MicroRNA-152-3p and MicroRNA-196a-5p Are Downregulated When Müller Cells Are Promoted by Components of the Internal Limiting Membrane: Implications for Macular Hole Healing
Abstract
:1. Introduction
2. Results
2.1. Effect of ILM Components in Müller Cell Migration
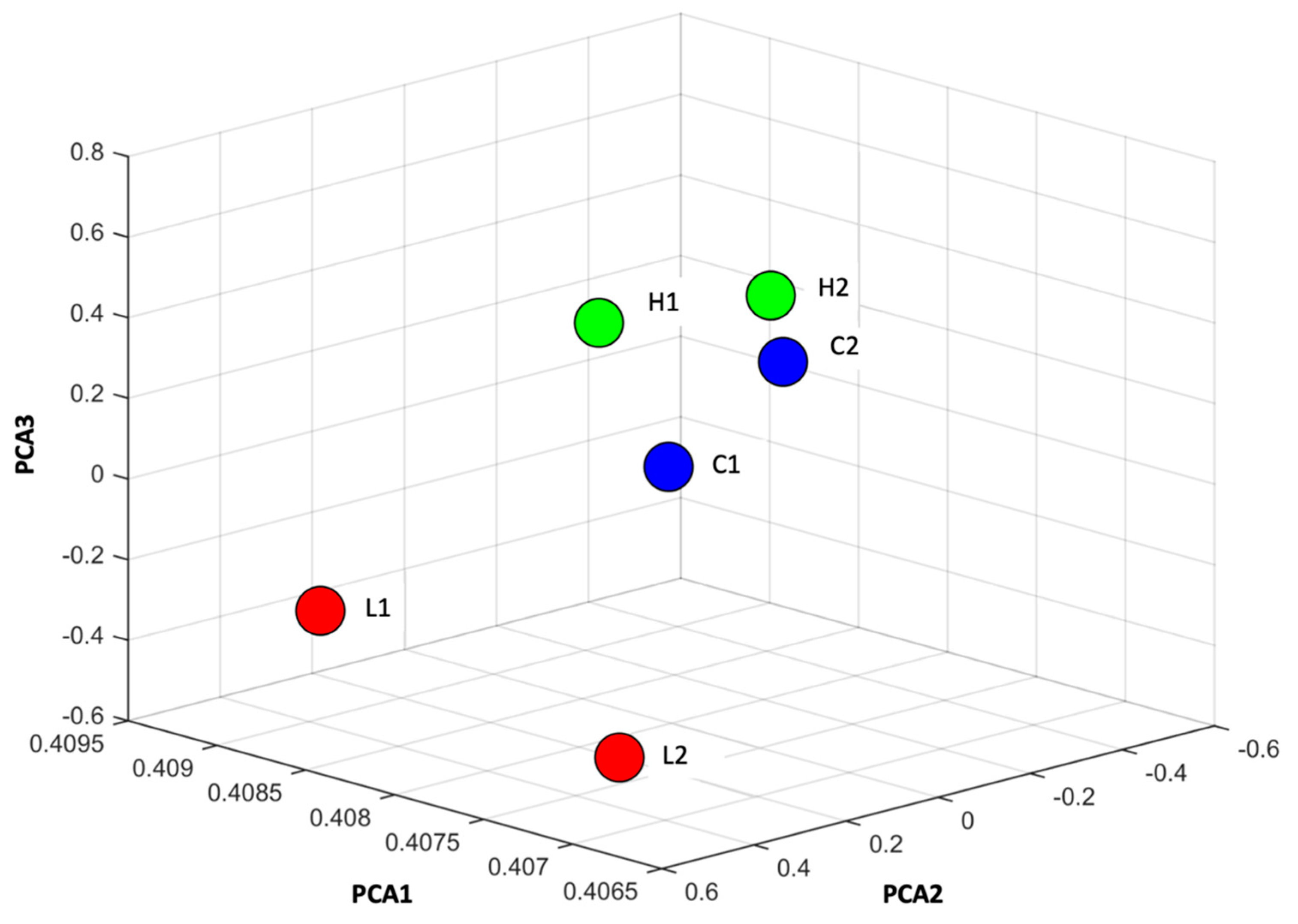
2.2. MiRNA Profiling of Müller Cell Stimulated by ILM Components
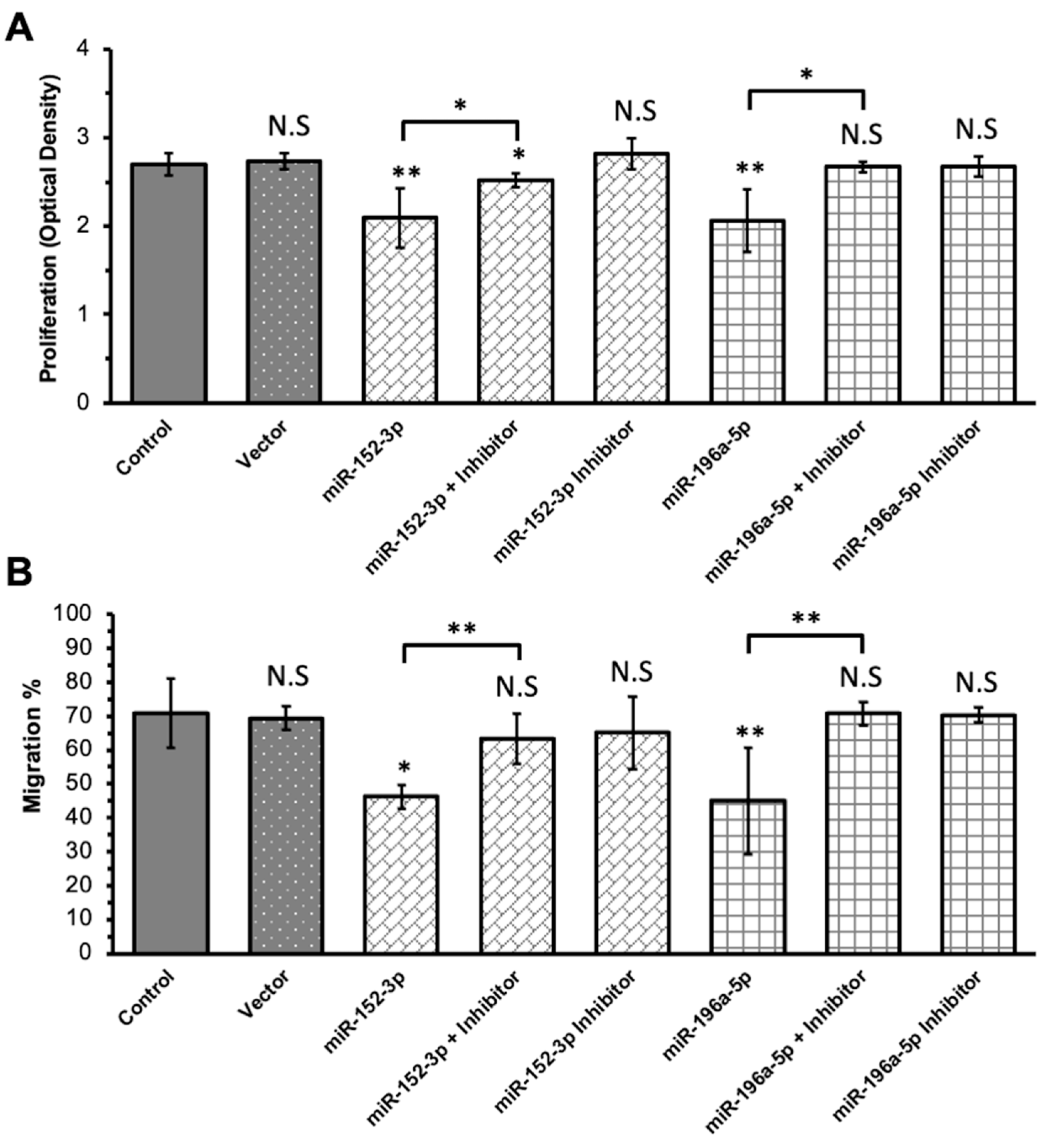
2.3. Transfection of MiRNA Mimics and Inhibitors
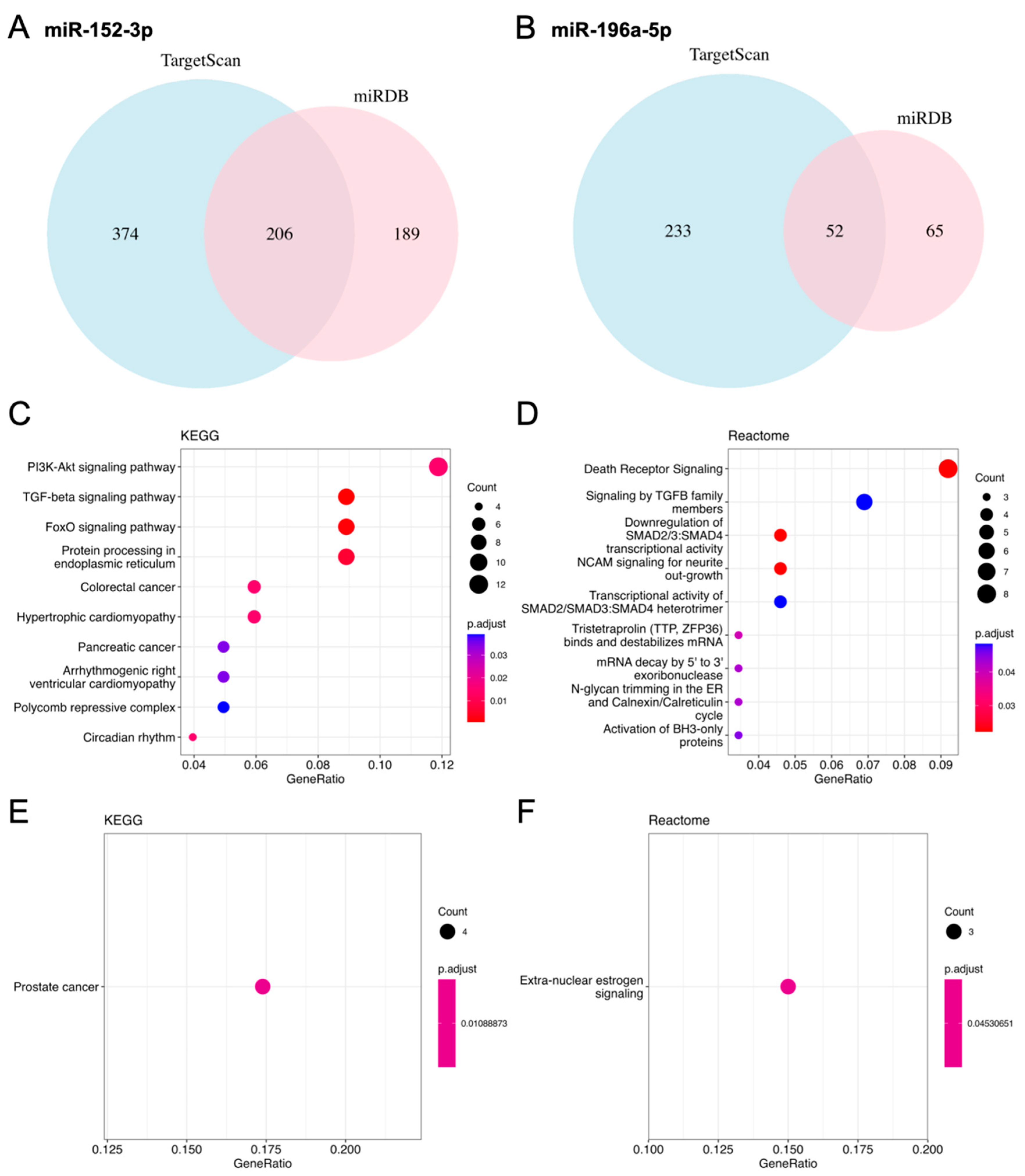
2.4. Prediction of Target Genes for Selected MiRNAs
3. Discussion
4. Materials and Methods
4.1. Cell Line
4.2. Cell Migration Assay
4.3. RNA Purification and MiRNA Profiling
4.4. Transfection of MiRNA Mimics and Inhibitors Followed by Migration Assay
4.5. Cell Proliferation Assay
4.6. Target Gene Prediction and Gene Ontology Enrichment Analysis
4.7. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gass, J.D. Idiopathic senile macular hole. Its early stages and pathogenesis. Arch. Ophthalmol. 1988, 106, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Hirata, Y.; Hayashi, A.; Fujikado, T.; Ohji, M.; Tano, Y. The visual performance and metamorphopsia of patients with macular holes. Arch. Ophthalmol. 2000, 118, 41–46. [Google Scholar] [CrossRef]
- Casuso, L.A.; Scott, I.U.; Flynn, H.W., Jr.; Gass, J.D.; Smiddy, W.E.; Lewis, M.L.; Schiffman, J. Long-term follow-up of unoperated macular holes. Ophthalmology 2001, 108, 1150–1155. [Google Scholar] [CrossRef]
- Bringmann, A.; Unterlauft, J.D.; Barth, T.; Wiedemann, R.; Rehak, M.; Wiedemann, P. Müller cells and astrocytes in tractional macular disorders. Prog. Retin. Eye Res. 2022, 86, 100977. [Google Scholar] [CrossRef]
- Bringmann, A.; Unterlauft, J.D.; Barth, T.; Wiedemann, R.; Rehak, M.; Wiedemann, P. Different modes of full-thickness macular hole formation. Exp. Eye Res. 2021, 202, 108393. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, A.; Pannicke, T.; Grosche, J.; Francke, M.; Wiedemann, P.; Skatchkov, S.N.; Osborne, N.N.; Reichenbach, A. Müller cells in the healthy and diseased retina. Prog. Retin. Eye Res. 2006, 25, 397–424. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, A.; Unterlauft, J.D.; Wiedemann, R.; Barth, T.; Rehak, M.; Wiedemann, P. Two different populations of Müller cells stabilize the structure of the fovea: An optical coherence tomography study. Int. Ophthalmol. 2020, 40, 2931–2948. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, S.; Tartaro, R.; Barca, F.; Caporossi, T.; Bacherini, D.; Giansanti, F. Internal Limiting Membrane Peeling Versus Inverted Flap Technique for Treatment of Full-Thickness Macular Holes: A Comparative Study in a Large Series of Patients. Retina 2018, 38 (Suppl. S1), S73–S78. [Google Scholar] [CrossRef]
- Yamashita, T.; Sakamoto, T.; Terasaki, H.; Iwasaki, M.; Ogushi, Y.; Okamoto, F.; Takeuchi, M.; Yasukawa, T.; Takamura, Y.; Ogata, N.; et al. Best surgical technique and outcomes for large macular holes: Retrospective multicentre study in Japan. Acta Ophthalmol. 2018, 96, e904–e910. [Google Scholar] [CrossRef]
- Baumann, C.; Kaye, S.; Iannetta, D.; Sultan, Z.; Dwivedi, R.; Pearce, I. Effect of Inverted Internal Limiting Membrane Flap on Closure Rate, Postoperative Visual Acuity, and Restoration of Outer Retinal Layers in Primary Idiopathic Macular Hole Surgery. Retina 2020, 40, 1955–1963. [Google Scholar] [CrossRef]
- Shiode, Y.; Morizane, Y.; Matoba, R.; Hirano, M.; Doi, S.; Toshima, S.; Takahashi, K.; Araki, R.; Kanzaki, Y.; Hosogi, M.; et al. The Role of Inverted Internal Limiting Membrane Flap in Macular Hole Closure. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4847–4855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.; Johnson, T.V. The internal limiting membrane: Roles in retinal development and implications for emerging ocular therapies. Exp. Eye Res. 2021, 206, 108545. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.L.; Liu, Y.T.; Chou, H.D.; Chuang, L.H.; Chen, K.J.; Chen, Y.P.; Liu, L.; Yeung, L.; Wang, N.K.; Hwang, Y.S.; et al. Role of growth factors and internal limiting membrane constituents in müller cell migration. Exp. Eye Res. 2021, 202, 108352. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, A.; Duncker, T.; Jochmann, C.; Barth, T.; Duncker, G.I.W.; Wiedemann, P. Spontaneous closure of small full-thickness macular holes: Presumed role of Müller cells. Acta Ophthalmol. 2020, 98, e447–e456. [Google Scholar] [CrossRef] [PubMed]
- Kitao, M.; Wakabayashi, T.; Nishida, K.; Sakaguchi, H.; Nishida, K. Long-term reconstruction of foveal microstructure and visual acuity after idiopathic macular hole repair: Three-year follow-up study. Br. J. Ophthalmol. 2019, 103, 238–244. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human diseases. J. Cell. Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Zhou, S.-S.; Jin, J.-P.; Wang, J.-Q.; Zhang, Z.-G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef]
- Roy, B.; Lee, E.; Li, T.; Rampersaud, M. Role of miRNAs in Neurodegeneration: From Disease Cause to Tools of Biomarker Discovery and Therapeutics. Genes 2022, 13, 425. [Google Scholar] [CrossRef]
- Kim, M.; Zhang, X. The Profiling and Role of miRNAs in Diabetes Mellitus. J. Diabetes Clin. Res. 2019, 1, 5–23. [Google Scholar]
- Wohl, S.G. View on microRNAs as a potential tool to fight blindness: Focus on Müller glia and gliosis. Neural Regen. Res. 2022, 17, 1501–1502. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.D.; Reibaldi, M.; Avitabile, T.; Bucolo, C.; Salomone, S.; Rejdak, R.; Nowomiejska, K.; Tripodi, S.; Posarelli, C.; Ragusa, M.; et al. MicroRNAs in the Vitreous Humor of Patients with Retinal Detachment and a Different Grading of Proliferative Vitreoretinopathy: A Pilot Study. Transl. Vis. Sci. Technol. 2020, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Ando, Y.; Keino, H.; Inoue, M.; Hirota, K.; Takahashi, H.; Sano, K.; Koto, T.; Sato, T.; Takeuchi, M.; Hirakata, A. Circulating Vitreous microRNA as Possible Biomarker in High Myopic Eyes with Macular Hole. Int. J. Mol. Sci. 2022, 23, 3647. [Google Scholar] [CrossRef] [PubMed]
- ElShelmani, H.; Wride, M.A.; Saad, T.; Rani, S.; Kelly, D.J.; Keegan, D. The Role of Deregulated MicroRNAs in Age-Related Macular Degeneration Pathology. Transl. Vis. Sci. Technol. 2021, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.; Peplow, P.V. MicroRNAs as biomarkers of diabetic retinopathy and disease progression. Neural Regen. Res. 2019, 14, 1858–1869. [Google Scholar] [PubMed]
- Wohl, S.G.; Jorstad, N.L.; Levine, E.M.; Reh, T.A. Müller glial microRNAs are required for the maintenance of glial homeostasis and retinal architecture. Nat. Commun. 2017, 8, 1603. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Bai, Z.; Wen, X.; Du, H.; Zhou, L.; Tang, Z.; Yang, Z.; Ma, W. MiR-152-3p regulates cell proliferation, invasion and extracellular matrix expression through by targeting FOXF1 in keloid fibroblasts. Life Sci. 2019, 234, 116779. [Google Scholar] [CrossRef] [PubMed]
- Desjarlais, M.; Rivera, J.C.; Lahaie, I.; Cagnone, G.; Wirt, M.; Omri, S.; Chemtob, S. MicroRNA expression profile in retina and choroid in oxygen-induced retinopathy model. PLoS ONE 2019, 14, e0218282. [Google Scholar] [CrossRef]
- Ménard, C.; Rezende, F.A.; Miloudi, K.; Wilson, A.; Tétreault, N.; Hardy, P.; SanGiovanni, J.P.; De Guire, V.; Sapieha, P. MicroRNA signatures in vitreous humour and plasma of patients with exudative AMD. Oncotarget 2016, 7, 19171–19184. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tang, Y.; Fan, Y.; Wu, D. MiR-196a-5p facilitates progression of estrogen-dependent endometrial cancer by regulating FOXO1. Histol. Histopathol. 2023, 38, 1157–1168. [Google Scholar] [PubMed]
- Guerriero, I.; D’Angelo, D.; Pallante, P.; Santos, M.; Scrima, M.; Malanga, D.; De Marco, C.; Ravo, M.; Weisz, A.; Laudanna, C.; et al. Analysis of miRNA profiles identified miR-196a as a crucial mediator of aberrant PI3K/AKT signaling in lung cancer cells. Oncotarget 2017, 8, 19172–19191. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Wang, C.; Chi, Y.; Liu, Z. MicroRNA-196b-5p promotes malignant progression of colorectal cancer by targeting ING5. Cancer Cell Int. 2020, 20, 119. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Qiu, O.; Huang, J.; Liu, J.; Wang, H. Upregulation of hsa-miR-196a-5p is associated with MIR196A2 methylation and affects the malignant biological behaviors of glioma. Genomics 2021, 113, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhou, T. Knockdown of lncRNA TUG1 protects lens epithelial cells from oxidative stress-induced injury by regulating miR-196a-5p expression in age-related cataracts. Exp. Ther. Med. 2021, 22, 1286. [Google Scholar] [CrossRef]
- Chou, H.D.; Liu, L.; Wang, C.T.; Chen, K.J.; Wu, W.C.; Hwang, Y.S.; Chen, Y.P.; Kang, E.Y.; Chen, Y.H.; Yeung, L.; et al. Single-Layer Inverted Internal Limiting Membrane Flap Versus Conventional Peel for Small- or Medium-Sized Full-Thickness Macular Holes. Am. J. Ophthalmol. 2022, 235, 111–119. [Google Scholar] [CrossRef]
- Bringmann, A.; Iandiev, I.; Pannicke, T.; Wurm, A.; Hollborn, M.; Wiedemann, P.; Osborne, N.N.; Reichenbach, A. Cellular signaling and factors involved in Müller cell gliosis: Neuroprotective and detrimental effects. Prog. Retin. Eye Res. 2009, 28, 423–451. [Google Scholar] [CrossRef]
- Eastlake, K.; Lamb, W.D.B.; Luis, J.; Khaw, P.T.; Jayaram, H.; Limb, G.A. Prospects for the application of Müller glia and their derivatives in retinal regenerative therapies. Prog. Retin. Eye Res. 2021, 85, 100970. [Google Scholar] [CrossRef]
- Mitra, S.; Devi, S.; Lee, M.S.; Jui, J.; Sahu, A.; Goldman, D. Vegf signaling between Müller glia and vascular endothelial cells is regulated by immune cells and stimulates retina regeneration. Proc. Natl. Acad. Sci. USA 2022, 119, e2211690119. [Google Scholar] [CrossRef]
- Tackenberg, M.A.; Tucker, B.A.; Swift, J.S.; Jiang, C.; Redenti, S.; Greenberg, K.P.; Flannery, J.G.; Reichenbach, A.; Young, M.J. Müller cell activation, proliferation and migration following laser injury. Mol. Vis. 2009, 15, 1886–1896. [Google Scholar]
- Peña, J.S.; Vazquez, M. Harnessing the Neuroprotective Behaviors of Müller Glia for Retinal Repair. Front. Biosci. (Landmark Ed.) 2022, 27, 169. [Google Scholar] [CrossRef]
- Rueda, E.M.; Hall, B.M.; Hill, M.C.; Swinton, P.G.; Tong, X.; Martin, J.F.; Poché, R.A. The Hippo Pathway Blocks Mammalian Retinal Müller Glial Cell Reprogramming. Cell Rep. 2019, 27, 1637–1649.e6. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Lin, X.; Wu, M. Nerve growth factor promotes the proliferation of Müller cells co-cultured with internal limiting membrane by regulating cell cycle via Trk-A/PI3K/Akt pathway. BMC Ophthalmol. 2019, 19, 130. [Google Scholar] [CrossRef] [PubMed]
- Morawski, K.; Kocemba-Pilarczyk, K.; Zarzycka, M.; Dudzik, P.; Trojan, S.E.; Laidler, P. In vitro culture Muller cell model to study the role of inverted internal limiting membrane flap technique in macular hole closure. J. Physiol. Pharmacol. 2021, 72. [Google Scholar]
- Li, Y.; Wang, X.; Wu, Q.; Liu, F.; Yang, L.; Gong, B.; Zhang, K.; Ma, Y.; Li, Y. miR-152-3p Represses the Proliferation of the Thymic Epithelial Cells by Targeting Smad2. Genes 2022, 13, 576. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Ma, R.; Tan, W.; Zhang, L. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp. Mol. Med. 2014, 46, e112. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, A.; Li, T.; Su, X.; Ding, H.; Li, H.; Qin, X.; Hou, L.; Zhao, Q.; Ge, X.; et al. MiR-152 reduces human umbilical vein endothelial cell proliferation and migration by targeting ADAM17. FEBS Lett. 2014, 588, 2063–2069. [Google Scholar] [CrossRef]
- Zhang, A.; Qian, Y.; Qian, J. MicroRNA-152-3p protects neurons from oxygen-glucose-deprivation/reoxygenation-induced injury through upregulation of Nrf2/ARE antioxidant signaling by targeting PSD-93. Biochem. Biophys. Res. Commun. 2019, 517, 69–76. [Google Scholar] [CrossRef]
- Ueki, Y.; Reh, T.A. EGF stimulates Müller glial proliferation via a BMP-dependent mechanism. Glia 2013, 61, 778–789. [Google Scholar] [CrossRef]
- Eastlake, K.; Banerjee, P.J.; Angbohang, A.; Charteris, D.G.; Khaw, P.T.; Limb, G.A. Müller glia as an important source of cytokines and inflammatory factors present in the gliotic retina during proliferative vitreoretinopathy. Glia 2016, 64, 495–506. [Google Scholar] [CrossRef]
- Nonaka, A.; Kiryu, J.; Tsujikawa, A.; Yamashiro, K.; Nishijima, K.; Kamizuru, H.; Ieki, Y.; Miyamoto, K.; Nishiwaki, H.; Honda, Y.; et al. Inflammatory Response after Scatter Laser Photocoagulation in Nonphotocoagulated Retina. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1204–1209. [Google Scholar]
- Allen, R.D.; Brown, J., Jr.; Zwick, H.; Schuschereba, S.T.; Lund, D.J.; Stuck, B.E. Laser-induced macular holes demonstrate impaired choroidal perfusion. Retina 2004, 24, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Yamana, T.; Kita, M.; Ozaki, S.; Negi, A.; Honda, Y. The process of closure of experimental retinal holes in rabbit eyes. Graefes Arch. Clin. Exp. Ophthalmol. 2000, 238, 81–87. [Google Scholar] [CrossRef]
- Pfeffer, B.A.; Xu, L.; Porter, N.A.; Rao, S.R.; Fliesler, S.J. Differential cytotoxic effects of 7-dehydrocholesterol-derived oxysterols on cultured retina-derived cells: Dependence on sterol structure, cell type, and density. Exp. Eye Res. 2016, 145, 297–316. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Gene Ontol. Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef] [PubMed]




| MicroRNA | Sequence |
|---|---|
| rno-miR-196a-5p | UAGGUAGUUUCAUGUUGUUGGG |
| rno-miR-152-3p | UCAGUGCAUGACAGAACUUGG |
| rno-miR-132-3p | UAACAGUCUACAGCCAUGGUCG |
| rno-miR-146b-5p | UGAGAACUGAAUUCCAUAGGCUGU |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chou, H.-D.; Shiah, S.-G.; Chuang, L.-H.; Wu, W.-C.; Hwang, Y.-S.; Chen, K.-J.; Kang, E.Y.-C.; Yeung, L.; Nien, C.-Y.; Lai, C.-C. MicroRNA-152-3p and MicroRNA-196a-5p Are Downregulated When Müller Cells Are Promoted by Components of the Internal Limiting Membrane: Implications for Macular Hole Healing. Int. J. Mol. Sci. 2023, 24, 17188. https://doi.org/10.3390/ijms242417188
Chou H-D, Shiah S-G, Chuang L-H, Wu W-C, Hwang Y-S, Chen K-J, Kang EY-C, Yeung L, Nien C-Y, Lai C-C. MicroRNA-152-3p and MicroRNA-196a-5p Are Downregulated When Müller Cells Are Promoted by Components of the Internal Limiting Membrane: Implications for Macular Hole Healing. International Journal of Molecular Sciences. 2023; 24(24):17188. https://doi.org/10.3390/ijms242417188
Chicago/Turabian StyleChou, Hung-Da, Shine-Gwo Shiah, Lan-Hsin Chuang, Wei-Chi Wu, Yih-Shiou Hwang, Kuan-Jen Chen, Eugene Yu-Chuan Kang, Ling Yeung, Chung-Yi Nien, and Chi-Chun Lai. 2023. "MicroRNA-152-3p and MicroRNA-196a-5p Are Downregulated When Müller Cells Are Promoted by Components of the Internal Limiting Membrane: Implications for Macular Hole Healing" International Journal of Molecular Sciences 24, no. 24: 17188. https://doi.org/10.3390/ijms242417188







