide:CpG adjuvant enhances protection elicited by a SARS-CoV-2 receptor binding domain vaccine in aged mice. Sci. Transl. Med. 2022, 14, eabj5305. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, X.; Sheng, W.; Liang, H.; Zhao, Y.; Zhu, X.; Yang, R.; Zhang, Y.; Dong, X.; Li, W.; et al. Alum/CpG Adjuvanted Inactivated COVID-19 Vaccine with Protective Efficacy against SARS-CoV-2 and Variants. Vaccines 2022, 10, 1208. [Google Scholar] [CrossRef]
- Caruthers, M.H. The chemical synthesis of DNA/RNA: Our gift to science. J. Biol. Chem. 2013, 288, 1420–1427. [Google Scholar] [CrossRef] [Green Version]
- Saady, A.; Böttner, V.; Meng, M.; Varon, E.; Shav-Tal, Y.; Ducho, C.; Fischer, B. An oligonucleotide probe incorporating the chromophore of green fluorescent protein is useful for the detection of HER-2 mRNA breast cancer marker. Eur. J. Med. Chem. 2019, 173, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Narang, S.A.; Dubuc, G.; Yao, F.L.; Michniewicz, J.J. “In vitro” method of assembling a synthetic gene. Biochem. Biophys. Res. Commun. 1986, 134, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Corman, J.M.; Hamorsky, K.T.; Shepherd, J.W.; Hiatt, E.; Fuqua, J.L.; Palmer, K.E. Stability of plasmid and viral banks supporting the cGMP manufacture of Q-Griffithsin from a TMV-based viral vector. J. Biotechnol. 2020, 20, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Adamopoulos, P.G.; Tsiakanikas, P.; Kontos, C.K.; Panagiotou, A.; Vassilacopoulou, D.; Scorilas, A. Identification of novel alternative splice variants of the human L-DOPA decarboxylase (DDC) gene in human cancer cells, using high-throughput sequencing approaches. Gene 2019, 30, 144075. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhania, R.R.; Patel, A.K.; Pandey, A.; Ganansounou, E. Genetic modification: A tool for enhancing beta-glucosidase production for biofuel application. Bioresour. Technol. 2017, 245, 1352–1361. [Google Scholar] [CrossRef]
- Crooke, S.T.; Liang, X.H.; Crooke, R.M.; Baker, B.F.; Geary, R.S. Antisense drug discovery and development technology considered in a pharmacological context. Biochem. Pharmacol. 2020, 189, 114196. [Google Scholar] [CrossRef]
- Potaczek, D.P.; Garn, H.; Unger, S.D.; Renz, H. Antisense molecules: A new class of drugs. J. Allergy Clin. Immunol. 2016, 137, 1334–1346. [Google Scholar] [CrossRef] [Green Version]
- Damha, M.J.; Giannaris, P.A.; Zabarylo, S.V. An improved procedure for derivatization of controlled-pore glass beads for solid-phase oligonucleotide synthesis. Nucleic Acids Res. 1990, 18, 3813–3821. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html (accessed on 12 December 2022).
- Oberemok, V.V.; Laikova, K.V.; Yurchenko, K.A.; Marochkin, N.A.; Fomochkina, I.I.; Kubyshkin, A.V. SARS-CoV-2 will constantlysweep its tracks: A vaccine containing CpG motifs in ‘lasso’ forthe multi-faced virus. Inflamm. Res. 2020, 69, 801–812. [Google Scholar] [CrossRef]
- He, Y.; Jiang, S. Vaccine design for severe acute respiratory syndrome coronavirus. Viral. Immunol. 2005, 18, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S. Antibody responses to DNA in normal immunity and aberrant immunity. Clin. Diagn Lab Immunol. 1998, 5, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Aarden, L.A.; Lakmaker, F.; De Groot, E.R. Immunology of DNA IV quantitative aspects of the Farr assay. J. Immunol. Methods 1976, 11, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Plumridge, A.; Meisburger, S.P.; Pollack, L. Visualizing single-stranded nucleic acids in solution. Nucleic Acids Res. 2016, 45, e66. [Google Scholar] [CrossRef]
- Pisetsky, D.S.; Reich, C.F. The binding of anti-DNA antibodies to phosphorothioate oligonucleotides in a solid phase immunoassay. Mol. Immunol. 1998, 35, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Kawarada, Y.; Miura, N.; Sugiyama, T. Antibody against single-stranded DNA useful for detecting apoptotic cells recognizes hexadeoxynucleotides with various base sequences. J. Biochem. 1998, 123, 492–498. [Google Scholar] [CrossRef] [Green Version]
- Oberemok, V.V.; Andreeva, O.A.; Laikova, K.V.; Novikov, I.A.; Kubyshkin, A.V. Post-genomic platform for development of oligonucleotide vaccines against RNA viruses: Diamond cuts diamond. Inflamm. Res. 2022, 71, 729–739. [Google Scholar] [CrossRef]
- Pisetsky, D.; Vrabie, I. Antibodies to DNA: Infection or genetics? Lupus 2009, 18, 1176–1180. [Google Scholar] [CrossRef]
- Tu, L.; Sun, X.; Yang, L.; Zhang, T.; Zhang, X.; Li, X.; Dong, B.; Liu, Y.; Yang, M.; Wang, L.; et al. TGF-β2 interfering oligonucleotides used as adjuvants for microbial vaccines. J. Leukoc. Biol. 2020, 108, 1673–1692. [Google Scholar] [CrossRef]
- Gursel, M.; Klinman, D.M. Chapter 62—Use of CpG Oligonucleotides as Mucosal Adjuvants. In Mucosal Immunology, 4th ed.; Mestecky, J., Ed.; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Muller, S.; Zhao, Y.; Brown, T.L.; Morgan, A.l.C.; Kohler, H. TransMabs: Cell-penetrating antibodies, the next generation. Expert Opin. Biol. Ther. 2005, 5, 237–241. [Google Scholar] [CrossRef]
- Lackey, C.A.; Press, O.W.; Hoffman, A.S.; Stayton, P.S. A biomimetic pH-responsive polymer directs endosomal release and intracellular delivery of an endocytosed antibody complex. Bioconjug. Chem. 2002, 13, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Noble, P.W.; Young, M.R.; Bernatsky, S.; Weisbart, R.H.; Hansen, J.E. A nucleolytic lupus autoantibody is toxic to BRCA2-deficient cancer cells. Sci. Rep. 2014, 4, 5958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisbart, R.H.; Chan, G.; Jordaan, G.; Noble, P.W.; Liu, Y.; Glazer, P.M.; Hansen, J.E. DNA-dependent targeting of cell nuclei by a lupus autoantibody. Sci. Rep. 2015, 5, 12022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dieli, F. Dendritic cells and the handling of antigen. Clin. Exp. Immunol. 2003, 134, 178–180. [Google Scholar] [CrossRef]
- Guermonprez, P.; Valladeau, J.; Zitvogel, L.; Théry, C.; Amigorena, S. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 2002, 20, 621–667. [Google Scholar] [CrossRef]
- an Langelaar, J.; Rijvers, L.; Smolders, J.; Van Luijn, M.M. B and T cells driving multiple sclerosis: Identity, mechanisms and potential triggers. Front. Immunol. 2020, 11, 760. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Andreeva, O.A.; Novikov, I.A.; Laikova, K.V. Oligonucleotide Vaccines: The Joker in the Vaccine Deck. Vitr. Cell. Dev. Biol.—Anim. 2022, 58, 29–35. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oberemok, V.V.; Andreeva, O.A.; Alieva, E.E. DNA Oligonucleotides as Antivirals and Vaccine Constituents against SARS Coronaviruses: A Prospective Tool for Immune System Tuning. Int. J. Mol. Sci. 2023, 24, 1553. https://doi.org/10.3390/ijms24021553
Oberemok VV, Andreeva OA, Alieva EE. DNA Oligonucleotides as Antivirals and Vaccine Constituents against SARS Coronaviruses: A Prospective Tool for Immune System Tuning. International Journal of Molecular Sciences. 2023; 24(2):1553. https://doi.org/10.3390/ijms24021553
Chicago/Turabian StyleOberemok, Volodymyr V., Oksana A. Andreeva, and Edie E. Alieva. 2023. "DNA Oligonucleotides as Antivirals and Vaccine Constituents against SARS Coronaviruses: A Prospective Tool for Immune System Tuning" International Journal of Molecular Sciences 24, no. 2: 1553. https://doi.org/10.3390/ijms24021553








