Transcriptome and Metabolome Analysis Reveals Salt-Tolerance Pathways in the Leaves and Roots of ZM-4 (Malus zumi) in the Early Stages of Salt Stress
Abstract
:1. Introduction
2. Results
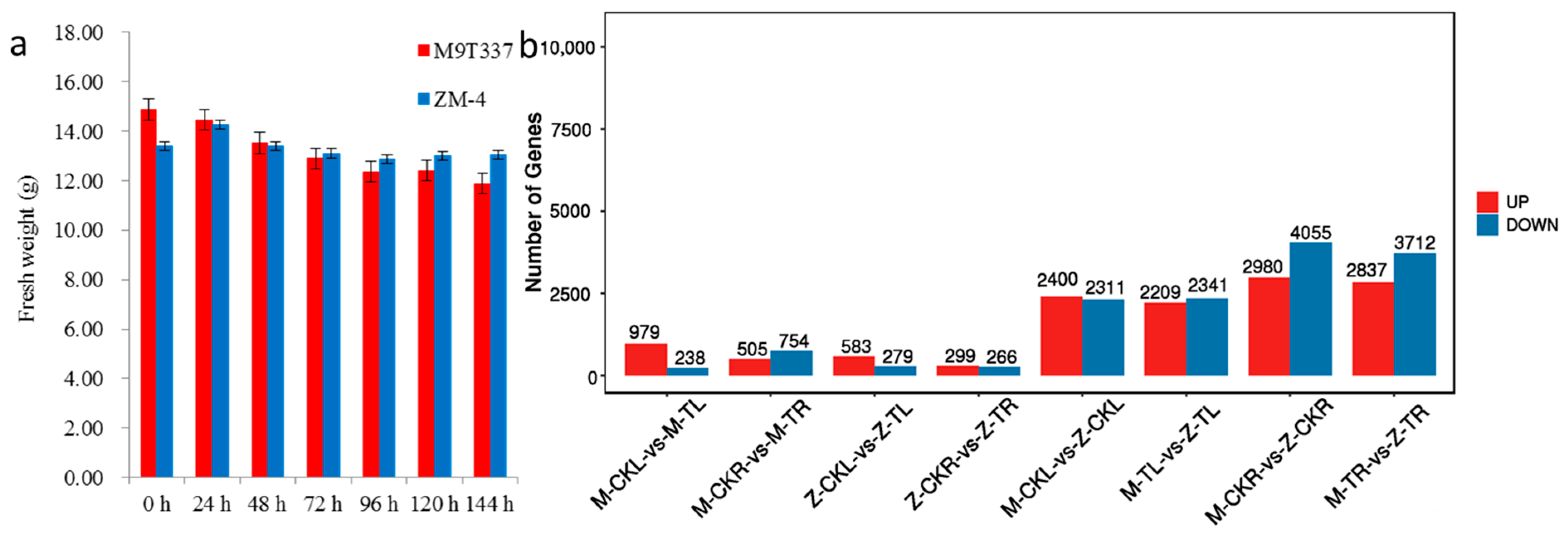
2.1. Comparison of Salt Tolerance between ZM-4 and M9T337 in Response to Salt Stress
2.2. Comparison of Transcriptional Profiling between ZM-4 and M9T337 in Response to Salt Stress
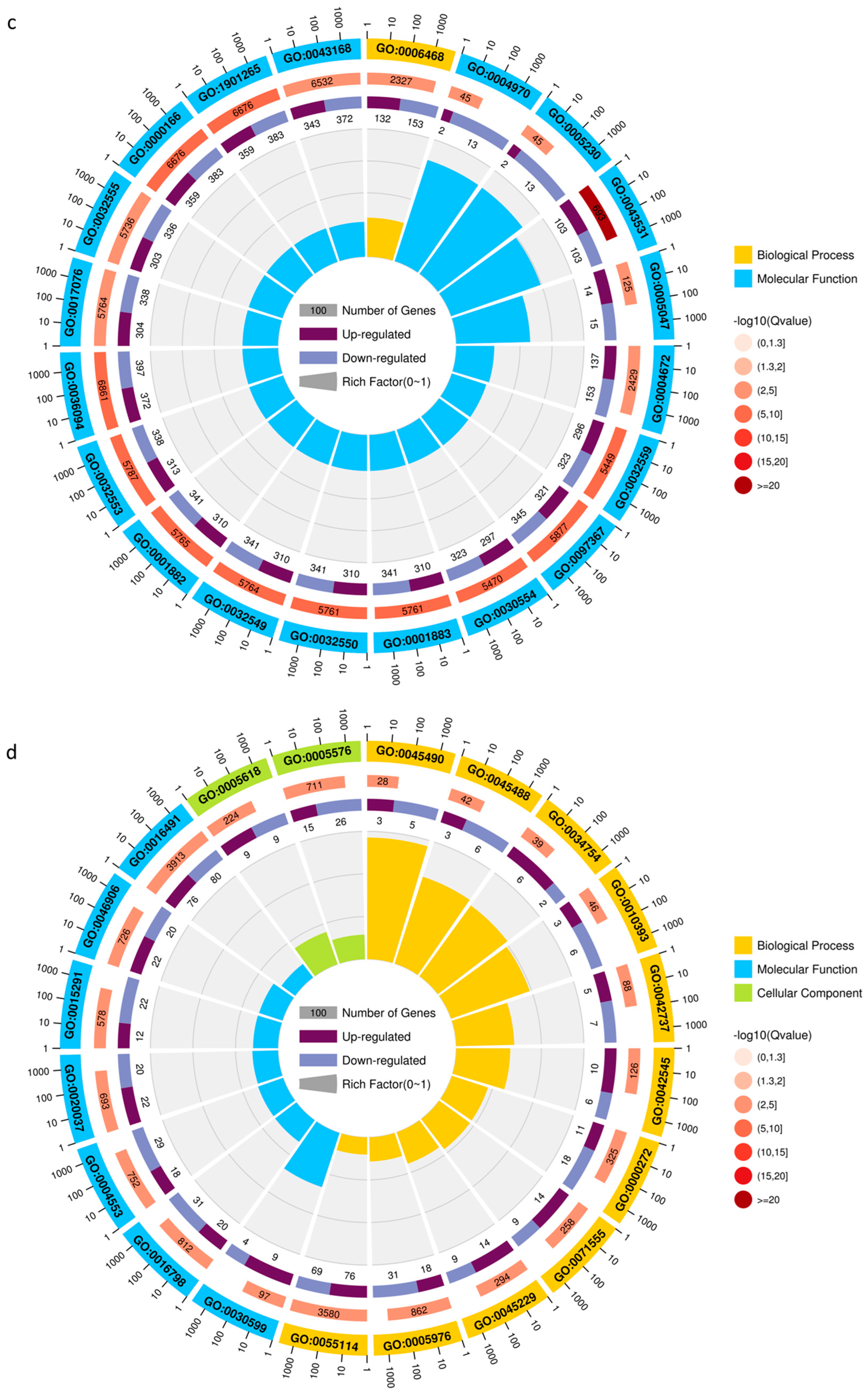
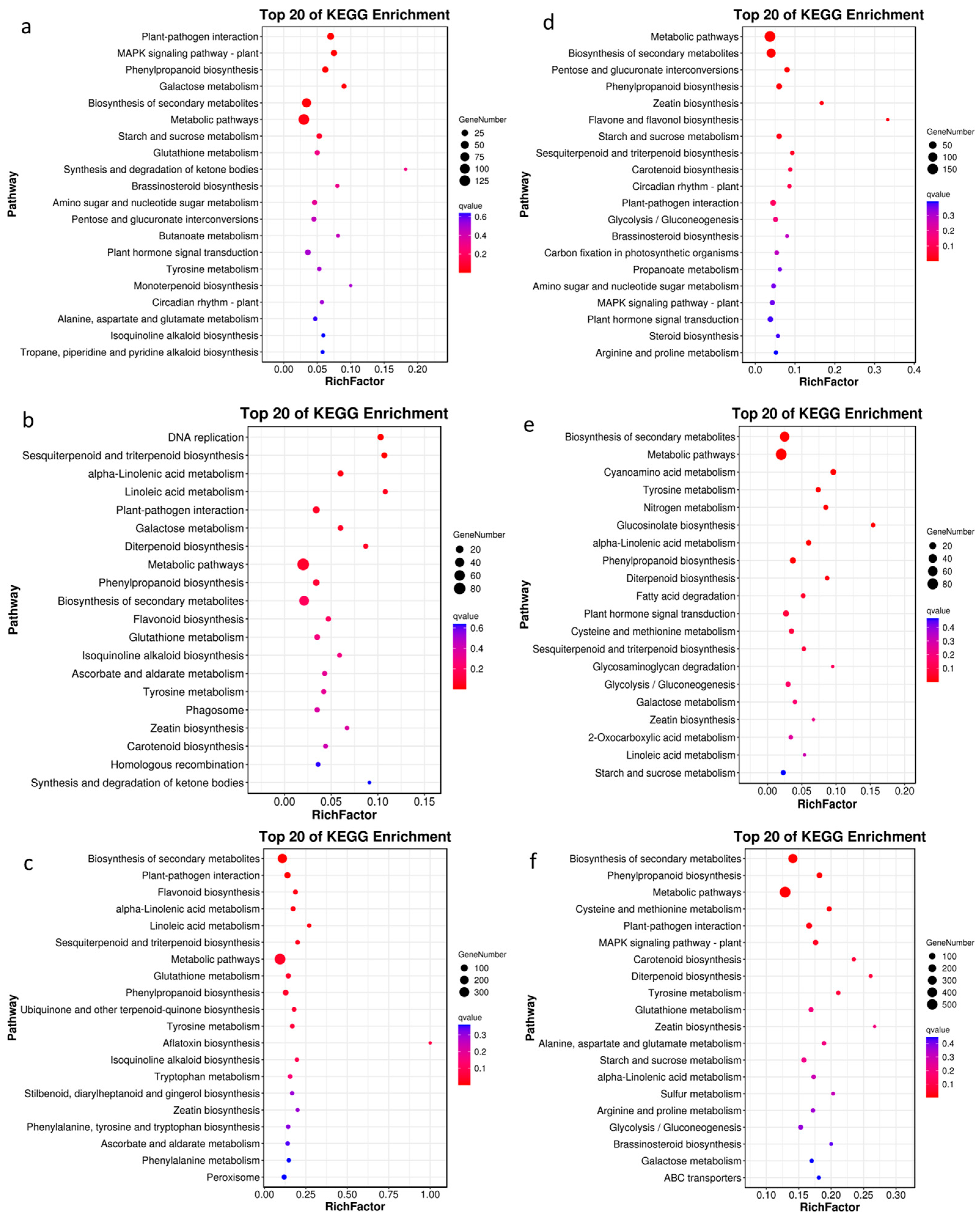
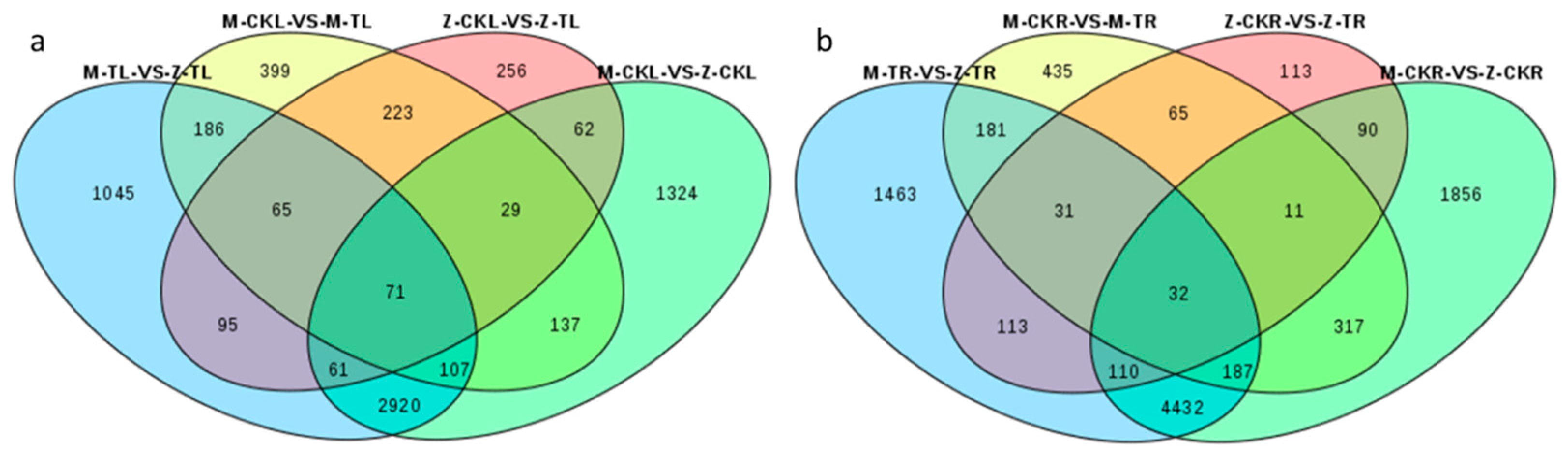
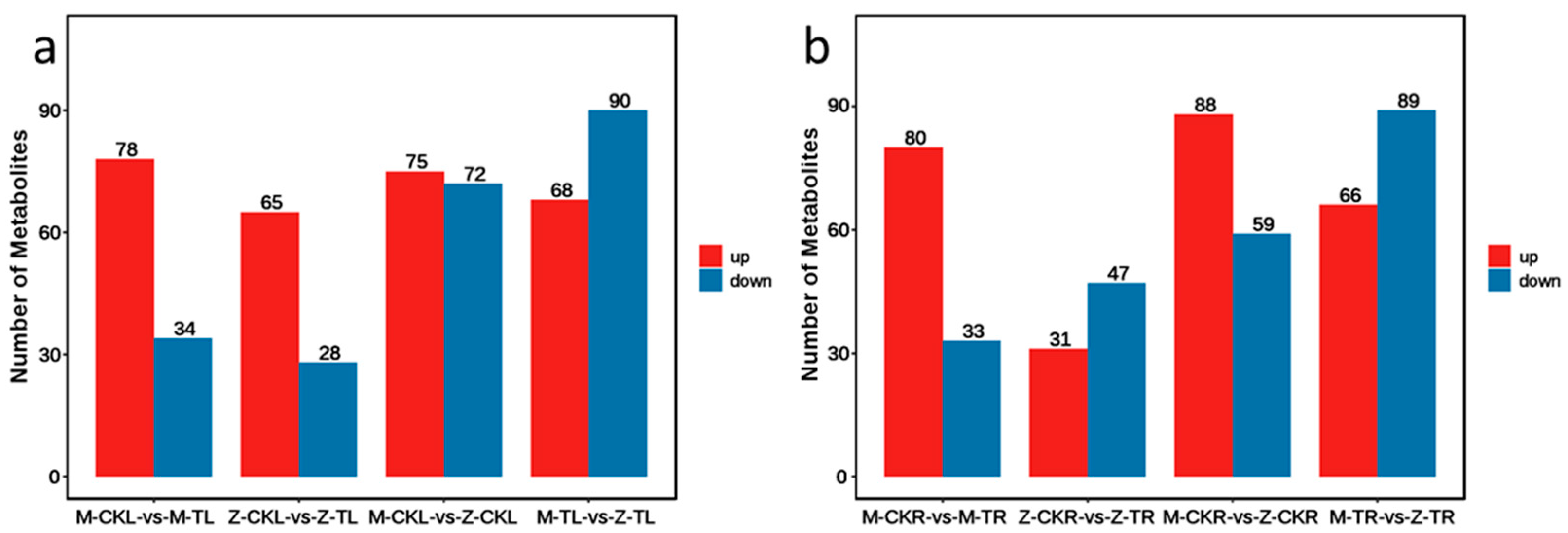
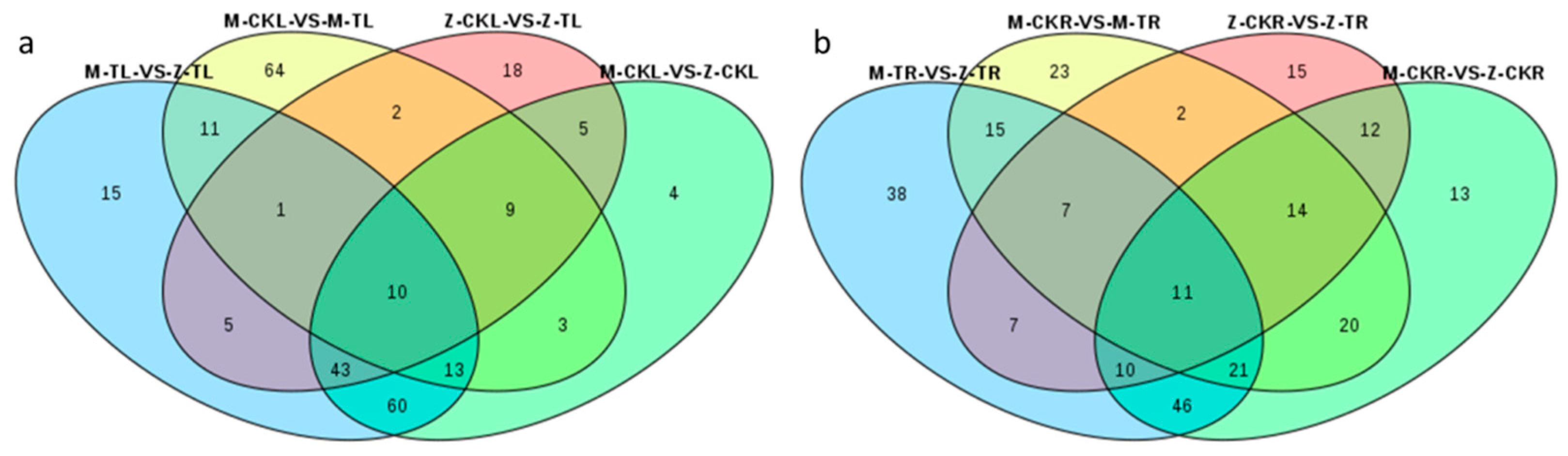
2.2.1. DEG Analysis in Leaves in Response to Salt Stress for ZM-4 and M9T337
2.2.2. DEG Analysis of Roots in Response to Salt Stress for ZM-4 and M9T337
2.3. Comparison of Metabolic Profiling between ZM-4 and M9T337 in Response to Salt Stress
2.3.1. DAM Analysis in Leaves in Response to Salt Stress for ZM-4 and M9T337
2.3.2. DAM Analysis in Roots in Response to Salt Stress for ZM-4 and M9T337
2.4. Integrated Analysis of the Transcriptome and Metabolome of ZM-4 and M9T337 Responsive to Salt Stress
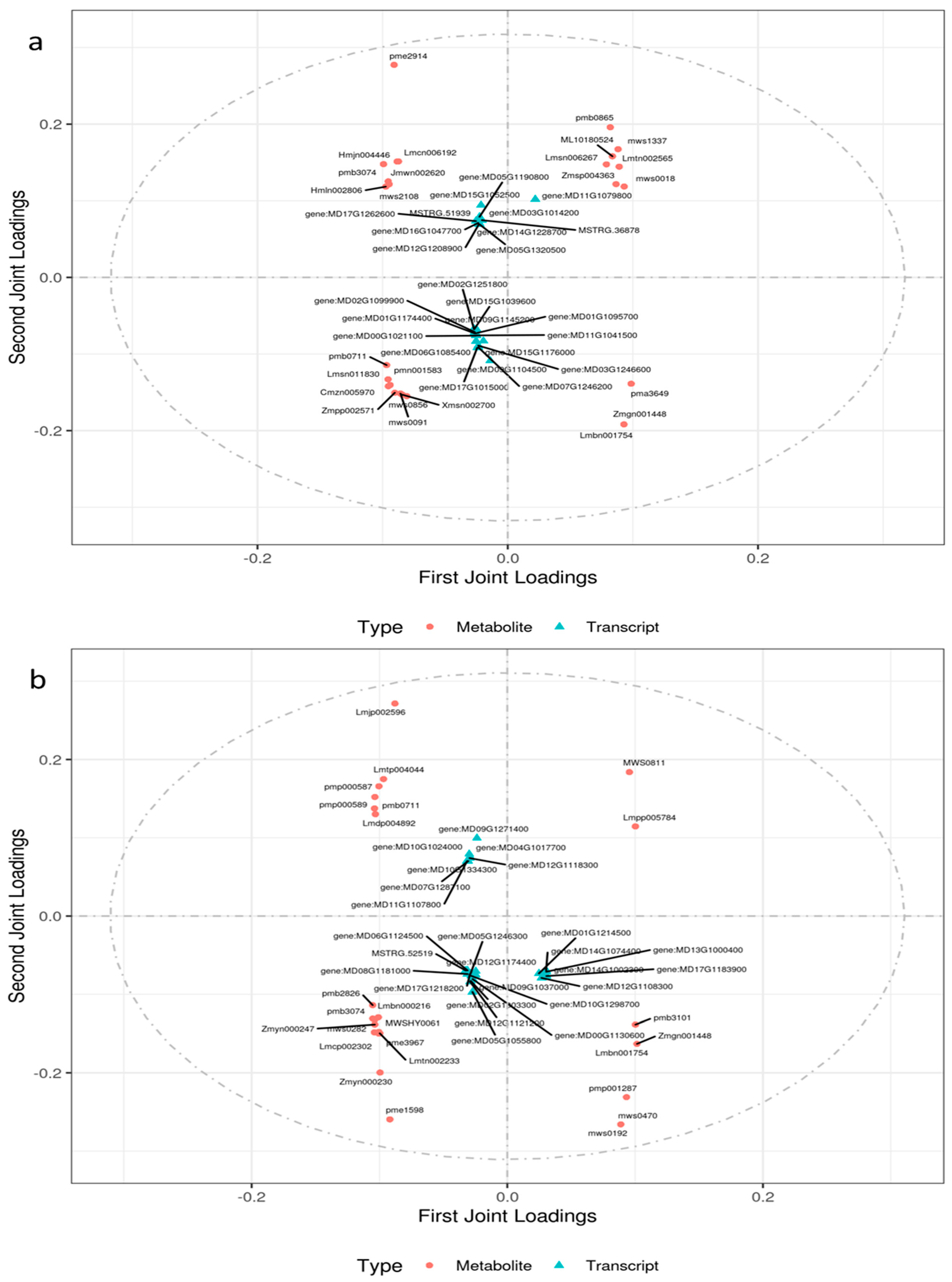
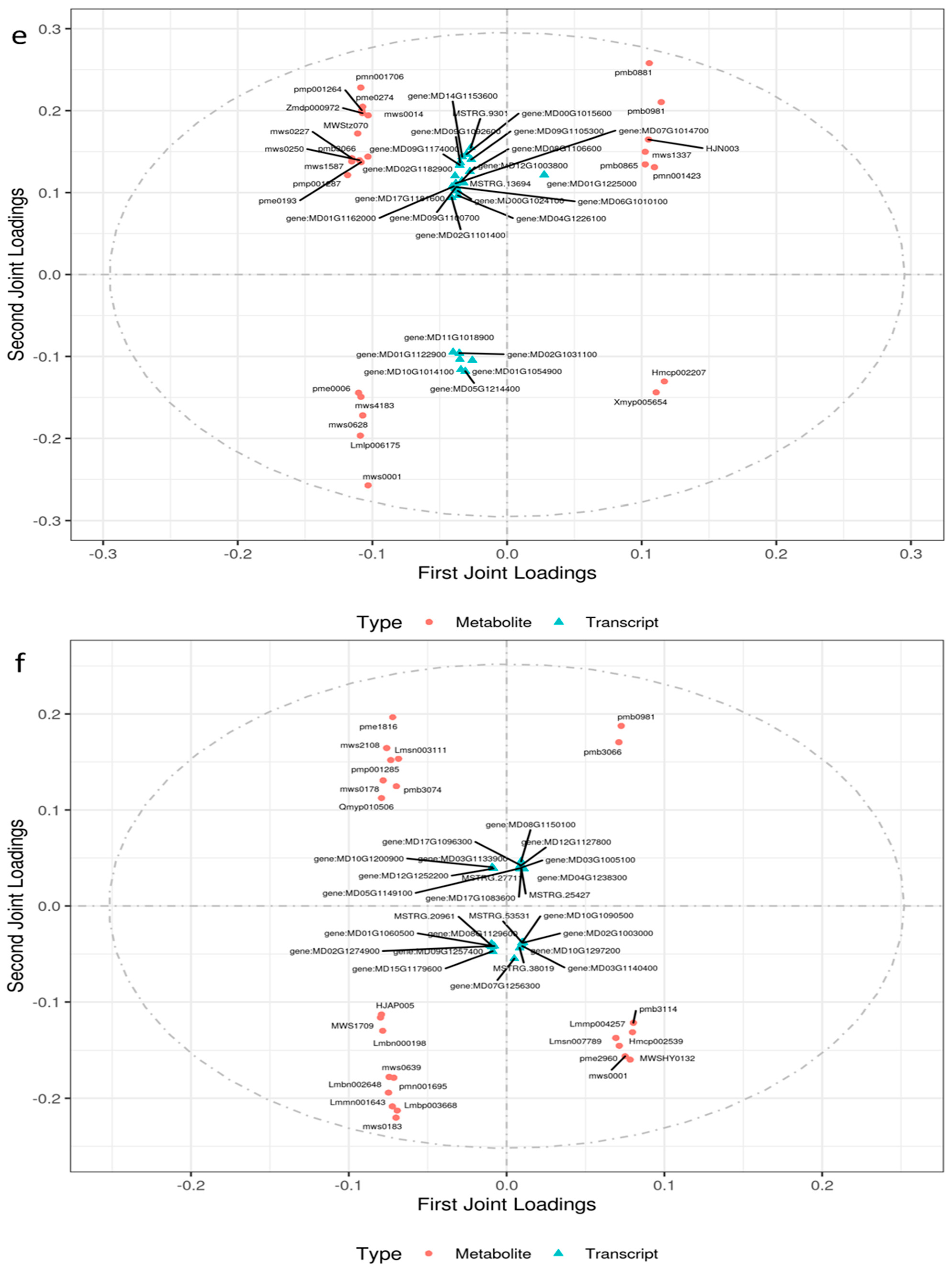
2.4.1. Integrated Analysis of the Transcriptome and Metabolome of Leaves Responsive to Salt Stress
2.4.2. Integrated Analysis of the Transcriptome and Metabolome of Roots Responsive to Salt Stress
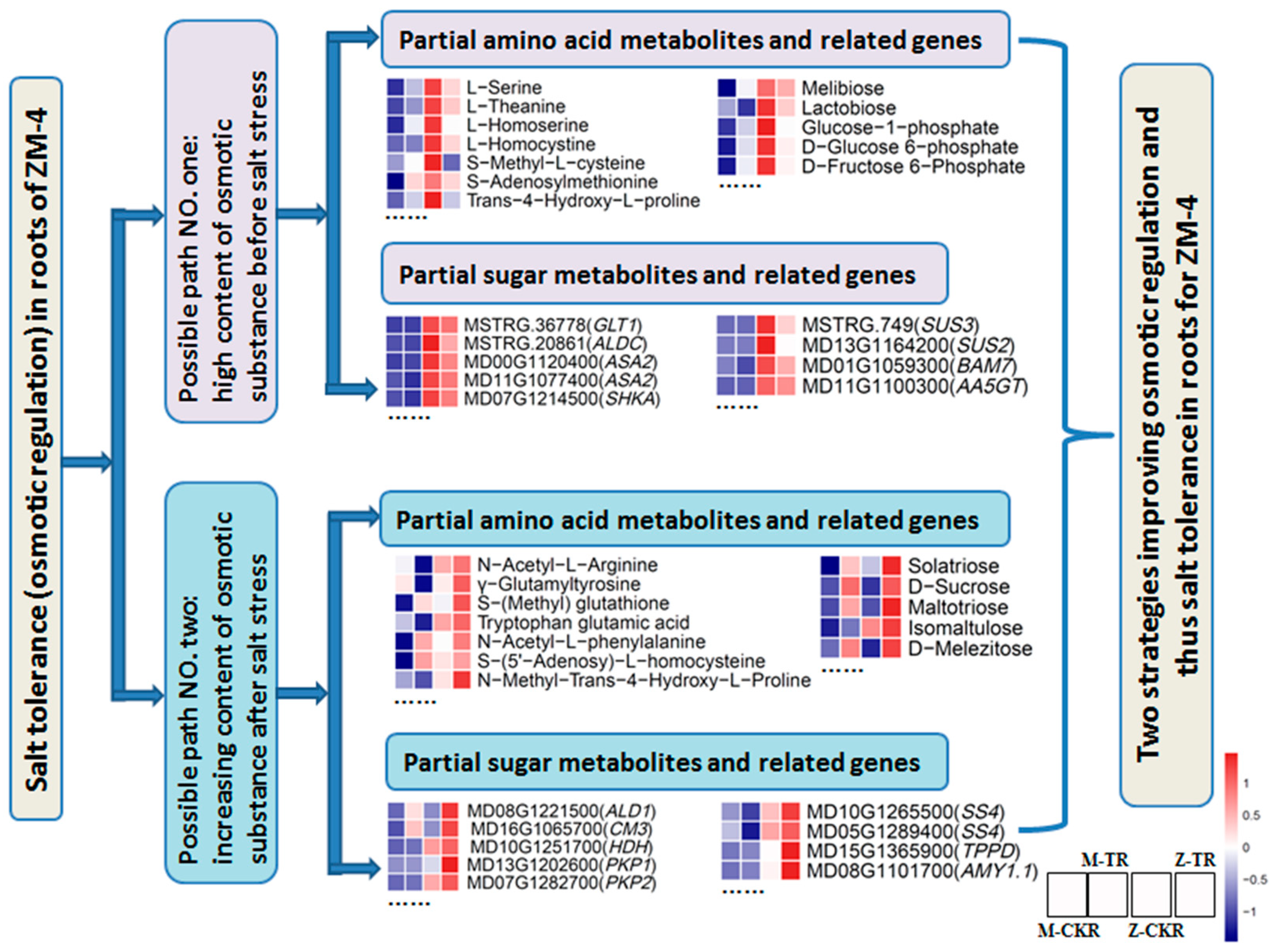
2.5. The Salt Tolerance Mechanisms Predicted in Leaves and Roots for ZM-4
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Phenotype of the Samples
4.3. Transcriptome Analysis
4.3.1. RNA Quantification and Qualification
4.3.2. Filtering of Clean Reads
4.3.3. Novel Transcript Identification and Annotation
4.3.4. Quantification of Transcript Abundance
4.3.5. Differentially Expressed Transcript (DEG) Analysis
4.4. Metabolic Analysis
4.4.1. Chemicals and Reagents
4.4.2. Sample Preparation and Extraction
4.4.3. AB Sciex QTRAP4500 (UPLC) Analysis
4.4.4. Data Pre-Processing and Metabolite Identification
4.4.5. Multivariate Statistical Analysis
4.4.6. Differential Metabolite Analysis
4.4.7. KEGG Pathway Analysis
4.5. Joint Analysis of Transcriptome and Metabolome
4.5.1. Pathway Model
4.5.2. O2PLS Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamil, A.; Riza, S.; Ashraf, M.; Foolad, M.R. Gene expression profiling of plants under salt stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Du, Z.J.; Zhai, H.; Luo, X.S.; Cheng, S.H.; Pan, Z.Y. Field identification of salt tolerance of apple rootstocks. China Fruits 2001, 2, 1–4. [Google Scholar]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Dai, W.S.; Wang, M.; Gong, X.Q.; Liu, J.H. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef]
- Sodani, R.; Mundiyara, R. Salinity stress: Its impact on plant growth and development. Agric. Food E-Newsl. 2021, 6, 535–537. [Google Scholar]
- Soltabayeva, A.; Ongaltay, A.; Omondi, J.O.; Srivastava, S. Morphological, physiological and molecular markers for salt-stressed plants. Plants 2021, 10, 243. [Google Scholar] [CrossRef]
- Richard, P.M.; Gennaro, F. Apple rootstocks: History, physiology, management, and breeding. Hortic. Rev. 2018, 45, 197–312. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savoure, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef]
- Wei, T.L.; Wang, Y.; Liu, J.H. Comparative transcriptome analysis reveals synergistic and disparate defense pathways in the leaves and roots of trifoliate orange (Poncirus trifoliata) autotetraploids with enhanced salt tolerance. Hortic. Res. 2020, 7, 88. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.C.; Zhao, X.; Xiao, Z.; Yin, X.H.; Xing, T.; Xia, G.M. A wheat superoxide dismutase gene TaSOD2 enhances salt resistance through modulating redox homeostasis by promoting NADPH oxidase activity. Plant Mol. Biol. 2016, 91, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000, 124, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 2001, 4, 401–406. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thalinan salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef]
- Pottosin, I.; Dobrovinskaya, O. Major vacuolar TPC1 channel in stress signaling: What matters, K+, Ca+ conductance or an ion-flux independent mechanism? Stress Biol. 2022, 2, 31. [Google Scholar] [CrossRef]
- Gong, Z.Z.; Xiong, L.M.; Shi, H.Z.; Yang, S.H.; Herrera-Estrella, L.R.; Xu, G.H.; Chao, D.Y.; Li, J.R.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 40. [Google Scholar] [CrossRef]
- Koprivova, A.; North, K.A.; Kopriva, S. Complex signaling network in regulation of adenosine 5’-phosphosulfate reducatase by salt stress in Arabidopsis roots. Plant Physiol. 2008, 146, 1408–1420. [Google Scholar] [CrossRef]
- Rus, A.; Baxter, I.; Muthukumar, B.; Gustin, J.; Lahner, B.; Yakubova, E.; Salt, D.E. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PloS Genet. 2006, 2, e210. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, X.W.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple MdERF4 negatively regulates salt tolerance by inhibiting MdERF3 transcription. Plant Sci. 2018, 276, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, J.; Zhai, L.M.; Gan, Z.Y.; Zhang, G.F.; Yang, S.H.; Wang, Y.; Wu, T.; Zhang, X.Z.; Xu, X.F.; et al. Natural variation in cytokinin maintenance improves salt tolerance in apple rootstocks. Plant Cell Environ. 2019, 42, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.M.; Zhu, Y.F.; Hu, Y.; Zhang, R.; Cheng, L.; Zhu, Z.L.; Zhao, T.; Zhang, X.Y.; Wang, Y.X. Integrated physiologic, proteomic, and metabolomic analyses of Malus halliana adaptation to saline–alkali stress. Hortic. Res. 2019, 6, 91. [Google Scholar] [CrossRef]
- Li, X.L.; Li, M.J.; Zhou, B.B.; Yang, Y.Z.; Wei, Q.P.; Zhang, J.K. Transcriptome analysis provides insights into the stress response crosstalk in apple (Malus × domestica) subjected to drought, cold and high salinity. Sci. Rep. 2019, 9, 9071. [Google Scholar] [CrossRef]
- Liu, J.H.; Inoue, H.; Moriguchi, T. Salt stress-mediated changes in free polyamine tiers and expression of genes responsible for polyamine biosynthesis of apple in vitro shoots. Environ. Exp. Bot. 2008, 62, 28–35. [Google Scholar] [CrossRef]
- Wang, L.; Li, Q.T.; Lei, Q.; Feng, C.; Gao, Y.N.; Zheng, X.D.; Zhao, Y.; Wang, Z.; Kong, J. MzPIP2;1: An aquaporin involved in radial water movement in both water uptake and transportation, altered the drought and salt tolerance of transgenic Arabidopsis. PLoS ONE 2015, 10, e0142446. [Google Scholar] [CrossRef]
- Wu, R.G.; Wang, Y.; Wu, T.; Xu, X.F.; Han, Z.H. MdMYB4, an R2R3-Type MYB transcription factor, play a crucial role in cold and salt stress in apple calli. J. Am. Soc. Hortic. Sci. 2017, 142, 209–216. [Google Scholar] [CrossRef]
- Yu, J.Q.; Wang, J.H.; Sun, C.H.; Zhang, Q.Y.; Hu, D.G.; Hao, Y.J. Ectopic expression of the apple nucleus-encoded thylakoid protein MdY3IP1triggers early-flowering and enhanced salt-tolerance in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 18. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Qu, S.C.; Qiao, Y.S.; Zhang, Z.; Guo, Z.R. Overexpression of the Malus hupehensis MhNPR1 gene increased tolerance to salt and osmotic stress in transgenic tobacco. Mol. Biol. Rep. 2014, 41, 1553–1561. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Luo, H.T.; Guo, Z.R. Overexpression of Malus hupehensis MhSHN1 gene enhances salt and osmotic stress tolerance in transgenic tobacco plants. Russ. J. Plant Physiol. 2018, 65, 857–864. [Google Scholar] [CrossRef]
- Liu, J.; Shen, F.; Xiao, Y.; Fang, H.C.; Qiu, C.P.; Li, W.; Wu, T.; Xu, X.F.; Wang, Y.; Zhang, X.Z.; et al. Genomics-assisted prediction of salt and alkali tolerances and functional marker development in apple rootstocks. BMC Genom. 2020, 21, 550. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.Q.; Song, M.R.; Guo, Y.N.; Liu, L.F.; Xue, H.; Dai, H.Y.; Zhang, Z.H. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 2019, 17, 2341–2355. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol. Plant. 2018, 164, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.L.; Zhao, H.X.; Ma, J.L.; Zhou, S.W. The adaption and application area on the salinity for Malus zumi Mats. J. Tianjin Agric. Coll. 1996, 3, 48–52. [Google Scholar]
- Wang, Y.Z.; Feng, X.Z.; Luo, J.L. Multi-purpose and salt-tolerant economic tree-Malus zumi. For. Sci. Technol. 1999, 24, 53–55. [Google Scholar]
- Fang, H.L.; Liu, G.H.; Kearney, M. Georelational analysis of soil type, soil salt content, landform, and land use in the Yellow River Delta, China. Environ. Manag. 2005, 35, 72–83. [Google Scholar] [CrossRef]
- Wang, Y.G.; Li, Y.; Xiao, D.N. Catchment scale spatial variability of soil salt content in agricultural oasis, Northwest China. Environ. Geol. 2008, 56, 439–446. [Google Scholar] [CrossRef]
- Li, F.Z.; Huang, Z.B.; Ma, Y.; Sun, Z.J. Improvement effects of different environmental materials on coastal saline-alkali soil in Yellow River Delta. Mater. Sci. Forum 2018, 913, 879–886. [Google Scholar] [CrossRef]
- Li, Q.T.; Liu, J.; Tan, D.X.; Allan, A.C.; Jiang, Y.Z.; Xu, X.F.; Han, Z.H.; Kong, J. A genome-wide expression profile of salt-responsive genes in the apple rootstock Malus zumi. Int. J. Mol. Sci. 2013, 14, 21053–21070. [Google Scholar] [CrossRef]
- Upadhyay, A.; Gaonkar, T.; Upadhyay, A.K.; Jogaiah, S.; Shinde, M.P.; Kadoo, N.Y.; Gupta, V.S. Global transcriptome analysis of grapevine (Vitis vinifera L.) leaves under salt stress reveals differential response at early and late stages of stress in table grape cv. Thompson Seedless. Plant Physiol. Biochem. 2018, 129, 168. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Shin, T.; Peter, N. Life and death under salt: Same players, different timing? J. Exp. Bot. 2014, 65, 2963–2979. [Google Scholar] [CrossRef]
- Zhang, H.; Li, D.S.; Zhou, Z.G.; Zahoor, R.; Chen, B.L.; Meng, Y.L. Soil water and salt affect cotton (Gossypium hirsutum L.) photosynthesis, yield and fiber quality in coastal saline soil. Agric. Water Manag. 2017, 187, 112–121. [Google Scholar] [CrossRef]
- Liang, W.J.; Ma, X.L.; Wan, P.; Liu, L.Y. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Hore, T.; Kaneko, T.; Sugimoto, G.; Sasano, S.; Panda, S.K.; Shibasaka, M.; Katsuhara, M. Mechanisms of Water Transport Mediated by PIP Aquaporins and Their Regulation Via Phosphorylation Events Under Salinity Stress in Barley Roots. Plant Cell Physiol. 2011, 52, 663–675. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; Henri, V.D.G.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Hu, J.; Han, X.L.; Li, J.J.; Gao, Y.; Richards, C.M.; Zhang, C.X.; Tian, Y.; Liu, G.M.; Gul, H.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1. [Google Scholar] [CrossRef]
- Sharp, R.E.; Hsiao, T.C.; Silk, W.K. Growth of the maize primary root at low water potentials: II. Role of growth and deposition of hexoes and potassium in osmotic adjustment. Plant Physiol. 1990, 93, 1337–1346. [Google Scholar] [CrossRef]
- Wang, D.J.; Gao, Y.; Sun, S.M.; Lu, X.; Li, Q.S.; Li, L.W.; Wang, K. Effects of salt stress on the antioxidant activity and malondialdehyde, solution protein, proline, and chlorophyll contents of three Malus species. Life 2022, 12, 1929. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Mori, T.; Saito, K. Alternation of flavonoid accumulation under drought stress in Arabidopsis thaliana. Plant Signal. Behav. 2014, 9, e29518. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by over accumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ren, Y.; Wei, W.; Yang, J.; Zhong, Q.; Li, Z. Metabolite analysis of jerusalem artichoke (Helianthus tuberosus L.) Seedlings in response to polyethylene glycol-simulated drought stress. Int. J. Mol. Sci. 2021, 22, 3294. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Wang, C.; Bi, S.T.; Liu, C.Y.; Li, M.Y.; Liu, X.Y.; Cheng, J.N.; Wang, L.; Lv, Y.L.; He, M. Arabidopsis ADF7 inhibits VLN1 to regulate actin filament dynamics and ROS accumulation in root hair development responses to osmotic stress. Authorea 2021, 2. [Google Scholar] [CrossRef]
- Shi, B.; Di, Y. Plant Polyphenols; Science Press: Beijing, China, 2000; pp. 134–135. [Google Scholar]
- Valifard, M.; Mohsenzadeh, S.; Kholdebarin, B.; Rowshan, V. Effects of salt stress on volatile compounds, total phenolic content and antioxidant activities of Salvia mirzayanii. S. Afr. J. Bot. 2014, 93, 92–97. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Salinity stress enhances color parameters, bioactive leaf pigments, vitamins, polyphenols, flavonoids and antioxidant activity in selected Amaranthus leafy vegetables. J. Sci. Food Agric. 2019, 5, 99. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef]
- Jiao, Y.; Zhang, J.H.; Pan, C. Integrated physiological, proteomic, and metabolomics analyses of pecan cultivar ‘Pawnee’ adaptation to salt stress. Sci. Rep. 2022, 12, 1841. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef]
- Silva, E.N.; Ferreira-Silva, S.L.; Viégas, R.A.; Silveira, J.A.G. The role of organic and inorganic solutes in the osmotic adjustment of drought-stressed Jatropha curcas plants. Environ. Exp. Bot. 2010, 69, 279–285. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. bioRxiv 2018, 274100. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzbery, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzbery, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Dewey, C.N.; Bo, L. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.L.; Wang, W.S.; Zhang, H.Y.; Liu, X.Q.; Yu, S.B.; Xiong, L.Z.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.F.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0-The human metabolome database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Zhu, Z.J.; Schultz, A.W.; Wang, J.H.; Johnson, C.H.; Yannone, S.M.; Patti, G.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Propocals 2013, 8, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Velzen, E.J.J.V.; Duijnhoven, J.P.M.V.; Dorsten, F.A.V. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36 (Suppl. S1), D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Bylesjö, M.; Eriksson, D.; Kusano, M.; Moritz, T.; Trygg, J. Data integration in plant biology: The O2PLS method for combined modeling of transcript and metabolite data. Plant J. 2007, 52, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddani, S.E.; Houwing-Duistermaat, J.; Salo, P.; Perola, M.; Jongbloed, G.; Uh, H. Evaluation of O2PLS in Omics data integration. BMC Bioinform. 2016, 17 (Suppl. S2), S11. [Google Scholar] [CrossRef] [PubMed]
- The genome sequence archive family: Toward explosive data growth and diverse data types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [CrossRef]
- Database resources of the national genomics data center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022, 50, D27–D38. [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Wang, K.; Sun, S.; Yan, P.; Lu, X.; Liu, Z.; Li, Q.; Li, L.; Gao, Y.; Liu, J. Transcriptome and Metabolome Analysis Reveals Salt-Tolerance Pathways in the Leaves and Roots of ZM-4 (Malus zumi) in the Early Stages of Salt Stress. Int. J. Mol. Sci. 2023, 24, 3638. https://doi.org/10.3390/ijms24043638
Wang D, Wang K, Sun S, Yan P, Lu X, Liu Z, Li Q, Li L, Gao Y, Liu J. Transcriptome and Metabolome Analysis Reveals Salt-Tolerance Pathways in the Leaves and Roots of ZM-4 (Malus zumi) in the Early Stages of Salt Stress. International Journal of Molecular Sciences. 2023; 24(4):3638. https://doi.org/10.3390/ijms24043638
Chicago/Turabian StyleWang, Dajiang, Kun Wang, Simiao Sun, Peng Yan, Xiang Lu, Zhao Liu, Qingshan Li, Lianwen Li, Yuan Gao, and Jihong Liu. 2023. "Transcriptome and Metabolome Analysis Reveals Salt-Tolerance Pathways in the Leaves and Roots of ZM-4 (Malus zumi) in the Early Stages of Salt Stress" International Journal of Molecular Sciences 24, no. 4: 3638. https://doi.org/10.3390/ijms24043638





