BMPR2 Variants Underlie Nonsyndromic Oligodontia
Abstract
:1. Introduction
2. Results
2.1. Clinical Findings and Variant Identification
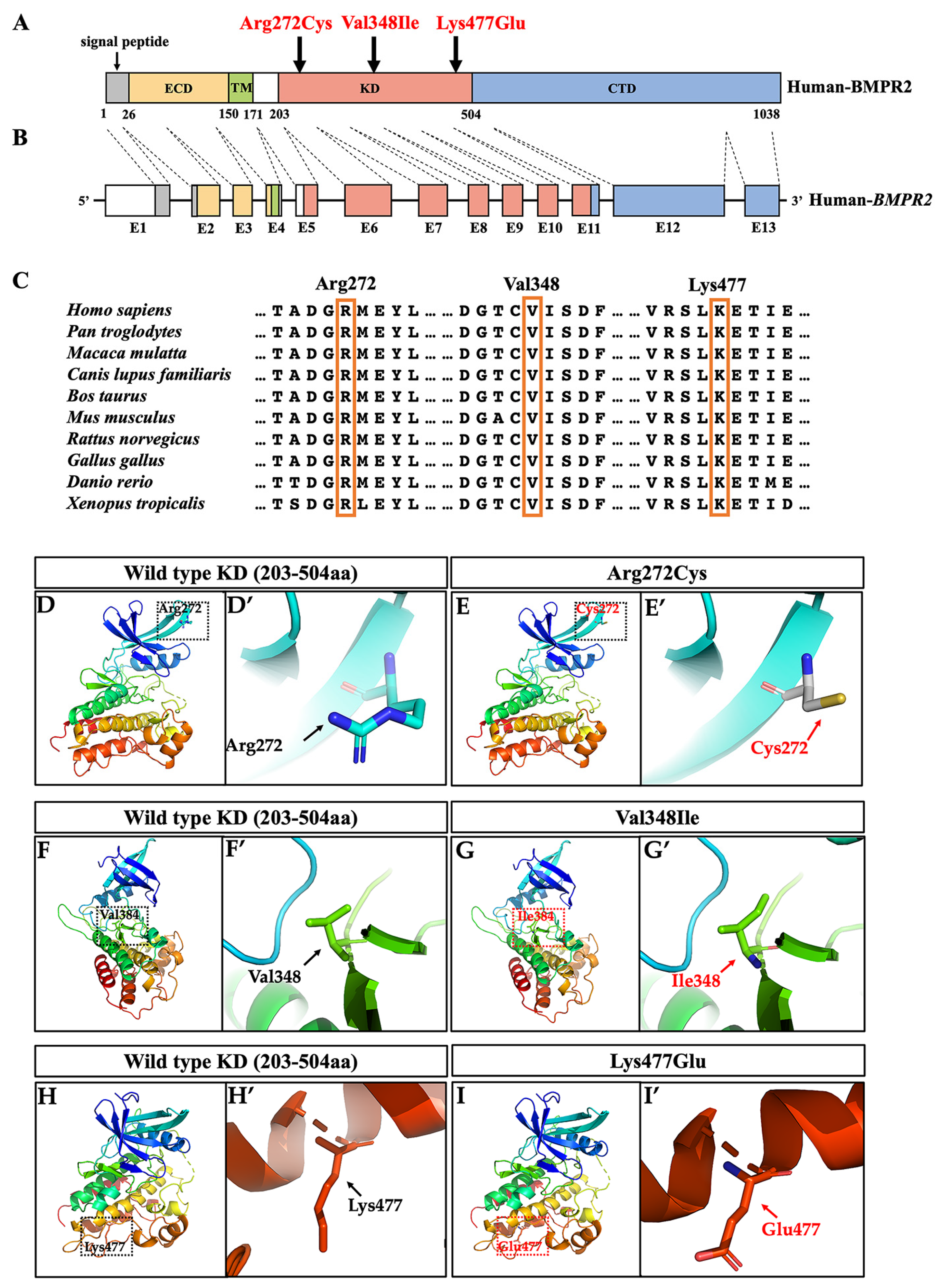
2.2. Bioinformatics and Conservation Analysis
2.3. Three-Dimensional Conformational Analysis
2.4. Genetic Mechanism Explorations of BMPR2 Variants in Oligodontia
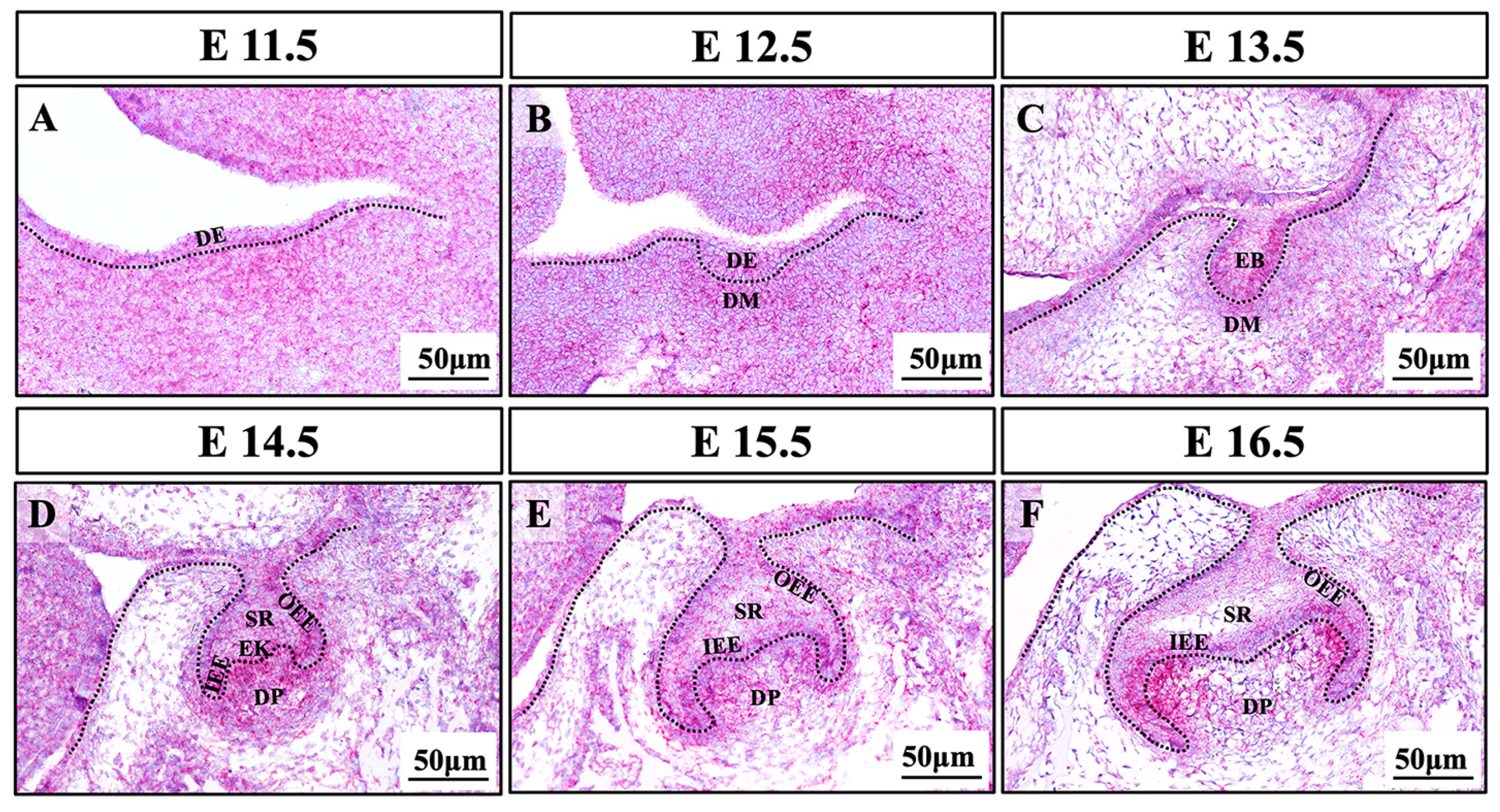
2.5. Expression Pattern of Bmpr2 during Murine Molar Development
3. Discussion
4. Materials and Methods
4.1. Recruitment of Subjects
4.2. WES
4.3. Sanger Sequencing and Genetic Screening
4.4. Bioinformatic and Conservative Analysis
4.5. Three-Dimensional Structural Modeling
4.6. Plasmid Construction
4.7. Cell Preparation and Western Blot Analysis
4.8. Tooth Germ Preparation and RNAscope In Situ Hybridization Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vastardis, H.; Karimbux, N.; Guthua, S.W.; Seidman, J.G.; Seidman, C.E. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 1996, 13, 417–421. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, H.; Camhi, H.; Seymen, F.; Koruyucu, M.; Kasimoglu, Y.; Kim, J.W.; Kim-Berman, H.; Yuson, N.M.R.; Benke, P.J.; et al. Analyses of oligodontia phenotypes and genetic etiologies. Int J. Oral Sci 2021, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- van den Boogaard, M.J.; Dorland, M.; Beemer, F.A.; van Amstel, H.K. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 2000, 24, 342–343. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Groppe, J.C.; Wu, J.; Ogawa, T.; Mues, G.; D’Souza, R.N.; Kapadia, H. Pathogenic mechanisms of tooth agenesis linked to paired domain mutations in human PAX9. Hum. Mol. Genet. 2009, 18, 2863–2874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincent, M.C.; Biancalana, V.; Ginisty, D.; Mandel, J.L.; Calvas, P. Mutational spectrum of the ED1 gene in X-linked hypohidrotic ectodermal dysplasia. Eur J. Hum. Genet. 2001, 9, 355–363. [Google Scholar] [CrossRef] [Green Version]
- Bohring, A.; Stamm, T.; Spaich, C.; Haase, C.; Spree, K.; Hehr, U.; Hoffmann, M.; Ledig, S.; Sel, S.; Wieacker, P.; et al. WNT10A mutations are a frequent cause of a broad spectrum of ectodermal dysplasias with sex-biased manifestation pattern in heterozygotes. Am. J. Hum. Genet. 2009, 85, 97–105. [Google Scholar] [CrossRef] [Green Version]
- Massink, M.P.; Creton, M.A.; Spanevello, F.; Fennis, W.M.; Cune, M.S.; Savelberg, S.M.; Nijman, I.J.; Maurice, M.M.; van den Boogaard, M.J.; van Haaften, G. Loss-of-Function Mutations in the WNT Co-receptor LRP6 Cause Autosomal-Dominant Oligodontia. Am. J. Hum. Genet. 2015, 97, 621–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, P.; Yang, W.; Han, D.; Wang, X.; Guo, S.; Li, J.; Li, F.; Zhang, X.; Wong, S.W.; Bai, B.; et al. Mutations in WNT10B Are Identified in Individuals with Oligodontia. Am. J. Hum. Genet. 2016, 99, 195–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kantaputra, P.N.; Kaewgahya, M.; Hatsadaloi, A.; Vogel, P.; Kawasaki, K.; Ohazama, A.; Ketudat Cairns, J.R. GREMLIN 2 Mutations and Dental Anomalies. J. Dent. Res. 2015, 94, 1646–1652. [Google Scholar] [CrossRef]
- Yu, M.; Wong, S.W.; Han, D.; Cai, T. Genetic analysis: Wnt and other pathways in nonsyndromic tooth agenesis. Oral Dis. 2019, 25, 646–651. [Google Scholar] [CrossRef]
- Yin, W.; Bian, Z. The Gene Network Underlying Hypodontia. J. Dent. Res. 2015, 94, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Park, S.Y.; Chang, H.R.; Jung, E.Y.; Munkhjargal, A.; Lim, J.S.; Lee, M.S.; Kim, Y. Clinical significance linked to functional defects in bone morphogenetic protein type 2 receptor, BMPR2. BMB Rep. 2017, 50, 308–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Puerto, M.C.; Iyengar, P.V.; Garcia de Vinuesa, A.; Ten Dijke, P.; Sanchez-Duffhues, G. Bone morphogenetic protein receptor signal transduction in human disease. J. Pathol. 2019, 247, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beppu, H.; Kawabata, M.; Hamamoto, T.; Chytil, A.; Minowa, O.; Noda, T.; Miyazono, K. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev. Biol. 2000, 221, 249–258. [Google Scholar] [CrossRef] [Green Version]
- International, P.P.H.C.; Lane, K.B.; Machado, R.D.; Pauciulo, M.W.; Thomson, J.R.; Phillips, J.A., 3rd; Loyd, J.E.; Nichols, W.C.; Trembath, R.C. Heterozygous germline mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension. Nat. Genet. 2000, 26, 81–84. [Google Scholar]
- Rigelsky, C.M.; Jennings, C.; Lehtonen, R.; Minai, O.A.; Eng, C.; Aldred, M.A. BMPR2 mutation in a patient with pulmonary arterial hypertension and suspected hereditary hemorrhagic telangiectasia. Am. J. Med. Genet. A 2008, 146, 2551–2556. [Google Scholar] [CrossRef]
- Lowery, J.W.; Intini, G.; Gamer, L.; Lotinun, S.; Salazar, V.S.; Ote, S.; Cox, K.; Baron, R.; Rosen, V. Loss of BMPR2 leads to high bone mass due to increased osteoblast activity. J. Cell Sci. 2015, 128, 1308–1315. [Google Scholar] [CrossRef] [Green Version]
- Machado, R.D.; Aldred, M.A.; James, V.; Harrison, R.E.; Patel, B.; Schwalbe, E.C.; Gruenig, E.; Janssen, B.; Koehler, R.; Seeger, W.; et al. Mutations of the TGF-beta type II receptor BMPR2 in pulmonary arterial hypertension. Hum. Mutat. 2006, 27, 121–132. [Google Scholar] [CrossRef]
- Newman, J.H.; Trembath, R.C.; Morse, J.A.; Grunig, E.; Loyd, J.E.; Adnot, S.; Coccolo, F.; Ventura, C.; Phillips, J.A., 3rd; Knowles, J.A.; et al. Genetic basis of pulmonary arterial hypertension: Current understanding and future directions. J. Am. Coll Cardiol. 2004, 43 (Suppl S12), 33–39. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.J. Pulmonary hypertension. Jama 2012, 308, 1366–1374. [Google Scholar] [CrossRef]
- Machado, R.D.; Eickelberg, O.; Elliott, C.G.; Geraci, M.W.; Hanaoka, M.; Loyd, J.E.; Newman, J.H.; Phillips, J.A., 3rd; Soubrier, F.; Trembath, R.C.; et al. Genetics and genomics of pulmonary arterial hypertension. J. Am. Coll Cardiol. 2009, 54 (Suppl S1), 32–42. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Hong, K.H.; Kim, Y.H.; Kim, M.J.; Song, C.; Kim, M.J.; Kim, S.J.; Raizada, M.K.; Oh, S.P. SMAD1 deficiency in either endothelial or smooth muscle cells can predispose mice to pulmonary hypertension. Hypertension 2013, 61, 1044–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez-Sanchez, G.; Childs, B.; Valle, D. Human disease genes. Nature 2001, 409, 853–855. [Google Scholar] [CrossRef] [Green Version]
- Veitia, R.A.; Caburet, S.; Birchler, J.A. Mechanisms of Mendelian dominance. Clin. Genet. 2018, 93, 419–428. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Wang, H.; Fan, Z.; Xie, C.; Liu, H.; Liu, Y.; Han, D.; Wong, S.W.; Feng, H. BMP4 mutations in tooth agenesis and low bone mass. Arch. Oral Biol 2019, 103, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lu, Y.; Mues, G.; Wang, S.; Bonds, J.; D’Souza, R. Functional evaluation of a novel tooth agenesis-associated bone morphogenetic protein 4 prodomain mutation. Eur. J. Oral Sci. 2013, 121, 313–318. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, L.; Zheng, Y.; Yuan, G.; Yang, G.; He, F.; Chen, Y. BMP activity is required for tooth development from the lamina to bud stage. J. Dent. Res. 2012, 91, 690–695. [Google Scholar] [CrossRef] [Green Version]
- Andl, T.; Ahn, K.; Kairo, A.; Chu, E.Y.; Wine-Lee, L.; Reddy, S.T.; Croft, N.J.; Cebra-Thomas, J.A.; Metzger, D.; Chambon, P.; et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development 2004, 131, 2257–2268. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Yang, G.; Yuan, G.; Gluhak-Heinrich, J.; Yang, W.; Wang, L.; Chen, Z.; Schulze McDaniel, J.; Donly, K.J.; Harris, S.E.; et al. Abnormalities in the enamel in bmp2-deficient mice. Cells Tissues Organs 2011, 194, 216–221. [Google Scholar] [CrossRef] [Green Version]
- Jia, S.; Zhou, J.; Gao, Y.; Baek, J.A.; Martin, J.F.; Lan, Y.; Jiang, R. Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development 2013, 140, 423–432. [Google Scholar] [CrossRef] [Green Version]
- Nadiri, A.; Kuchler-Bopp, S.; Perrin-Schmitt, F.; Lesot, H. Expression patterns of BMPRs in the developing mouse molar. Cell Tissue Res. 2006, 324, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]


| Patient | Exon | Nucleotide Change | Protein Change | Variation Type | dbSNP | gnomAD (MAF) | SIFT | PolyPhen-2 | Mutation Taster | ACMG Classification (Evidence of Pathogenicity) |
|---|---|---|---|---|---|---|---|---|---|---|
| #537 | 6 | c.814C > T | p.Arg272Cys | Missense | rs773655445 | 0.00003186 | 0.008 (damaging) | 0.999 (probably damaging) | Disease causing | Likely pathogenic (PS3 + PM2 + PP1 + PP3) |
| #615 | 8 | c.1042G > A | p.Val348Ile | Missense | rs201067849 | 0.0005339 | 0.023 (damaging) | 0.715 (possibly damaging) | Disease causing | Pathogenic (PS1 + PS2 + PS3 + PP3) |
| #531 | 11 | c.1429A > G | p.Lys477Glu | Missense | — 1 | — 1 | 0.060 (tolerated) | 1.000 (probably damaging) | Disease causing | Pathogenic (PS2 + PS3 + PM2 + PP3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Liu, H.; Yu, M.; Lin, B.; Sun, K.; Liu, H.; Feng, H.; Liu, Y.; Han, D. BMPR2 Variants Underlie Nonsyndromic Oligodontia. Int. J. Mol. Sci. 2023, 24, 1648. https://doi.org/10.3390/ijms24021648
Zheng J, Liu H, Yu M, Lin B, Sun K, Liu H, Feng H, Liu Y, Han D. BMPR2 Variants Underlie Nonsyndromic Oligodontia. International Journal of Molecular Sciences. 2023; 24(2):1648. https://doi.org/10.3390/ijms24021648
Chicago/Turabian StyleZheng, Jinglei, Haochen Liu, Miao Yu, Bichen Lin, Kai Sun, Hangbo Liu, Hailan Feng, Yang Liu, and Dong Han. 2023. "BMPR2 Variants Underlie Nonsyndromic Oligodontia" International Journal of Molecular Sciences 24, no. 2: 1648. https://doi.org/10.3390/ijms24021648






