Mismatch Repair Protein Msh6Tt Is Necessary for Nuclear Division and Gametogenesis in Tetrahymena thermophila
Abstract
:1. Introduction
2. Results
2.1. Characterization of MSH6Tt in T. thermophila
2.2. Msh6Tt-3HA Localizes in the MIC and MAC during Vegetative Growth and Starvation
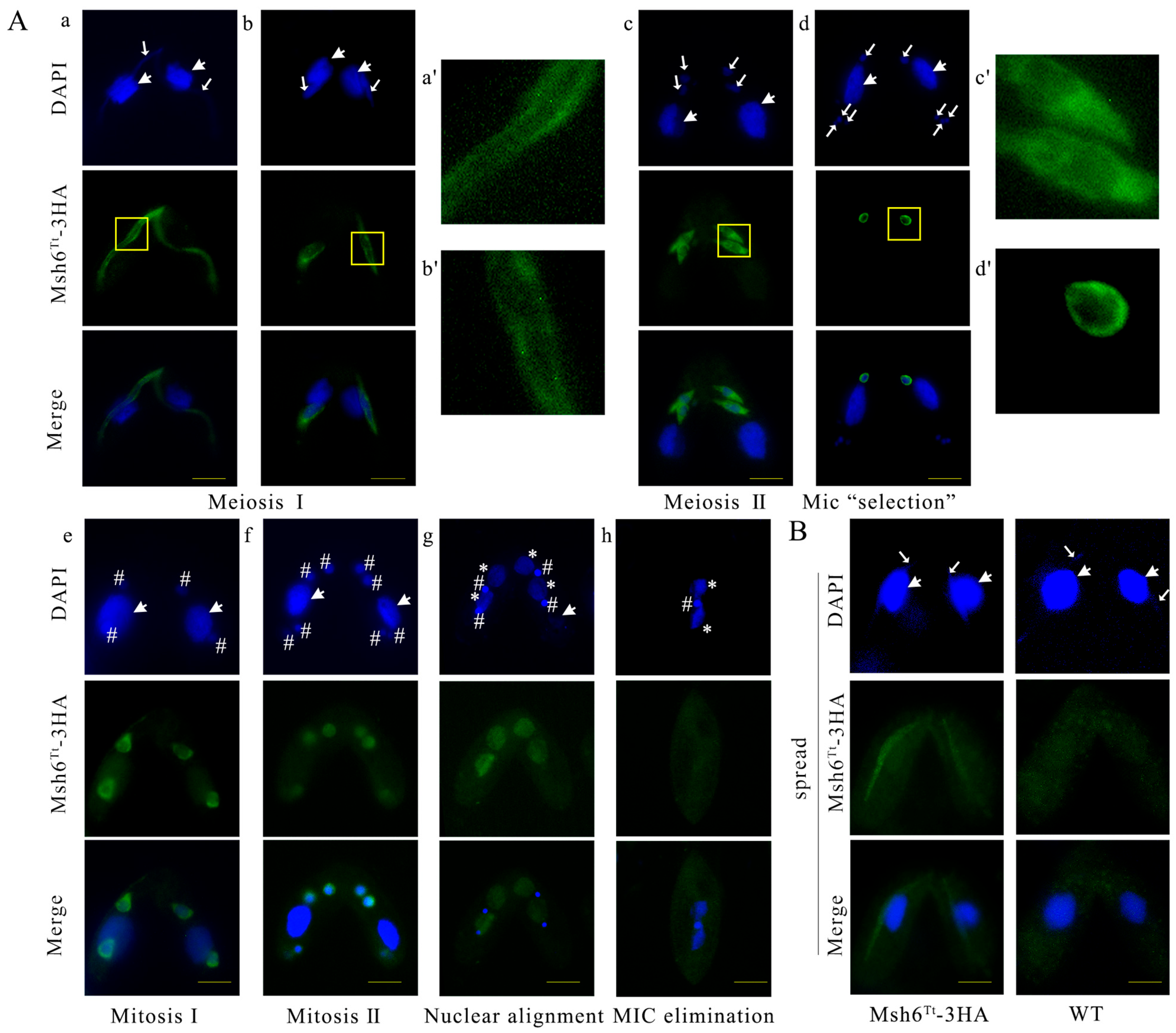
2.3. Msh6Tt-3HA Localizes in the MIC and New MAC during Conjugation
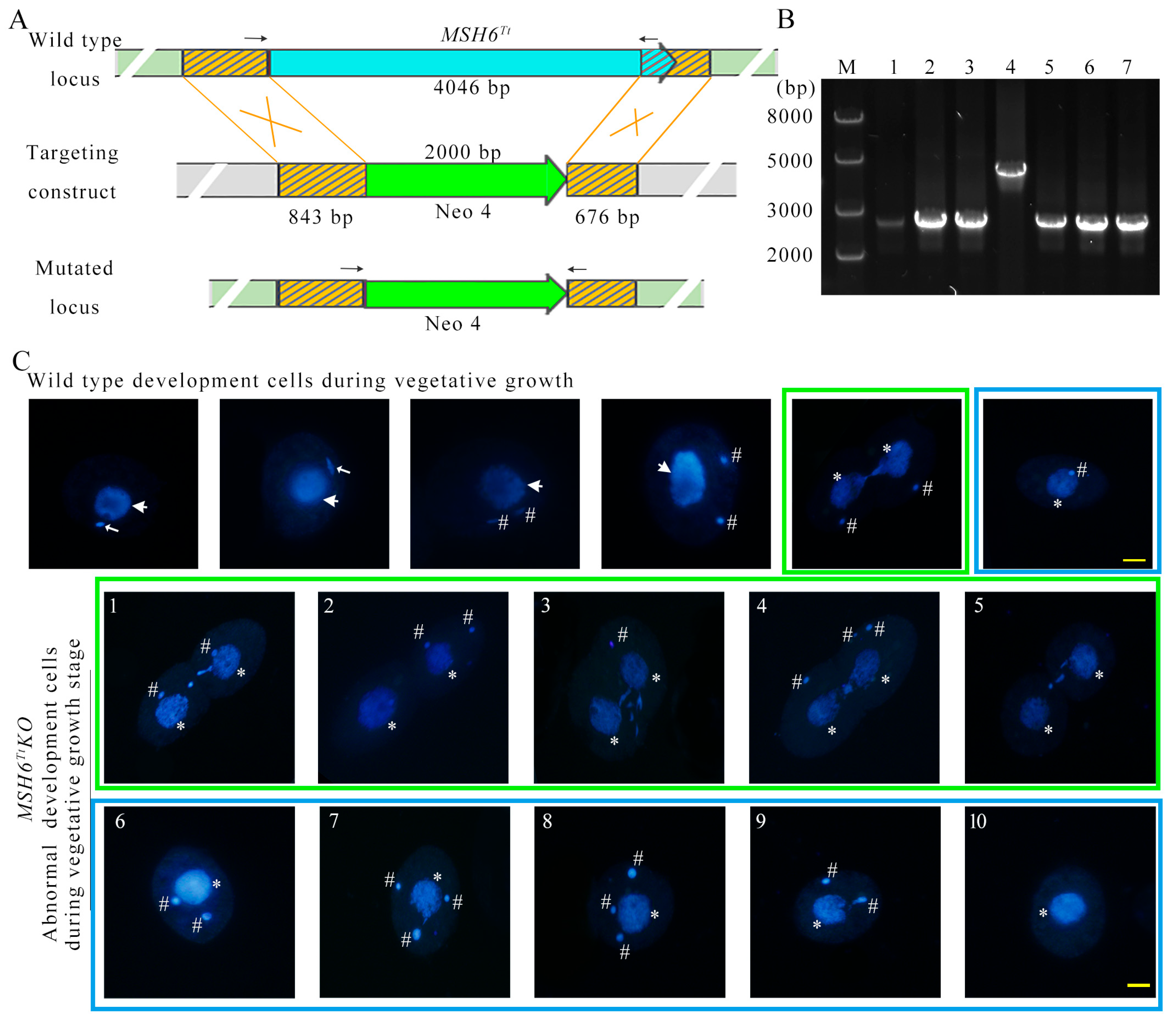
2.4. MSH6Tt Knockout Affects Nuclear Divisions during the Vegetative Growth Stage
2.5. MSH6Tt Knockout Hinders Micronuclear Meiosis and Gametic Selection during the Sexual Development Stage
2.6. Msh6Tt Interacts with Msh2Tt and MMR-Independent Factors
2.7. Msh2Tt Maintains the Stability of Msh6Tt
2.8. Expression of Redundant MSH6Tt Family Genes
3. Discussion
4. Materials and Methods
4.1. Cell Growth and Conjugation
4.2. Identification of Msh6Tt
4.3. Construction of Msh6Tt-3HA Mutants
4.4. Immunofluorescent Localization
4.5. Construction of MSH6Tt-Knockout Mutants
4.6. Synchronization of Cell Division
4.7. qRT-PCR Analysis
4.8. Nuclear Development
4.9. Coimmunoprecipitation and Mass Spectrometry (Co-IP-MS)
4.10. Protein–Protein Docking
4.11. Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boland, C.R.; Luciani, M.G.; Gasche, C.; Goel, A. Infection, inflammation, and gastrointestinal cancer. Gut 2005, 54, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, S.D.; Kunkel, T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008, 18, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Gruz, P.; Kamiya, H.; Kim, S.R.; Pisani, F.M.; Masutani, C.; Kanke, Y.; Harashima, H.; Hanaoka, F.; Nohmi, T. Erroneous incorporation of oxidized DNA precursors by Y-family DNA polymerases. EMBO Rep. 2003, 4, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A. Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009, 361, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Reyes, G.X.; Schmidt, T.T.; Kolodner, R.D.; Hombauer, H. New insights into the mechanism of DNA mismatch repair. Chromosoma 2015, 124, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.R.; Pluciennik, A.; Burdett, V.; Modrich, P.L. DNA mismatch repair: Functions and mechanisms. Chem. Rev. 2006, 106, 302–323. [Google Scholar] [CrossRef]
- Cooper, D.L.; Lahue, R.S.; Modrich, P. Methyl-directed mismatch repair is bidirectional. J. Biol. Chem. 1993, 268, 11823–11829. [Google Scholar] [CrossRef]
- Kolodner, R.D.; Marsischky, G.T. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 1999, 9, 89–96. [Google Scholar] [CrossRef]
- Obmolova, G.; Ban, C.; Hsieh, P.; Yang, W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 2000, 407, 703–710. [Google Scholar] [CrossRef]
- Lamers, M.H.; Perrakis, A.; Enzlin, J.H.; Winterwerp, H.H.; de Wind, N.; Sixma, T.K. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 2000, 407, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.H.; Winterwerp, H.H.; Sixma, T.K. The alternating ATPase domains of MutS control DNA mismatch repair. EMBO J. 2003, 22, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Genschel, J.; Littman, S.J.; Drummond, J.T.; Modrich, P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 1998, 273, 19895–19901. [Google Scholar] [CrossRef] [PubMed]
- Heinen, C.D.; Cyr, J.L.; Cook, C.; Punja, N.; Sakato, M.; Forties, R.A.; Lopez, J.M.; Hingorani, M.M.; Fishel, R. Human MSH2 (hMSH2) protein controls ATP processing by hMSH2-hMSH6. J. Biol. Chem. 2011, 286, 40287–40295. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A.; Erie, D.A. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu. Rev. Genet. 2015, 49, 291–313. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, S.D.; Gu, L.; Li, G.M. Bi-directional processing of DNA loops by mismatch repair-dependent and -independent pathways in human cells. J. Biol. Chem. 2003, 278, 3891–3896. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, M.L.; Smith, J.; Snowden, T.; Kim, M.; Fishel, R.; Poulos, B.K.; Cohen, P.E. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am. J. Hum. Genet. 2005, 76, 112–127. [Google Scholar] [CrossRef]
- Lam, I.; Keeney, S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 2014, 7, a016634. [Google Scholar] [CrossRef]
- Spies, M.; Fishel, R. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 2015, 7, a022657. [Google Scholar] [CrossRef]
- Datta, A.; Hendrix, M.; Lipsitch, M.; Jinks-Robertson, S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 9757–9762. [Google Scholar] [CrossRef]
- Stambuk, S.; Radman, M. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics 1998, 150, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Hum, Y.F.; Jinks-Robertson, S. Mismatch recognition and subsequent processing have distinct effects on mitotic recombination intermediates and outcomes in yeast. Nucleic Acids Res. 2019, 47, 4554–4568. [Google Scholar] [CrossRef] [PubMed]
- Snowden, T.; Acharya, S.; Butz, C.; Berardini, M.; Fishel, R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 2004, 15, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Fink, D.; Aebi, S.; Howell, S.B. The role of DNA mismatch repair in drug resistance. Clin. Cancer Res. 1998, 4, 1–6. [Google Scholar] [PubMed]
- Zhang, M.; Xiang, S.; Joo, H.Y.; Wang, L.; Williams, K.A.; Liu, W.; Hu, C.; Tong, D.; Haakenson, J.; Wang, C.; et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSα. Mol. Cell 2014, 55, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Gangavarapu, V.; Johnson, R.; Prakash, L.; Prakash, S. Mismatch repair operates at the replication fork in direct competition with mismatch extension by DNA polymerase delta. J. Biol. Chem. 2023, 299, 104598. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, U.; Shen, Z.J.; Tyler, J. Chaperoning histones at the DNA repair dance. DNA Repair 2021, 108, 103240. [Google Scholar] [CrossRef]
- Kadyrova, L.Y.; Blanko, E.R.; Kadyrov, F.A. CAF-I-dependent control of degradation of the discontinuous strands during mismatch repair. Proc. Natl. Acad. Sci. USA 2011, 108, 2753–2758. [Google Scholar] [CrossRef]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G.M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 2013, 153, 590–600. [Google Scholar] [CrossRef]
- Edelbrock, M.A.; Kaliyaperumal, S.; Williams, K.J. Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat. Res. 2013, 743–744, 53–66. [Google Scholar] [CrossRef]
- Kolodner, R.D. A personal historical view of DNA mismatch repair with an emphasis on eukaryotic DNA mismatch repair. DNA Repair 2016, 38, 3–13. [Google Scholar] [CrossRef]
- Orias, E.; Cervantes, M.D.; Hamilton, E.P. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res. Microbiol. 2011, 162, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Eisen, J.A.; Coyne, R.S.; Wu, M.; Wu, D.; Thiagarajan, M.; Wortman, J.R.; Badger, J.H.; Ren, Q.; Amedeo, P.; Jones, K.M.; et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006, 4, e286. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xue, Y.; Yang, S.; Bo, T.; Xu, J.; Wang, W. Mismatch Repair Protein Msh2 Is Necessary for Macronuclear Stability and Micronuclear Division in Tetrahymena thermophila. Int. J. Mol. Sci. 2023, 24, 10559. [Google Scholar] [CrossRef] [PubMed]
- Cole, E.; Sugai, T. Chapter 7—Developmental progression of Tetrahymena through the cell cycle and conjugation. In Methods in Cell Biology; Collins, K., Ed.; Academic Press: New York, NY, USA, 2012; Volume 109, pp. 177–236. [Google Scholar]
- Yao, N.Y.; O’Donnell, M. The RFC clamp loader: Structure and function. Subcell. Biochem. 2012, 62, 259–279. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Shimomura, T.; Hashimoto, K.; Araki, H.; Sugino, A.; Matsumoto, K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 7048–7052. [Google Scholar] [CrossRef] [PubMed]
- Barroso-Gonzalez, J.; Garcia-Exposito, L.; Galaviz, P.; Lynskey, M.L.; Allen, J.A.M.; Hoang, S.; Watkins, S.C.; Pickett, H.A.; O’Sullivan, R.J. Anti-recombination function of MutSalpha restricts telomere extension by ALT-associated homology-directed repair. Cell Rep. 2021, 37, 110088. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.P.; Sevi, L.A.; Feldt, M.C.; Rose, M.D.; Gammie, A.E. Reciprocal regulation of nuclear import of the yeast MutSalpha DNA mismatch repair proteins Msh2 and Msh6. DNA Repair 2009, 8, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Marsischky, G.T.; Filosi, N.; Kane, M.F.; Kolodner, R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996, 10, 407–420. [Google Scholar] [CrossRef]
- Brian, D.H.; Jinks-Robertson, S. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 2000, 34, 359–399. [Google Scholar] [CrossRef]
- Huang, Y.Q.; Yuan, Y.; Wang, Y.P.; Zhu, M.; Zhang, S.Z.; Zheng, S. Mutation detection of mismatch repair genes in hereditary nonpolyposis colorectal cancer by denaturing high-performance liquid chromatography. Zhonghua Wai Ke Za Zhi 2005, 43, 317–320. [Google Scholar] [PubMed]
- Chen, Y.; Liu, P.; Sun, P.; Jinag, J.; Zhu, Y.; Dong, T.; Cui, Y.; Tian, Y.; An, T.; Zhang, J.; et al. Oncogenic MSH6-CXCR4-TGFB1 Feedback Loop: A Novel Therapeutic Target of Photothermal Therapy in Glioblastoma Multiforme. Theranostics 2019, 9, 1453–1473. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.E.; Jackson, S.A.; Susswein, L.R.; Zeinomar, N.; Ma, X.; Marshall, M.L.; Stettner, A.R.; Milewski, B.; Xu, Z.; Solomon, B.D.; et al. MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet. Med. 2018, 20, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Wilczak, W.; Rashed, S.; Hube-Magg, C.; Kluth, M.; Simon, R.; Büscheck, F.; Clauditz, T.S.; Grupp, K.; Minner, S.; Tsourlakis, M.C.; et al. Up-regulation of mismatch repair genes MSH6, PMS2 and MLH1 parallels development of genetic instability and is linked to tumor aggressiveness and early PSA recurrence in prostate cancer. Carcinogenesis 2017, 38, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Cederquist, K.; Emanuelsson, M.; Göransson, I.; Holinski-Feder, E.; Müller-Koch, Y.; Golovleva, I.; Grönberg, H. Mutation analysis of the MLH1, MSH2 and MSH6 genes in patients with double primary cancers of the colorectum and the endometrium: A population-based study in northern Sweden. Int. J. Cancer 2004, 109, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.J.; Pohlhaus, T.J.; Changela, A.; Iyer, R.R.; Modrich, P.L.; Beese, L.S. Structure of the human MutSalpha DNA lesion recognition complex. Mol. Cell 2007, 26, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, X.; Feng, J.; Leng, H.; Li, S.; Xiao, J.; Liu, S.; Xu, Z.; Xu, J.; Li, D.; et al. The Histone Chaperone FACT Contributes to DNA Replication-Coupled Nucleosome Assembly. Cell Rep. 2016, 14, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Midic, U.; Xie, H.; Xue, B.; Vucetic, S.; Iakoucheva, L.M.; Obradovic, Z.; Dunker, A.K. Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genom. 2009, 10 (Suppl. S1), S7. [Google Scholar] [CrossRef]
- Bowman, G.D.; O’Donnell, M.; Kuriyan, J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 2004, 429, 724–730. [Google Scholar] [CrossRef]
- Tsurimoto, T. PCNA binding proteins. Front. Biosci.-Landmark 1999, 4, D849–D858. [Google Scholar] [CrossRef]
- Olaisen, C.; Kvitvang, H.F.N.; Lee, S.; Almaas, E.; Bruheim, P.; Drabløs, F.; Otterlei, M. The role of PCNA as a scaffold protein in cellular signaling is functionally conserved between yeast and humans. FEBS Open Bio 2018, 8, 1135–1145. [Google Scholar] [CrossRef]
- Gassman, N.R.; Clodfelter, J.E.; McCauley, A.K.; Bonin, K.; Salsbury, F.R., Jr.; Scarpinato, K.D. Cooperative nuclear localization sequences lend a novel role to the N-terminal region of MSH6. PLoS ONE 2011, 6, e17907. [Google Scholar] [CrossRef] [PubMed]
- Hombauer, H.; Srivatsan, A.; Putnam, C.D.; Kolodner, R.D. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science 2011, 334, 1713–1716. [Google Scholar] [CrossRef] [PubMed]
- Sirbu, B.M.; McDonald, W.H.; Dungrawala, H.; Badu-Nkansah, A.; Kavanaugh, G.M.; Chen, Y.; Tabb, D.L.; Cortez, D. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem. 2013, 288, 31458–31467. [Google Scholar] [CrossRef] [PubMed]
- Lundin, C.; North, M.; Erixon, K.; Walters, K.; Jenssen, D.; Goldman, A.S.; Helleday, T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005, 33, 3799–3811. [Google Scholar] [CrossRef] [PubMed]
- Doerder, F.P.; Debault, L.E. Cytofluorimetric analysis of nuclear DNA during meiosis, fertilization and macronuclear development in the ciliate Tetrahymena pyriformis, syngen 1. J. Cell Sci. 1975, 17, 471–493. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.K.; Park, D.; Xu, L.; Kleckner, N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 1992, 69, 439–456. [Google Scholar] [CrossRef] [PubMed]
- Pittman, D.L.; Cobb, J.; Schimenti, K.J.; Wilson, L.A.; Cooper, D.M.; Brignull, E.; Handel, M.A.; Schimenti, J.C. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1998, 1, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Howard-Till, R.A.; Lukaszewicz, A.; Loidl, J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genet. 2011, 7, e1001359. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Erie, D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005, 74, 681–710. [Google Scholar] [CrossRef]
- Gorovsky, M.A.; Yao, M.C.; Keevert, J.B.; Pleger, G.L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975, 9, 311–327. [Google Scholar] [CrossRef] [PubMed]
- Bruns, P.J.; Brussard, T.B. Pair formation in Tetrahymena pyriformis, an inducible developmental system. J. Exp. Zool. 1974, 188, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, A.K.; Stefans, M.; Christopher, M.Y.; Mark, N.W.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar]
- Xu, J.; Li, X.; Song, W.; Wang, W.; Gao, S. Cyclin Cyc2p is required for micronuclear bouquet formation in Tetrahymena thermophila. Sci China Life Sci. 2019, 62, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Holz, G.G.; Scherbaum, O.H.; Williams, N. The arrest of mitosis and stomatogenesis during temperature-induction of synchronous division in Tetrahymena pyriformis, mating type 1, variety 1. Exp. Cell Res. 1957, 13, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef] [PubMed]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef]
- Nabeel-Shah, S.; Garg, J.; Saettone, A.; Ashraf, K.; Lee, H.; Wahab, S.; Ahmed, N.; Fine, J.; Derynck, J.; Pu, S.; et al. Functional characterization of RebL1 highlights the evolutionary conservation of oncogenic activities of the RBBP4/7 orthologue in Tetrahymena thermophila. Nucleic Acids Res. 2021, 49, 6196–6212. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- González-Arzola, K.; Díaz-Quintana, A.; Rivero-Rodríguez, F.; Velázquez-Campoy, A.; De la Rosa, M.A.; Díaz-Moreno, I. Histone chaperone activity of Arabidopsis thaliana NRP1 is blocked by cytochrome c. Nucleic Acids Res. 2017, 45, 2150–2165. [Google Scholar] [CrossRef]





| Gene Model Identifier | Gene Name | GO_Term |
|---|---|---|
| TTHERM_00372688 | - | hypothetical protein |
| TTHERM_00268040 | NUP185 | |
| TTHERM_00112690 | - | |
| TTHERM_00047280 | - | |
| TTHERM_00941540 | RAB11B (RAB GTPase 11B) | GTPase activity |
| TTHERM_00079520 | GTU1 (Gamma-TUbulin 1) | Microtubule nucleation |
| TTHERM_01289180 | - | Integral component of membrane |
| TTHERM_00355660 | - | |
| TTHERM_000079948 | - | |
| TTHERM_01099050 | RRM53 (RNA recognition motif-containing protein 53) | RNA binding |
| TTHERM_01076960 | - | mRNA processing |
| TTHERM_00085290 | - | Oxidoreductase activity |
| TTHERM_00194810 | MSH6 | Mismatch repair |
| TTHERM_00295920 | MSH2 | |
| TTHERM_00418100 | - | P28 protein |
| TTHERM_00357080 | - | Superoxide dismutase |
| Gene Model Identifier and Standard Name | GO_Term |
|---|---|
| TTHERM_00295920_MSH2 | Mismatch repair |
| TTHERM_00194810_MSH6 | |
| TTHERM_00772030 | Protein modification and processing |
| TTHERM_00043890 | |
| TTHERM_00622710 | Oxidation-reduction process |
| TTHERM_00151470 | |
| TTHERM_00387080 | Cation transport/metabolic process |
| TTHERM_00245100_TPA1 | |
| TTHERM_01276420_DYH3 | Microtubule-based movement |
| TTHERM_00849320 | Small GTPase mediated signal transduction |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Yang, S.; Xue, Y.; Bo, T.; Xu, J.; Wang, W. Mismatch Repair Protein Msh6Tt Is Necessary for Nuclear Division and Gametogenesis in Tetrahymena thermophila. Int. J. Mol. Sci. 2023, 24, 17619. https://doi.org/10.3390/ijms242417619
Wang L, Yang S, Xue Y, Bo T, Xu J, Wang W. Mismatch Repair Protein Msh6Tt Is Necessary for Nuclear Division and Gametogenesis in Tetrahymena thermophila. International Journal of Molecular Sciences. 2023; 24(24):17619. https://doi.org/10.3390/ijms242417619
Chicago/Turabian StyleWang, Lin, Sitong Yang, Yuhuan Xue, Tao Bo, Jing Xu, and Wei Wang. 2023. "Mismatch Repair Protein Msh6Tt Is Necessary for Nuclear Division and Gametogenesis in Tetrahymena thermophila" International Journal of Molecular Sciences 24, no. 24: 17619. https://doi.org/10.3390/ijms242417619





