Strain-Specific Interactions between the Viral Capsid Proteins VP4, VP7 and VP6 Influence Rescue of Rotavirus Reassortants by Reverse Genetics
Abstract
:1. Introduction
2. Results
2.1. Generation of VP7-Wa- and VP6-Wa-Encoding Plasmids
2.2. Generation of Mono-, Double-, and Triple-Reassortants
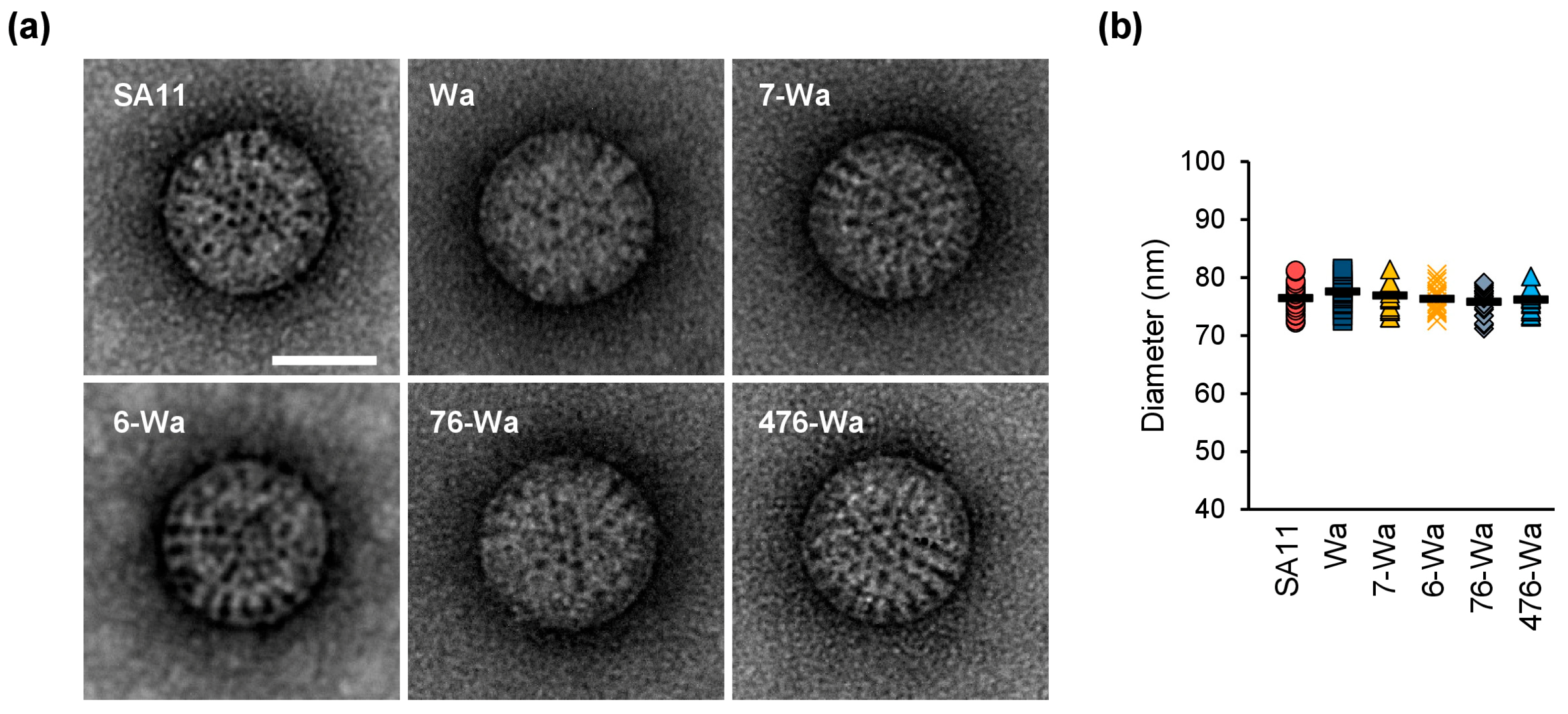
2.3. Transmission Electron Microscopy (TEM) Analyses
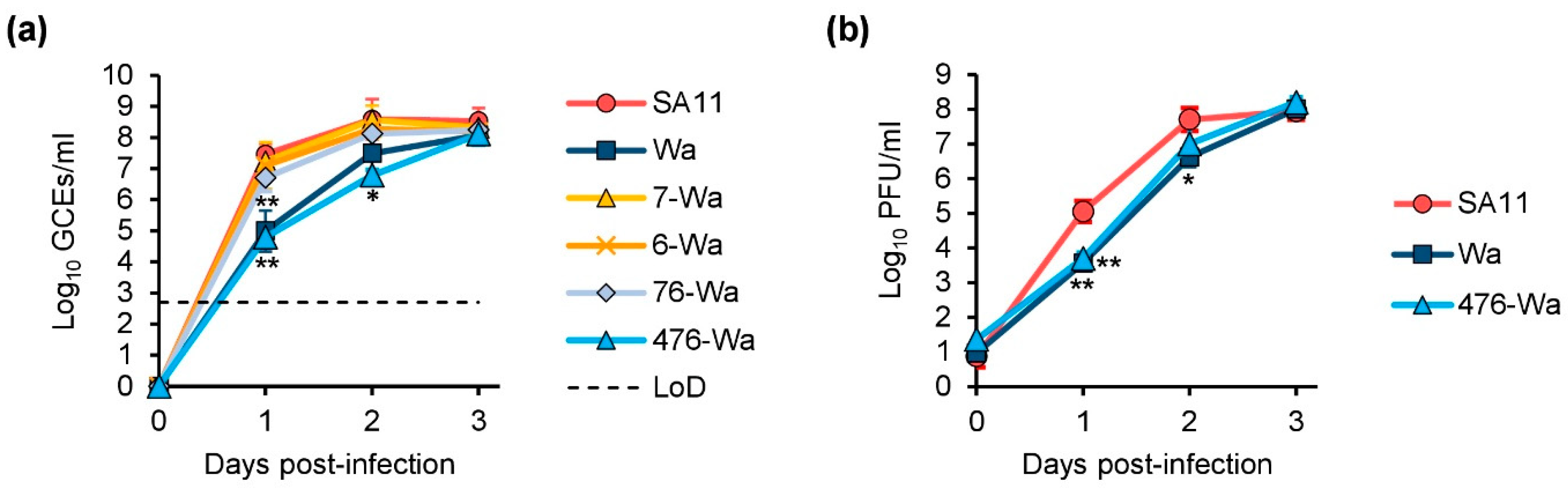
2.4. Replication Kinetics
2.5. Untranslated Region (UTR) Sequence Analyses
2.6. Protein Interface Analyses
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Viruses
4.2. Plasmids
4.3. Plasmid-Based RG System
4.4. Passaging of Reassortant Virus
4.5. qRT-PCR and RT-PCR
4.6. Replication Kinetics
4.7. Plaque Assays
4.8. Transmission Electron Microscope (TEM) Analyses
4.9. Sequence Analyses and Protein Structure Visualization
4.10. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matthijnssens, J.; Attoui, H.; Banyai, K.; Brussaard, C.P.D.; Danthi, P.; Del Vas, M.; Dermody, T.S.; Duncan, R.; Fang, Q.; Johne, R.; et al. ICTV Virus Taxonomy Profile: Sedoreoviridae 2022. J. Gen. Virol. 2022, 103, 001782. [Google Scholar] [CrossRef]
- Desselberger, U. Differences of Rotavirus Vaccine Effectiveness by Country: Likely Causes and Contributing Factors. Pathogens 2017, 6, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, A.L.; A Platts-Mills, J.; Nakamura, T.; Operario, D.J.; Antoni, S.; Mwenda, J.M.; Weldegebriel, G.; Rey-Benito, G.; de Oliveira, L.H.; Ortiz, C.; et al. Aetiology and incidence of diarrhoea requiring hospitalisation in children under 5 years of age in 28 low-income and middle-income countries: Findings from the Global Pediatric Diarrhea Surveillance network. BMJ Glob. Health 2022, 7, e009548. [Google Scholar] [CrossRef]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef] [Green Version]
- Settembre, E.C.; Chen, J.Z.; Dormitzer, P.R.; Grigorieff, N.; Harrison, S.C. Atomic model of an infectious rotavirus particle. EMBO J. 2011, 30, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.; Both, G. Completion of the genomic sequence of the simian rotavirus SA11: Nucleotide sequences of segments 1, 2, and 3. Virology 1990, 177, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Iturriza-Gómara, M.; Maes, P.; Patton, J.; et al. Full Genome-Based Classification of Rotaviruses Reveals a Common Origin between Human Wa-Like and Porcine Rotavirus Strains and Human DS-1-Like and Bovine Rotavirus Strains. J. Virol. 2008, 82, 3204–3219. [Google Scholar] [CrossRef] [Green Version]
- RCWG Rotavirus Classification Working Group. List of Accepted Genotypes. Laboratory of Viral Metagenomics. Available online: https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg (accessed on 3 November 2022).
- Johne, R.; Tausch, S.H.; Schilling-Loeffler, K.; Ulrich, R.G. Genome sequence analysis of a novel rotavirus strain indicates a broad genetic diversity of rotavirus A in shrews. Infect. Genet. Evol. 2023, 107, 105392. [Google Scholar] [CrossRef]
- Offit, P.A.; Shaw, R.D.; Greenberg, H.B. Passive protection against rotavirus-induced diarrhea by monoclonal antibodies to surface proteins vp3 and vp7. J. Virol. 1986, 58, 700–703. [Google Scholar] [CrossRef] [Green Version]
- Menchaca, G.; Padilla-Noriega, L.; Méndez-Toss, M.; Contreras, J.F.; Puerto, F.I.; Guiscafré, H.; Mota, F.; Herrera, I.; Cedillo, R.; Muñoz, O.; et al. Serotype Specificity of the Neutralizing-Antibody Response Induced by the Individual Surface Proteins of Rotavirus in Natural Infections of Young Children. Clin. Diagn. Lab. Immunol. 1998, 5, 328–334. [Google Scholar] [CrossRef] [Green Version]
- Nair, N.; Feng, N.; Blum, L.K.; Sanyal, M.; Ding, S.; Jiang, B.; Sen, A.; Morton, J.M.; He, X.-S.; Robinson, W.H.; et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci. Transl. Med. 2017, 9, eaam5434. [Google Scholar] [CrossRef] [Green Version]
- Caddy, S.L.; Vaysburd, M.; Wing, M.; Foss, S.; Andersen, J.T.; O’Connell, K.; Mayes, K.; Higginson, K.; Iturriza-Gómara, M.; Desselberger, U.; et al. Intracellular neutralisation of rotavirus by VP6-specific IgG. PLoS Pathog. 2020, 16, e1008732. [Google Scholar] [CrossRef]
- Martella, V.; Bányai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010, 140, 246–255. [Google Scholar] [CrossRef] [Green Version]
- Angel, J.; Franco, M.A.; Greenberg, H. Rotavirus vaccines: Recent developments and future considerations. Nat. Rev. Genet. 2007, 5, 529–539. [Google Scholar] [CrossRef]
- McDonald, S.M.; Nelson, M.I.; Turner, P.E.; Patton, J.T. Reassortment in segmented RNA viruses: Mechanisms and outcomes. Nat. Rev. Genet. 2016, 14, 448–460. [Google Scholar] [CrossRef] [Green Version]
- Gentsch, J.R.; Laird, A.R.; Bielfelt, B.; Griffin, D.D.; Bányai, K.; Ramachandran, M.; Jain, V.; Cunliffe, N.; Nakagomi, O.; Kirkwood, C.D.; et al. Serotype Diversity and Reassortment between Human and Animal Rotavirus Strains: Implications for Rotavirus Vaccine Programs. J. Infect. Dis. 2005, 192, S146–S159. [Google Scholar] [CrossRef] [Green Version]
- McDonald, S.M.; Patton, J.T. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 2011, 19, 136–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patzina-Mehling, C.; Falkenhagen, A.; Trojnar, E.; Gadicherla, A.K.; Johne, R. Potential of avian and mammalian species A rotaviruses to reassort as explored by plasmid only-based reverse genetics. Virus Res. 2020, 286, 198027. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Tacuba, L.; Feng, N.; Meade, N.J.; Mellits, K.H.; Jaïs, P.H.; Yasukawa, L.L.; Resch, T.K.; Jiang, B.; López, S.; Ding, S.; et al. An Optimized Reverse Genetics System Suitable for Efficient Recovery of Simian, Human, and Murine-Like Rotaviruses. J. Virol. 2020, 94, e01294-20. [Google Scholar] [CrossRef]
- Falkenhagen, A.; Tausch, S.H.; Labutin, A.; Grützke, J.; Heckel, G.; Ulrich, R.G.; Johne, R. Genetic and biological characteristics of species A rotaviruses detected in common shrews suggest a distinct evolutionary trajectory. Virus Evol. 2022, 8, veac004. [Google Scholar] [CrossRef] [PubMed]
- Mingo, R.; Zhang, S.; Long, C.P.; LaConte, L.E.W.; McDonald, S.M. Genetic determinants restricting the reassortment of heterologous NSP2 genes into the simian rotavirus SA11 genome. Sci. Rep. 2017, 7, 9301. [Google Scholar] [CrossRef] [Green Version]
- McDonald, S.M.; Matthijnssens, J.; McAllen, J.K.; Hine, E.; Overton, L.; Wang, S.; Lemey, P.; Zeller, M.; Van Ranst, M.; Spiro, D.J.; et al. Evolutionary Dynamics of Human Rotaviruses: Balancing Reassortment with Preferred Genome Constellations. PLoS Pathog. 2009, 5, e1000634. [Google Scholar] [CrossRef]
- Heiman, E.M.; McDonald, S.M.; Barro, M.; Taraporewala, Z.F.; Bar-Magen, T.; Patton, J.T. Group A Human Rotavirus Genomics: Evidence that Gene Constellations Are Influenced by Viral Protein Interactions. J. Virol. 2008, 82, 11106–11116. [Google Scholar] [CrossRef] [Green Version]
- Kanai, Y.; Komoto, S.; Kawagishi, T.; Nouda, R.; Nagasawa, N.; Onishi, M.; Matsuura, Y.; Taniguchi, K.; Kobayashi, T. Entirely plasmid-based reverse genetics system for rotaviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 2349–2354. [Google Scholar] [CrossRef] [Green Version]
- Komoto, S.; Fukuda, S.; Kugita, M.; Hatazawa, R.; Koyama, C.; Katayama, K.; Murata, T.; Taniguchi, K. Generation of Infectious Recombinant Human Rotaviruses from just 11 Cloned cDNAs Encoding the Rotavirus Genome. J. Virol. 2019, 93, e02207-18. [Google Scholar] [CrossRef] [Green Version]
- Kawagishi, T.; Nurdin, J.A.; Onishi, M.; Nouda, R.; Kanai, Y.; Tajima, T.; Ushijima, H.; Kobayashi, T. Reverse Genetics System for a Human Group A Rotavirus. J. Virol. 2020, 94, e00963-19. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Onishi, M.; Kawagishi, T.; Pannacha, P.; Nurdin, J.A.; Nouda, R.; Yamasaki, M.; Lusiany, T.; Khamrin, P.; Okitsu, S.; et al. Reverse Genetics Approach for Developing Rotavirus Vaccine Candidates Carrying VP4 and VP7 Genes Cloned from Clinical Isolates of Human Rotavirus. J. Virol. 2020, 95, e01374-20. [Google Scholar] [CrossRef] [PubMed]
- Falkenhagen, A.; Patzina-Mehling, C.; Gadicherla, A.K.; Strydom, A.; O’Neill, H.G.; Johne, R. Generation of Simian Rotavirus Reassortants with VP4- and VP7-Encoding Genome Segments from Human Strains Circulating in Africa Using Reverse Genetics. Viruses 2020, 12, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkenhagen, A.; Patzina-Mehling, C.; Rückner, A.; Vahlenkamp, T.W.; Johne, R. Generation of simian rotavirus reassortants with diverse VP4 genes using reverse genetics. J. Gen. Virol. 2019, 100, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Falkenhagen, A.; Huyzers, M.; van Dijk, A.; Johne, R. Rescue of Infectious Rotavirus Reassortants by a Reverse Genetics System Is Restricted by the Receptor-Binding Region of VP4. Viruses 2021, 13, 363. [Google Scholar] [CrossRef]
- Prasad, B.; Wang, G.; Clerx, J.P.; Chiu, W. Three-dimensional structure of rotavirus. J. Mol. Biol. 1988, 199, 269–275. [Google Scholar] [CrossRef]
- Boudreaux, C.E.; Kelly, D.F.; McDonald, S.M. Electron microscopic analysis of rotavirus assembly-replication intermediates. Virology 2015, 477, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, M.; Petitpas, I.; Navaza, J.; Lepault, J.; Kohli, E.; Pothier, P.; Prasad, B.; Cohen, J.; Rey, F.A. Atomic structure of the major capsid protein of rotavirus: Implications for the architecture of the virion. EMBO J. 2001, 20, 1485–1497. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.Z.; Settembre, E.C.; Aoki, S.T.; Zhang, X.; Bellamy, A.R.; Dormitzer, P.R.; Harrison, S.C.; Grigorieff, N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc. Natl. Acad. Sci. USA 2009, 106, 10644–10648. [Google Scholar] [CrossRef] [Green Version]
- Uprety, T.; Wang, D.; Li, F. Recent advances in rotavirus reverse genetics and its utilization in basic research and vaccine development. Arch. Virol. 2021, 166, 2369–2386. [Google Scholar] [CrossRef] [PubMed]
- Borodavka, A.; Dykeman, E.C.; Schrimpf, W.; Lamb, D.C. Protein-mediated RNA folding governs sequence-specific interactions between rotavirus genome segments. Elife 2017, 6, 27453. [Google Scholar] [CrossRef]
- Fajardo, T., Jr.; Sung, P.-Y.; Celma, C.C.; Roy, P. Rotavirus Genomic RNA Complex Forms via Specific RNA–RNA Interactions: Disruption of RNA Complex Inhibits Virus Infectivity. Viruses 2017, 9, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesavento, J.B.; Billingsley, A.M.; Roberts, E.J.; Ramig, R.F.; Prasad, B.V.V. Structures of Rotavirus Reassortants Demonstrate Correlation of Altered Conformation of the VP4 Spike and Expression of Unexpected VP4-Associated Phenotypes. J. Virol. 2003, 77, 3291–3296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altenburg, B.C.; Graham, D.Y.; Estes, M.K. Ultrastructural Study of Rotavirus Replication in Cultured Cells. J. Gen. Virol. 1980, 46, 75–85. [Google Scholar] [CrossRef]
- Ludert, J.; Michelangeli, F.; Gil, F.; Liprandi, F.; Esparza, J. Penetration and Uncoating of Rotaviruses in Cultured Cells. Intervirology 1987, 27, 95–101. [Google Scholar] [CrossRef]
- González, S.; Affranchino, J.L. Assembly of double-layered virus-like particles in mammalian cells by coexpression of human rotavirus VP2 and VP6. J. Gen. Virol. 1995, 76, 2357–2360. [Google Scholar] [CrossRef]
- Johne, R.; Reetz, J.; Kaufer, B.B.; Trojnar, E. Generation of an Avian-Mammalian Rotavirus Reassortant by Using a Helper Virus-Dependent Reverse Genetics System. J. Virol. 2016, 90, 1439–1443. [Google Scholar] [CrossRef] [Green Version]
- Otto, P.H.; Rosenhain, S.; Elschner, M.C.; Hotzel, H.; Machnowska, P.; Trojnar, E.; Hoffmann, K.; Johne, R. Detection of rotavirus species A, B and C in domestic mammalian animals with diarrhoea and genotyping of bovine species A rotavirus strains. Vet. Microbiol. 2015, 179, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.; Matthijnssens, J.; Rahman, M.; Van Ranst, M. RotaC: A web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009, 9, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Sundaram, J.P.; Spiro, D. VIGOR, an annotation program for small viral genomes. BMC Bioinform. 2010, 11, 451. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Sundaram, J.P.; Stockwell, T.B. VIGOR extended to annotate genomes for additional 12 different viruses. Nucleic Acids Res. 2012, 40, W186–W192. [Google Scholar] [CrossRef] [Green Version]
- Tsugawa, T.; Tsutsumi, H. Genomic changes detected after serial passages in cell culture of virulent human G1P[8] rotaviruses. Infect. Genet. Evol. 2016, 45, 6–10. [Google Scholar] [CrossRef]
- Taniguchi, K.; Nishikawa, K.; Kobayashi, N.; Urasawa, T.; Wu, H.; Gorziglia, M.; Urasawa, S. Differences in Plaque Size and VP4 Sequence Found in SA11 Virus Clones Having Simian Authentic VP4. Virology 1994, 198, 325–330. [Google Scholar] [CrossRef] [PubMed]




| Strain | Genotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | |
| SA11 | G3 | P[2] | I2 | R2 | C5 | M5 | A5 | N5 | T5 | E2 | H5 |
| Wa | G1 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valusenko-Mehrkens, R.; Gadicherla, A.K.; Johne, R.; Falkenhagen, A. Strain-Specific Interactions between the Viral Capsid Proteins VP4, VP7 and VP6 Influence Rescue of Rotavirus Reassortants by Reverse Genetics. Int. J. Mol. Sci. 2023, 24, 5670. https://doi.org/10.3390/ijms24065670
Valusenko-Mehrkens R, Gadicherla AK, Johne R, Falkenhagen A. Strain-Specific Interactions between the Viral Capsid Proteins VP4, VP7 and VP6 Influence Rescue of Rotavirus Reassortants by Reverse Genetics. International Journal of Molecular Sciences. 2023; 24(6):5670. https://doi.org/10.3390/ijms24065670
Chicago/Turabian StyleValusenko-Mehrkens, Roman, Ashish K. Gadicherla, Reimar Johne, and Alexander Falkenhagen. 2023. "Strain-Specific Interactions between the Viral Capsid Proteins VP4, VP7 and VP6 Influence Rescue of Rotavirus Reassortants by Reverse Genetics" International Journal of Molecular Sciences 24, no. 6: 5670. https://doi.org/10.3390/ijms24065670





