Mechanistic Insights into the Anti-Proliferative Action of Gut Microbial Metabolites against Breast Adenocarcinoma Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Anti-Proliferative Activity of the Three Gut Microbial Metabolites against the MCF7 and MDA-MB-231 Human Breast Adenocarcinoma Cells
2.2. Cytotoxicity of Gut Metabolites against Normal MCF10A Human Breast Epithelial Cells
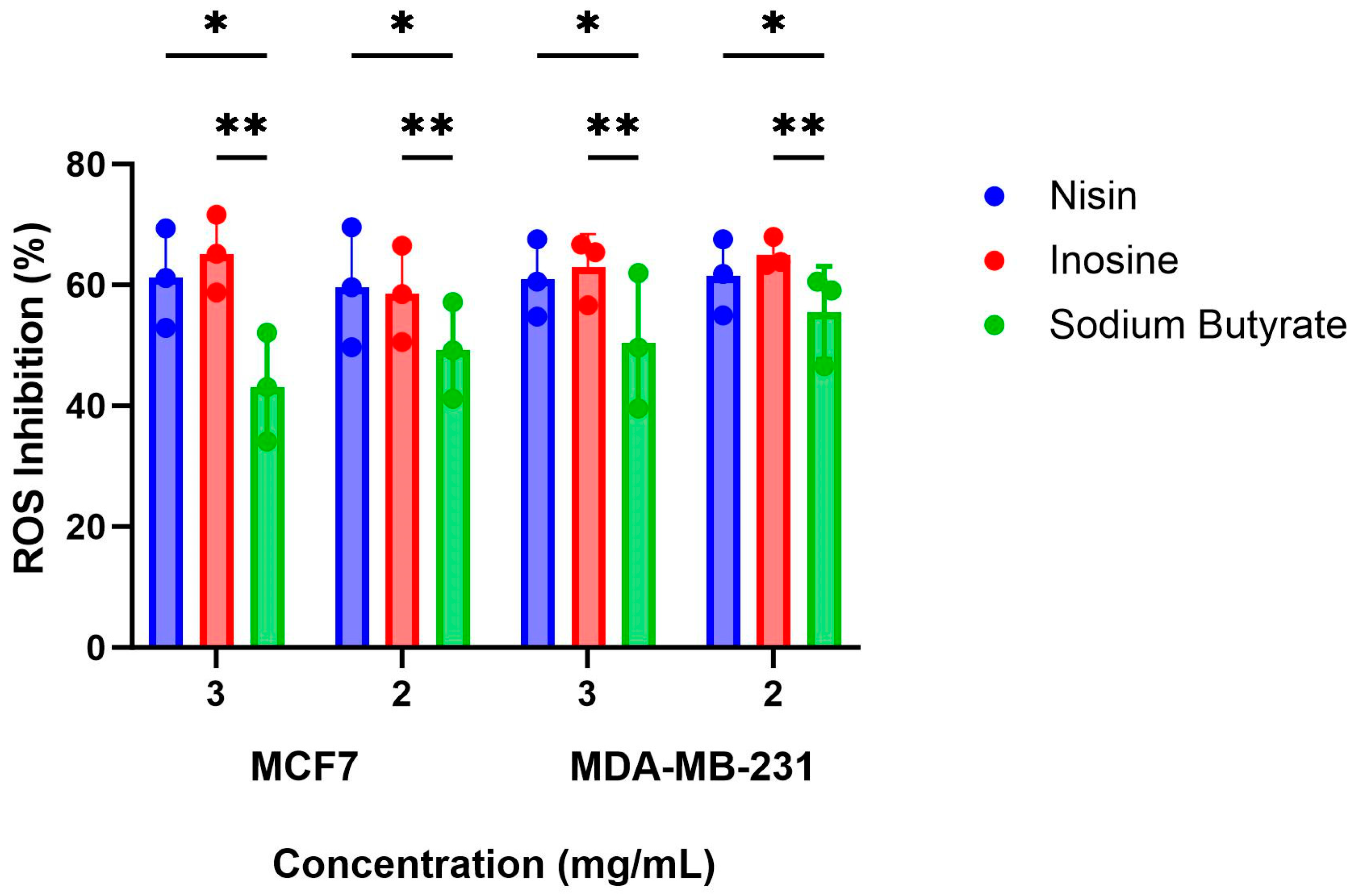
2.3. Inhibition of ROS Production by the Gut Metabolites in the MCF7 and MDA-MB-231 Cells
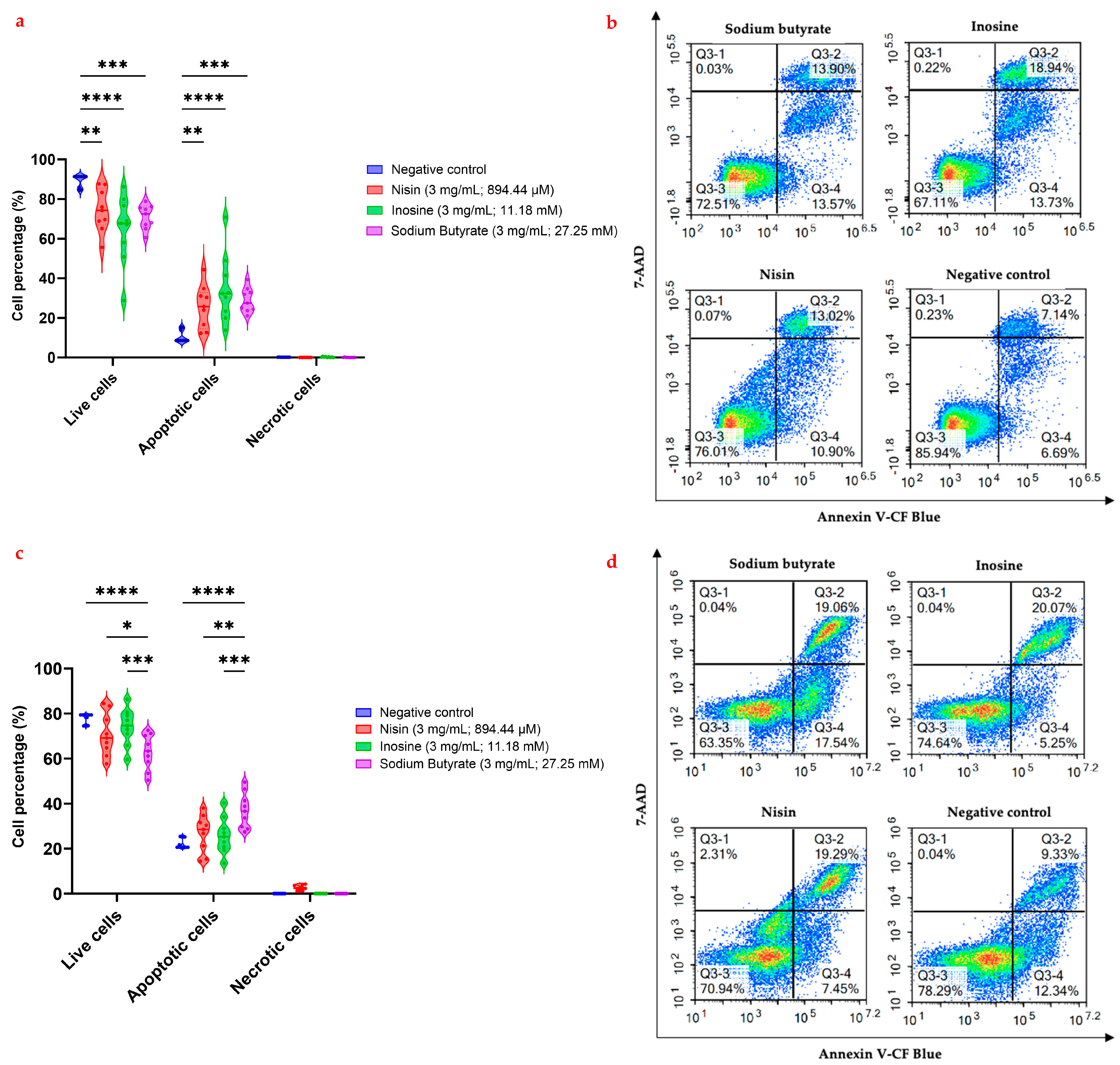
2.4. Flow Cytometric Analysis of Apoptosis in the MCF7 and MDA-MB-231 Cells Using Annexin V-CF Blue and 7-AAD
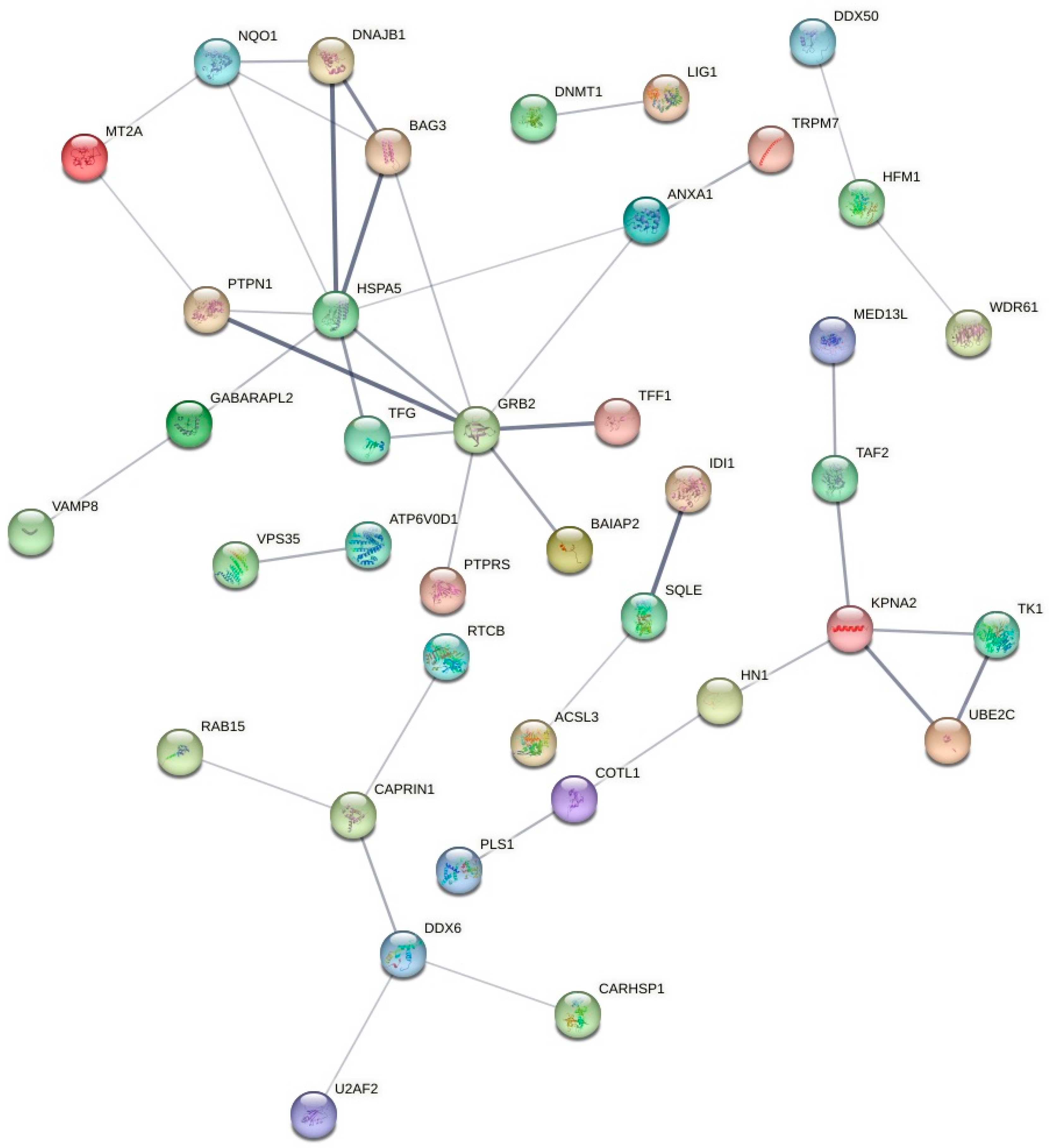
2.5. Proteomic Label-Free Bottom-up Quantification via Nano-UPLC-qTOF-MS
2.5.1. Protein Identification and Quantification in the MCF7 Cells after the Treatment with Sodium Butyrate
2.5.2. Protein Identification and Quantification in the MCF7 Cells after the Treatment with Inosine
2.5.3. Protein Identification and Quantification in the MCF7 Cells after the Treatment with Nisin
3. Materials and Methods
3.1. Preparation of Gut Microbial Metabolite
3.2. Determination of Cancer Cell Viability against MCF7 and MDA-MB-231 Cells, and Potential Cytotoxicity on the MCF10A Human Breast Epithelial Cells
3.3. Reactive Oxygen Species (ROS) Assay
3.4. Flow Cytometric Analysis Evaluation of the Apoptotic Profiles of the Cells
3.5. Proteomics Analyses
3.5.1. Cell Culture, Treatment, and Protein Extraction
3.5.2. Protein Quantification
3.5.3. Peptide Preparation and Clean up
3.5.4. Label-Free Bottom-Up Quantification via Nano-Ultra High-Performance Liquid Chromatography Coupled with Quadruple Time of Flight Mass Spectrometry (Nano-UPLC-qTOF-MS)
3.5.5. Data Processing and Availability
3.5.6. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaye, K.; Chang, D.; Li, C.G.; Bhuyan, D.J. Gut Metabolites and Breast Cancer: The Continuum of Dysbiosis, Breast Cancer Risk, and Potential Breast Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 9490. [Google Scholar] [CrossRef] [PubMed]
- Jaye, K.; Li, C.G.; Chang, D.; Bhuyan, D.J. The role of key gut microbial metabolites in the development and treatment of cancer. Gut Microbes 2022, 14, 2038865. [Google Scholar] [CrossRef]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Bedford, A.; Gong, J. Implications of butyrate and its derivatives for gut health and animal production. Anim. Nutr. 2018, 4, 151–159. [Google Scholar] [CrossRef]
- Steliou, K.; Boosalis, M.S.; Perrine, S.P.; Sangerman, J.; Faller, D.V. Butyrate histone deacetylase inhibitors. BioResearch Open Access 2012, 1, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.V.; Oesterreich, S.; Davidson, N.E. MCF-7 cells—Changing the course of breast cancer research and care for 45 years. J. Natl. Cancer Inst. 2015, 107, djv073. [Google Scholar] [CrossRef]
- Vantangoli, M.M.; Madnick, S.J.; Huse, S.M.; Weston, P.; Boekelheide, K. MCF-7 human breast cancer cells form differentiated microtissues in scaffold-free hydrogels. PLoS ONE 2015, 10, e0135426. [Google Scholar] [CrossRef]
- Dubois, C.; Dufour, R.; Daumar, P.; Aubel, C.; Szczepaniak, C.; Blavignac, C.; Mounetou, E.; Penault-Llorca, F.; Bamdad, M. Development and cytotoxic response of two proliferative MDA-MB-231 and non-proliferative SUM1315 three-dimensional cell culture models of triple-negative basal-like breast cancer cell lines. Oncotarget 2017, 8, 95316. [Google Scholar] [CrossRef]
- Huang, Z.; Yu, P.; Tang, J. Characterization of triple-negative breast cancer MDA-MB-231 cell spheroid model. OncoTargets Ther. 2020, 13, 5395. [Google Scholar] [CrossRef]
- Jenkins, D.E.; Hornig, Y.S.; Oei, Y.; Dusich, J.; Purchio, T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivodetection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res. 2005, 7, R444. [Google Scholar] [CrossRef]
- Coradini, D.; Biffi, A.; Costa, A.; Pellizzaro, C.; Pirronello, E.; DiFronzo, G. Effect of sodium butyrate on human breast cancer cell lines. Cell Prolif. 1997, 30, 149–159. [Google Scholar] [CrossRef]
- Andrade, F.O.; Nagamine, M.K.; De Conti, A.; Chaible, L.M.; Fontelles, C.C.; Jordão, A.A., Jr.; Vannucchi, H.; Dagli, M.L.Z.; Bassoli, B.K.; Moreno, F.S.; et al. Efficacy of the dietary histone deacetylase inhibitor butyrate alone or in combination with vitamin A against proliferation of MCF-7 human breast cancer cells. Braz. J. Med. Biol. Res. 2012, 45, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Semaan, J.; El-Hakim, S.; Ibrahim, J.N.; Safi, R.; Einar, A.A.; El Boustany, C. Comparative effect of sodium butyrate and sodium propionate on proliferation, cell cycle and apoptosis in human breast cancer cells MCF-7. Breast Cancer 2020, 27, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Chopin, V.; Toillon, R.A.; Jouy, N.; Bourhis, X.L. Sodium butyrate induces P53-independent, Fas-mediated apoptosis in MCF-7 human breast cancer cells. Br. J. Pharmacol. 2002, 135, 79–86. [Google Scholar] [CrossRef]
- Hamid, R.; Rotshteyn, Y.; Rabadi, L.; Parikh, R.; Bullock, P. Comparison of alamar blue and MTT assays for high through-put screening. Toxicol. Vitr. 2004, 18, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- He, C.; Liu, Y.; Ye, S.; Yin, S.; Gu, J. Changes of intestinal microflora of breast cancer in premenopausal women. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 503–513. [Google Scholar] [CrossRef]
- Nomura, M.; Nagatomo, R.; Doi, K.; Shimizu, J.; Baba, K.; Saito, T.; Matsumoto, S.; Inoue, K.; Muto, M. Association of short-chain fatty acids in the gut microbiome with clinical response to treatment with nivolumab or pembrolizumab in patients with solid cancer tumors. JAMA Netw. Open 2020, 3, e202895. [Google Scholar] [CrossRef]
- Yusuf, F.; Adewiah, S.; Syam, A.F.; Fatchiyah, F. Altered profile of gut microbiota and the level short chain fatty acids in colorectal cancer patients. J. Phys. Conf. Ser. 2019, 1146, 012037. [Google Scholar] [CrossRef]
- Smith, D.M. Inosine Production and Cytoprotective Activity in a Gradient Model of Breast Cancer Hypoxia Nutrient Exclusion (CHyNE). Ph.D. Thesis, Texas Tech University, Lubbock, TX, USA, 2018. [Google Scholar]
- Akbari, V.; Avand, A. In Vitro Cytotoxic Activity of a Lactococcus Lactis Antimicrobial Peptide against Breast Cancer Cells. Iran. J. Pharm. Sci. 2017, 13, 82–83. [Google Scholar]
- Paiva, A.D.; de Oliveira, M.D.; de Paula, S.O.; Baracat-Pereira, M.C.; Breukink, E.; Mantovani, H.C. Toxicity of bovicin HC5 against mammalian cell lines and the role of cholesterol in bacteriocin activity. Microbiology 2012, 158, 2851–2858. [Google Scholar] [CrossRef] [PubMed]
- Lulla, R.R.; Goldman, S.; Yamada, T.; Beattie, C.W.; Bressler, L.; Pacini, M.; Pollack, I.F.; Fisher, P.G.; Packer, R.J.; Dunkel, I.J. Phase I trial of p28 (NSC745104), a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in pediatric patients with recurrent or progressive central nervous system tumors: A Pediatric Brain Tumor Consortium Study. Neuro-Oncol. 2016, 18, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Warso, M.; Richards, J.; Mehta, D.; Christov, K.; Schaeffer, C.; Bressler, L.R.; Yamada, T.; Majumdar, D.; Kennedy, S.; Beattie, C. A first-in-class, first-in-human, phase I trial of p28, a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in patients with advanced solid tumours. Br. J. Cancer 2013, 108, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Tuli, H.S.; Varol, A.; Thakral, F.; Yerer, M.B.; Sak, K.; Varol, M.; Jain, A.; Khan, M.A.; Sethi, G. Role of reactive oxygen species in cancer progression: Molecular mechanisms and recent advancements. Biomolecules 2019, 9, 735. [Google Scholar] [CrossRef]
- Tobar, N.; Cáceres, M.; Santibáñez, J.F.; Smith, P.C.; Martínez, J. RAC1 activity and intracellular ROS modulate the migratory potential of MCF-7 cells through a NADPH oxidase and NFκB-dependent mechanism. Cancer Lett. 2008, 267, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Salimi, V.; Shahsavari, Z.; Safizadeh, B.; Hosseini, A.; Khademian, N.; Tavakoli-Yaraki, M. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis. 2017, 16, 208. [Google Scholar] [CrossRef] [PubMed]
- Ogura, J.; Kuwayama, K.; Sasaki, S.; Kaneko, C.; Koizumi, T.; Yabe, K.; Tsujimoto, T.; Takeno, R.; Takaya, A.; Kobayashi, M. Reactive oxygen species derived from xanthine oxidase interrupt dimerization of breast cancer resistance protein, resulting in suppression of uric acid excretion to the intestinal lumen. Biochem. Pharmacol. 2015, 97, 89–98. [Google Scholar] [CrossRef]
- Pfeffer, C.M.; Singh, A.T. Apoptosis: A target for anticancer therapy. Int. J. Mol. Sci. 2018, 19, 448. [Google Scholar] [CrossRef]
- Lallemand, F.; Courilleau, D.; Buquet-Fagot, C.; Atfi, A.; Montagne, M.-N.; Mester, J. Sodium butyrate induces G2 arrest in the human breast cancer cells MDA- MB-231 and renders them competent for DNA rereplication. Exp. Cell Res. 1999, 247, 432–440. [Google Scholar] [CrossRef]
- Liu, J.; Ma, L.; Li, L.; Sun, Y.; Wu, X.; Chen, L.; Wu, P. Histone deacetylase inhibitor sodium butyrate suppresses DNA double strand break repair induced by etoposide more effectively in MCF-7 cells than in HEK293 cells. BMC Biochem. 2015, 16, 2. [Google Scholar] [CrossRef]
- Salimi, V.; Shabani, M.; Tavakoli-Yaraki, M.; Nourbakhsh, M. Involvement of 15-lipoxygenase-1 in the regulation of breast cancer cell death induced by sodium butyrate. Cytotechnology 2016, 68, 2519–2528. [Google Scholar] [CrossRef] [PubMed]
- Louis, M.; Rosato, R.R.; Brault, L.; Osbild, S.; Battaglia, E.; Yang, X.-H.; Grant, S.; Bagrel, D. The histone deacetylase inhibitor sodium butyrate induces breast cancer cell apoptosis through diverse cytotoxic actions including glutathione depletion and oxidative stress. Int. J. Oncol. 2004, 25, 1701–1711. [Google Scholar] [CrossRef] [PubMed]
- Chopin, V.; Toillon, R.-A.; Le Bourhis, X.; Jouy, N. P21WAF1/CIP1 is dispensable for G1 arrest, but indispensable for apoptosis induced by sodium butyrate in MCF-7 breast cancer cells. Oncogene 2004, 23, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Chopin, V.; Hondermarck, H.; Le Bourhis, X.; Slomianny, C. Synergistic induction of apoptosis in breast cancer cells by cotreatment with butyrate and TNF-alpha, TRAIL, or anti-Fas agonist antibody involves enhancement of death receptors’ signaling and requires P21waf1. Exp. Cell Res. 2004, 298, 560–573. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, P.-C.; Ma, Y.-B.; Fan, R.; Gao, F.-F.; Wei, L.; Zhang, J.-W. Sodium butyrate-induced apoptosis and ultrastructural changes in MCF-7 breast cancer cells. Ultrastruct. Pathol. 2016, 40, 200–204. [Google Scholar] [CrossRef]
- Rodrigues, M.F.; Carvalho, E.; Rumjanek, F.D.; Amoedo, N.D.; Pezzuto, P. Reciprocal modulation of histone deacetylase inhibitors sodium butyrate and trichostatin A on the energy metabolism of breast cancer cells. J. Cell. Biochem. 2015, 116, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Rattanapornsompong, K.; Khattiya, J.; Phannasil, P.; Phaonakrop, N.; Roytrakul, S.; Jitrapakdee, S.; Akekawatchai, C. Impaired G2/M cell cycle arrest induces apoptosis in pyruvate carboxylase knockdown MDA-MB-231 cells. Biochem. Biophys. Rep. 2021, 25, 100903. [Google Scholar] [CrossRef]
- Price, M.T.; Olney, J.W.; Samson, L.; Labruyere, J. Calcium influx accompanies but does not cause excitotoxin-induced neuronal necrosis in retina. Brain Res. Bull. 1985, 14, 369–376. [Google Scholar] [CrossRef]
- Alsherbiny, M.A.; Bhuyan, D.J.; Radwan, I.; Chang, D.; Li, C.-G. Metabolomic identification of anticancer metabolites of Australian propolis and proteomic elucidation of its synergistic mechanisms with doxorubicin in the MCF7 cells. Int. J. Mol. Sci. 2021, 22, 7840. [Google Scholar] [CrossRef]
- Abe, M.; Kufe, D.W. Effect of Sodium-Butyrate on Human-Breast Carcinoma (MCF-7) Cellular Proliferation, Morphology, and Cea Production. Breast Cancer Res. Treat. 1984, 4, 269–274. [Google Scholar] [CrossRef]
- Graham, K.A.; Buick, R.N. Sodium-Butyrate Induces Differentiation in Breast-Cancer Cell-Lines Expressing the Estrogen-Receptor. J. Cell. Physiol. 1988, 136, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Janson, W.; Brandner, G.; Siegel, J. Butyrate modulates DNA-damage-induced p53 response by induction of p53-independent differentiation and apoptosis. Oncogene 1997, 15, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, V.; Le Bourhis, X.; Toillon, R.-A.; Boilly, B.; Hondermarck, H. Alterations in both heparan sulfate proteoglycans and mitogenic activity of fibroblast growth factor-2 are triggered by inhibitors of proliferation in normal and breast cancer epithelial cells. Exp. Cell Res. 1998, 245, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Kumar, R. BcI-2 expression regulates sodium butyrate-induced apoptosis in human MCF-7 breast cancer cells. Cell Growth Differ 1996, 7, 311–318. [Google Scholar] [PubMed]
- Stevens, M.S.; Aliabadi, Z.; Moore, M.R. Associated Effects of Sodium-Butyrate on Histone Acetylation and Estrogen-Receptor in the Human-Breast Cancer Cell-Line MCF-7. Biochem. Biophys. Res. Commun. 1984, 119, 132–138. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, L.; Zou, W.; Xu, J.; Liu, H.; Wang, W.; Yun, X.; Gu, J. Prosaposin, a regulator of estrogen receptor alpha, promotes breast cancer growth. Cancer Sci. 2012, 103, 1820–1825. [Google Scholar] [CrossRef]
- Kang, S.-Y.; Halvorsen, O.J.; Gravdal, K.; Bhattacharya, N.; Lee, J.M.; Liu, N.W.; Johnston, B.T.; Johnston, A.B.; Haukaas, S.A.; Aamodt, K. Prosaposin inhibits tumor metastasis via paracrine and endocrine stimulation of stromal p53 and Tsp-1. Proc. Natl. Acad. Sci. USA 2009, 106, 12115–12120. [Google Scholar] [CrossRef]
- Daly, R.J.; Binder, M.D.; Sutherland, R.L. Overexpression of the Grb2 gene in human breast cancer cell lines. Oncogene 1994, 9, 2723–2727. [Google Scholar]
- Tari, A.M.; Lopez-Berestein, G. GRB2: A pivotal protein in signal transduction. Semin. Oncol. 2001, 28 (Suppl. S16), 142–147. [Google Scholar] [CrossRef]
- Chen, D.; Si, W.; Shen, J.; Du, C.; Lou, W.; Bao, C.; Zheng, H.; Pan, J.; Zhong, G.; Xu, L. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018, 9, 188. [Google Scholar] [CrossRef]
- Braicu, C.; Raduly, L.; Morar-Bolba, G.; Cojocneanu, R.; Jurj, A.; Pop, L.-A.; Pileczki, V.; Ciocan, C.; Moldovan, A.; Irimie, A. Aberrant miRNAs expressed in HER-2 negative breast cancers patient. J. Exp. Clin. Cancer Res. 2018, 37, 257. [Google Scholar] [CrossRef]
- Murphy, E.; Steenbergen, C. Estrogen regulation of protein expression and signaling pathways in the heart. Biol. Sex Differ. 2014, 5, 6. [Google Scholar] [CrossRef]
- Kahl, I.; Mense, J.; Finke, C.; Boller, A.-L.; Lorber, C.; Győrffy, B.; Greve, B.; Götte, M.; Espinoza-Sánchez, N.A. The cell cycle-related genes RHAMM, AURKA, TPX2, PLK1, and PLK4 are associated with the poor prognosis of breast cancer patients. J. Cell. Biochem. 2022, 123, 581–600. [Google Scholar] [CrossRef] [PubMed]
- Ergul, M.; Bakar-Ates, F. RO3280: A novel PLK1 inhibitor, suppressed the proliferation of MCF-7 breast cancer cells through the induction of cell cycle arrest at G2/M point. Anti-Cancer Agents Med. Chem. 2019, 19, 1846–1854. [Google Scholar] [CrossRef] [PubMed]
- Grolmusz, V.K.; Karászi, K.; Micsik, T.; Tóth, E.A.; Mészáros, K.; Karvaly, G.; Barna, G.; Szabó, P.M.; Baghy, K.; Matkó, J. Cell cycle dependent RRM2 may serve as proliferation marker and pharmaceutical target in adrenocortical cancer. Am. J. Cancer Res. 2016, 6, 2041. [Google Scholar] [PubMed]
- Ma, J.; Zhang, F.; Sun, P. miR-140-3p impedes the proliferation of human cervical cancer cells by targeting RRM2 to induce cell-cycle arrest and early apoptosis. Bioorg. Med. Chem. 2020, 28, 115283. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Mahfouz, M.; Habashy, H.O. RRM2 expression in different molecular subtypes of breast cancer and its prognostic significance. Diagn. Pathol. 2022, 17, 1. [Google Scholar] [CrossRef]
- Chen, W.-x.; Yang, L.-g.; Xu, L.-y.; Cheng, L.; Qian, Q.; Sun, L.; Zhu, Y.-l. Bioinformatics analysis revealing prognostic significance of RRM2 gene in breast cancer. Biosci. Rep. 2019, 39, BSR20182062. [Google Scholar] [CrossRef]
- Bagegni, N.; Thomas, S.; Liu, N.; Luo, J.; Hoog, J.; Northfelt, D.W.; Goetz, M.P.; Forero, A.; Bergqvist, M.; Karen, J. Serum thymidine kinase 1 activity as a pharmacodynamic marker of cyclin-dependent kinase 4/6 inhibition in patients with early-stage breast cancer receiving neoadjuvant palbociclib. Breast Cancer Res. 2017, 19, 123. [Google Scholar] [CrossRef]
- Del Re, M.; Bertolini, I.; Crucitta, S.; Fontanelli, L.; Rofi, E.; De Angelis, C.; Diodati, L.; Cavallero, D.; Gianfilippo, G.; Salvadori, B. Overexpression of TK1 and CDK9 in plasma-derived exosomes is associated with clinical resistance to CDK4/6 inhibitors in metastatic breast cancer patients. Breast Cancer Res. Treat. 2019, 178, 57–62. [Google Scholar] [CrossRef]
- Dasgupta, T.; Antony, J.; Braithwaite, A.W.; Horsfield, J.A. HDAC8 inhibition blocks SMC3 deacetylation and delays cell cycle progression without affecting cohesin-dependent transcription in MCF7 cancer cells. J. Biol. Chem. 2016, 291, 12761–12770. [Google Scholar] [CrossRef]
- Kaindl, A.M.; Passemard, S.; Kumar, P.; Kraemer, N.; Issa, L.; Zwirner, A.; Gerard, B.; Verloes, A.; Mani, S.; Gressens, P. Many roads lead to primary autosomal recessive microcephaly. Prog. Neurobiol. 2010, 90, 363–383. [Google Scholar] [CrossRef]
- Stander, B.; Marais, S.; Vorster, C.; Joubert, A. In vitro effects of 2-methoxyestradiol on morphology, cell cycle progression, cell death and gene expression changes in the tumorigenic MCF-7 breast epithelial cell line. J. Steroid Biochem. Mol. Biol. 2010, 119, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Hung, L.-Y.; Chen, H.-L.; Chang, C.-W.; Li, B.-R.; Tang, T.K. Identification of a novel microtubule-destabilizing motif in CPAP that binds to tubulin heterodimers and inhibits microtubule assembly. Mol. Biol. Cell 2004, 15, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.D.; Kim, B.; Jung, S.; Kim, H.; Kim, M.K.; Kwon, J.-O.; Song, M.-K.; Lee, Z.H.; Kim, H.-H. The dynactin subunit DCTN1 controls osteoclastogenesis via the Cdc42/PAK2 pathway. Exp. Mol. Med. 2020, 52, 514–528. [Google Scholar] [CrossRef]
- Li, W.; Chen, J.; Xiong, Z.; Zhou, H.; Huang, S.; Ren, J.; Liu, B.; Zhou, T.; Hu, K. Dynactin 2 acts as an oncogene in hepatocellular carcinoma through promoting cell cycle progression. Liver Res. 2022, 6, 155–166. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Q.; Zhang, X.; Liao, X.; Wang, G.; Yu, L.; Zhang, W.; Zhou, Q.; Hu, S.; Yuan, W. Distinct prognostic value of dynactin subunit 4 (DCTN4) and diagnostic value of DCTN1, DCTN2, and DCTN4 in colon adenocarcinoma. Cancer Manag. Res. 2018, 10, 5807. [Google Scholar] [CrossRef]
- Mundt, K.E.; Golsteyn, R.M.; Lane, H.A.; Nigg, E.A. On the regulation and function of human polo-like kinase 1 (PLK1): Effects of overexpression on cell cycle progression. Biochem. Biophys. Res. Commun. 1997, 239, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Bhuyan, D.J.; Alsherbiny, M.A.; Low, M.N.; Zhou, X.; Kaur, K.; Li, G.; Li, C.G. Broad-spectrum pharmacological activity of Australian propolis and metabolomic-driven identification of marker metabolites of propolis samples from three continents. Food Funct. 2021, 12, 2498–2519. [Google Scholar] [CrossRef]
- Bhuyan, D.J.; Vuong, Q.V.; Bond, D.R.; Chalmers, A.C.; Bowyer, M.C.; Scarlett, C.J. Eucalyptus microcorys leaf extract derived HPLC-fraction reduces the viability of MIA PaCa-2 cells by inducing apoptosis and arresting cell cycle. Biomed. Pharmacother. 2018, 105, 449–460. [Google Scholar] [CrossRef]
- Alsherbiny, M.A.; Bhuyan, D.J.; Low, M.N.; Chang, D.; Li, C.G. Synergistic interactions of cannabidiol with chemotherapeutic drugs in mcf7 cells: Mode of interaction and proteomics analysis of mechanisms. Int. J. Mol. Sci. 2021, 22, 10103. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B. The reactome pathway knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Kamburov, A.; Cavill, R.; Ebbels, T.M.; Herwig, R.; Keun, H.C. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 2011, 27, 2917–2918. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]


| Concentration (mg/mL) | Cell Growth Inhibition (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| MCF7 | MDA-MB-231 | MCF10A | |||||||
| Sodium Butyrate | Inosine | Nisin | Sodium Butyrate | Inosine | Nisin | Sodium Butyrate | Inosine | Nisin | |
| 3 | 81.73 ± 4.81 a | 47.78 ± 8.89 b | 14.34 ± 15.66 c | 80.80 ± 1.06 a | 48.85 ± 1.11 b | 24.46 ± 6.79 c | 89.23 ± 0.92 a | 87.64 ± 5.51 a | 69.43 ± 10.28 a |
| 2 | 70.86 ± 7.24 a | 39.88 ± 7.04 b | 13.31 ± 7.43 c | 67.70 ± 6.28 a | 36.09 ± 3.32 b | 12.29 ± 7.97 c | 83.96 ± 4.65 a | 84.47 ± 7.38 a | 54.76 ± 15.69 a |
| 1 | 58.15 ± 2.90 a | 37.25 ± 5.07 b | 13.97 ± 9.09 c | 71.44 ± 6.84 a | 27.80 ± 6.40 b | 12.01 ± 4.13 c | 66.53 ± 3.85 a | 69.49 ± 13.77 a | 34.57 ± 15.22 b |
| 0.5 | 43.84 ± 3.49 a | 19.12 ± 5.39 b | 8.86 ± 5.47 b | 51.05 ± 2.71 a | 13.67 ± 3.55 b | 10.18 ± 5.13 b | 39.46 ± 12.27 a | 31.29 ± 18.71 a | 27.20 ± 12.77 a |
| 0.25 | 35.79 ± 3.44 a | 4.04 ± 5.61 b | 6.92 ± 6.47 b | 31.33 ± 3.83 a | 6.38 ± 1.67 b | 6.95 ± 5.39 b | 18.47 ± 17.41 a | 20.94 ± 14.73 a | 2.80 ± 29.91 a |
| 0.125 | 25.98 ± 4.62 a | - | 5.89 ± 4.10 b | 18.45 ± 5.87 a | 4.01 ± 0.75 b | 6.33 ± 7.06 b | 8.39 ± 18.78 a | 12.11 ± 19.81 a | 18.27 ± 13.50 a |
| 0.0625 | 17.62 ± 3.88 a | - | 2.48 ± 5.71 b | 7.57 ± 5.49 a | 2.08 ± 0.41 a | 2.41 ± 4.27 a | 3.76 ± 7.58 a | 18.71 ± 15.44 a | 21.05 ± 10.93 a |
| 0.03125 | 10.78 ± 2.08 a | - | 0.98 ± 7.78 a | 4.93 ± 5.23 a | 3.52 ± 0.92 a | 3.90 ± 1.87 a | 5.67 ± 8.31 a | 3.57 ± 17.48 a | 15.98 ± 6.31 a |
| IC50 | 0.576 mg/mL (5.23 mM) | 2.743 mg/mL (10.23 mM) | ND | 0.557 mg/mL (5.06 mM) | 3.262 mg/mL (12.16 mM) | ND | 0.655 mg/mL (5.95 mM) | 0.655 mg/mL (2.44 mM) | 1.63 mg/mL (485.98 μM) |
| Gut Metabolite | Regulation | Protein(s) | Gene(s) | Molecular Pathway | Mechanism of Action |
|---|---|---|---|---|---|
| Sodium butyrate | ↑ | C9JIZ6 | PSAP | Metabolism of proteins | Surfactant metabolism |
| Disease | Defective CSF2RA causes SMDP4 | ||||
| Defective CSF2RB causes SMDP5 | |||||
| Diseases associated with surfactant metabolism | |||||
| Defective SFTPA2 causes IPF | |||||
| ↑ | E7EVQ6 Q13907 | SQLE IDI1 | Metabolism | Activation of gene expression by SREBF (SREBP) | |
| Regulation of cholesterol biosynthesis by SREBP (SREBF) | |||||
| ↑ | C9JIZ6 P62993 | PSAP GRB2 | Cell-cell communication | Signal regulatory protein family interactions | |
| ↑ | P02795 P21291 | MT2A CSRP1 | Cellular responses to stimuli | Response to metal ions | |
| ↑ ↓ | P18031 P62993 F5GZ28 | PTPN1 GRB2 LIG1 | Signal transduction | Negative regulation of MET activity | |
| ↑ | P21291 | CSRP1 | Cellular responses to stimuli | MTF1 activates gene expression | |
| ↑ | P18031 Q9UHQ9 P11021 C9JIZ6 P62993 E9PI30 P04899 | PTPN1 CYB5R1 HSPA5 PSAP GRB2 CTSW GNAI2 | Hemostasis | Platelet activation, signalling and aggregation | |
| ↑ ↓ | P52292 P04155 | KPNA2 TFF1 | Signal transduction | Oestrogen-dependent gene expression | |
| ↑ | P11021 | HSPA5 | Cellular responses to stimuli | ATF6 (ATF6-alpha) activates chaperone genes | |
| ATF6 (ATF6-alpha) activates chaperones | |||||
| ↑ ↓ | P11216 Q9UHQ9 C9JIZ6 Q99519 Q14019 Q9BV40 Q9ULZ3 Q12912 Q92882 | PYGB CYB5R1 PSAP NEU1 COTL1 VAMP8 PYCARD IRAG2 OSTF1 | Immune system | Neutrophil degranulation | |
| ↑ ↓ | P25685 P11021 O95817 | DNAJB1 HSPA5 BAG3 | Cellular responses to stimuli | Regulation of HSF1-mediated heat shock response, cellular response to heat stress | |
| ↑ ↓ | P52292 P04899 P04155 | KPNA2 GNAI2 TFF1 | Signal transduction | ESR-mediated signalling | |
| ↑ ↓ | P25685 P02795 P11498 P11021 P21291 F5GYQ1 O00762 O95817 P15559 | DNAJB1 MT2A PC HSPA5 CSRP1 ATP6V0D1 UBE2C BAG3 NQO1 | Cellular responses to stimuli | Cellular responses to stimuli | |
| ↑ | P11216 | PYGB | Metabolism | Glycogen breakdown (glycogenolysis) | |
| ↑ | C9JIZ6 P62993 Q15404 | PSAP GRB2 RSU1 | Cell-cell communication | Cell-cell communication | |
| ↑ | Q13332 P62993 | PTPRS GRB2 | Signal transduction | Signalling by NTRK3 (TRKC) | |
| ↑ | Q9UHQ9 P11021 C9JIZ6 E9PI30 | CYB5R1 HSPA5 PSAP CTSW | Hemostasis | Platelet degranulation | |
| Response to elevated platelet cytosolic Ca2+ | |||||
| Inosine | ↑ | P31350 P04183 | RRM2 TK1 | Cell cycle | G1/S Transition and G1/S-Specific Transcription |
| Mitotic G1 phase and G1/S transition | |||||
| ↑ | P08243 | ASNS | Cellular responses to stimuli | Response of EIF2AK1 (HRI) to heme deficiency | |
| ATF4 activates genes in response to endoplasmic reticulum stress | |||||
| PERK regulates gene expression | |||||
| ↑ | P08727 P05783 P05787 | KRT19 KRT18 KRT8 | Developmental biology | Formation of the cornified envelope | |
| Keratinisation | |||||
| ↓ | P53350 Q9UQE7 | PLK1 SMC3 | Cell cycle | Mitotic Telephase/Cytokinesis | |
| ↑ ↓ | P21281 P08243 F2Z388 | ATP6V1B2 ASNS RPL35 | Cellular responses to stimuli | Cellular response to starvation | |
| ↑ | Q9Y2A7 H0Y6R4 I3L4C2 | NCKAP1 RAPGEF1 BAIAP2 | Signal transduction | RAC3 GTPase cycle | |
| ↑ | P30613 | PKLR | Developmental biology | Regulation of gene expression in beta cells | |
| ↑ ↓ | P08243 F2Z388 | ASNS RPL35 | Cellular responses to stimuli | Response of EIF2AK4 (GCN2) to amino acid deficiency | |
| ↑ | Q9Y2A7 I3L4C2 | NCKAP1 BAIAP2 | Signal transduction | RHO GTPases activate WASPs and WAVEs | |
| ↑ ↓ | P31350 P04183 Q9UQE7 P53350 | RRM2 TK1 SMC3 PLK1 | Cell cycle | Cell Cycle, Mitotic | |
| ↑ | P51572 Q9Y2A7 P06239 H0Y6R4 Q5VT25 I3L4C2 | BCAP31 NCKAP1 LCK RAPGEF1 CDC42BPA BAIAP2 | Signal transduction | RHO GTPase cycle | |
| ↑ | P31350 | RRM2 | Gene expression (transcription) | Transcriptional regulation by E2F6 | |
| ↑ | H0YAV1 P00403 | RRM2B MT-CO2 | TP53 Regulates metabolic genes | ||
| ↓ | P53350 | PLK1 | Cell cycle | Polo-like kinase-mediated events | |
| Mitotic metaphase/anaphase transition | |||||
| Nisin | ↑ ↓ | Q9HC77 A0A7P0Z4C3 P53350 | CENPJ DCTN1 PLK1 | Cell cycle | Loss of proteins required for interphase microtubule organization from the centrosome |
| Loss of Nlp from mitotic centrosomes | |||||
| AURKA activation by TPX2 | |||||
| Recruitment of mitotic centrosome proteins and complexes | |||||
| Centrosome maturation | |||||
| Regulation of PLK1 activity at G2/M transition | |||||
| Recruitment of NuMA to mitotic centrosomes | |||||
| Organelle biogenesis and maintenance | Anchoring of the basal body to the plasma membrane | ||||
| Cell cycle | G2/M transition | ||||
| Mitotic G2-G2/M phases | |||||
| ↓ | P53350 | PLK1 | Cell cycle | Polo-like kinase-mediated events | |
| Mitotic metaphase/anaphase transition | |||||
| ↑ ↓ | Q9Y6G9 A0A7P0Z4C3 Q9HC77 P53350 | DYNC1LI1 DCTN1 CENPJ PLK1 | Cell cycle | Mitotic prometaphase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaye, K.; Alsherbiny, M.A.; Chang, D.; Li, C.-G.; Bhuyan, D.J. Mechanistic Insights into the Anti-Proliferative Action of Gut Microbial Metabolites against Breast Adenocarcinoma Cells. Int. J. Mol. Sci. 2023, 24, 15053. https://doi.org/10.3390/ijms242015053
Jaye K, Alsherbiny MA, Chang D, Li C-G, Bhuyan DJ. Mechanistic Insights into the Anti-Proliferative Action of Gut Microbial Metabolites against Breast Adenocarcinoma Cells. International Journal of Molecular Sciences. 2023; 24(20):15053. https://doi.org/10.3390/ijms242015053
Chicago/Turabian StyleJaye, Kayla, Muhammad A. Alsherbiny, Dennis Chang, Chun-Guang Li, and Deep Jyoti Bhuyan. 2023. "Mechanistic Insights into the Anti-Proliferative Action of Gut Microbial Metabolites against Breast Adenocarcinoma Cells" International Journal of Molecular Sciences 24, no. 20: 15053. https://doi.org/10.3390/ijms242015053







