Up-Regulation of microRNA-424 Causes an Imbalance in AKT Phosphorylation and Impairs Enteric Neural Crest Cell Migration in Hirschsprung Disease
Abstract
1. Introduction
2. Results
2.1. RICTOR Correlates Inversely with miR-424 in HSCR
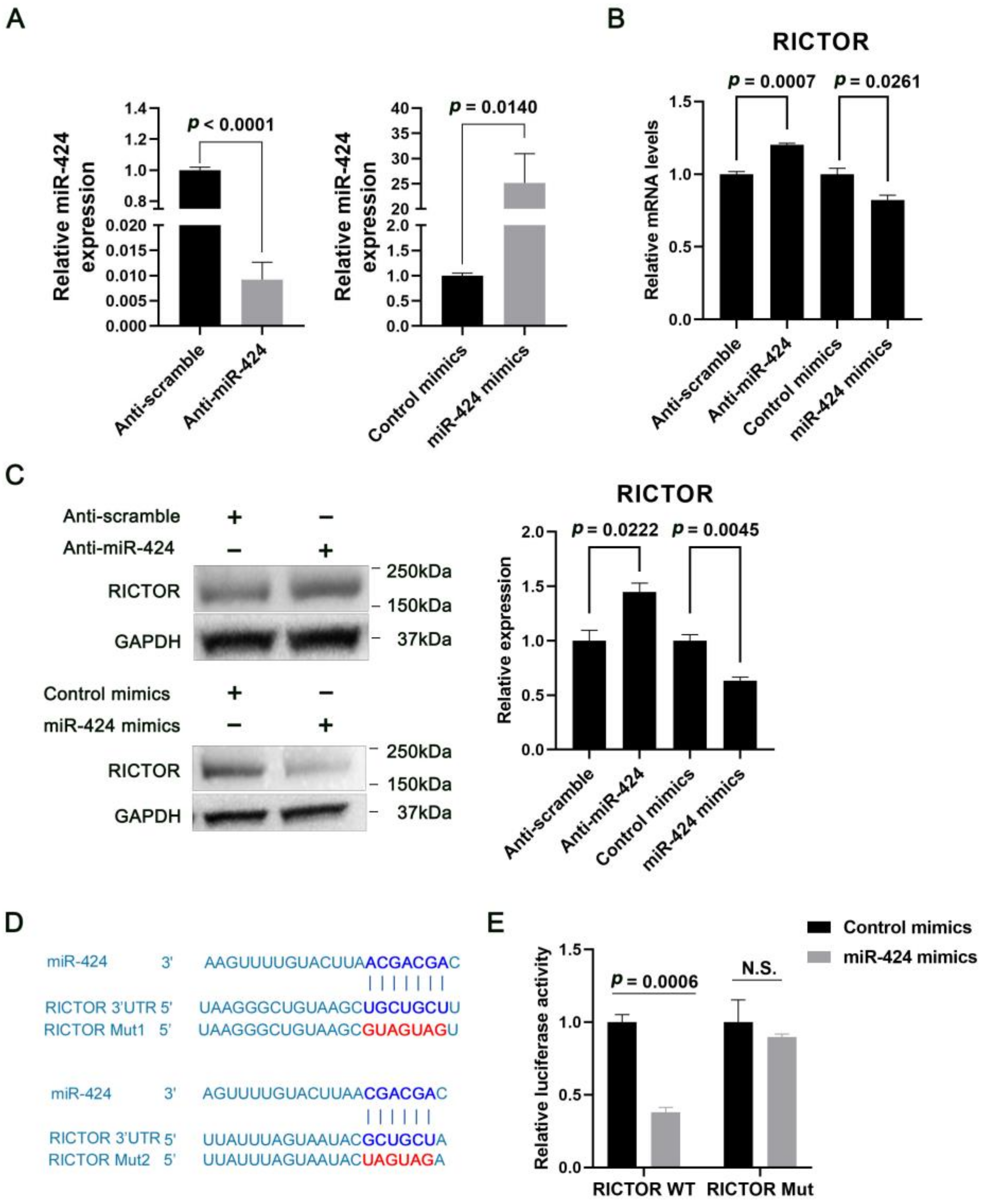
2.2. MiR-424 Directly Targets RICTOR in Human Neural Cells
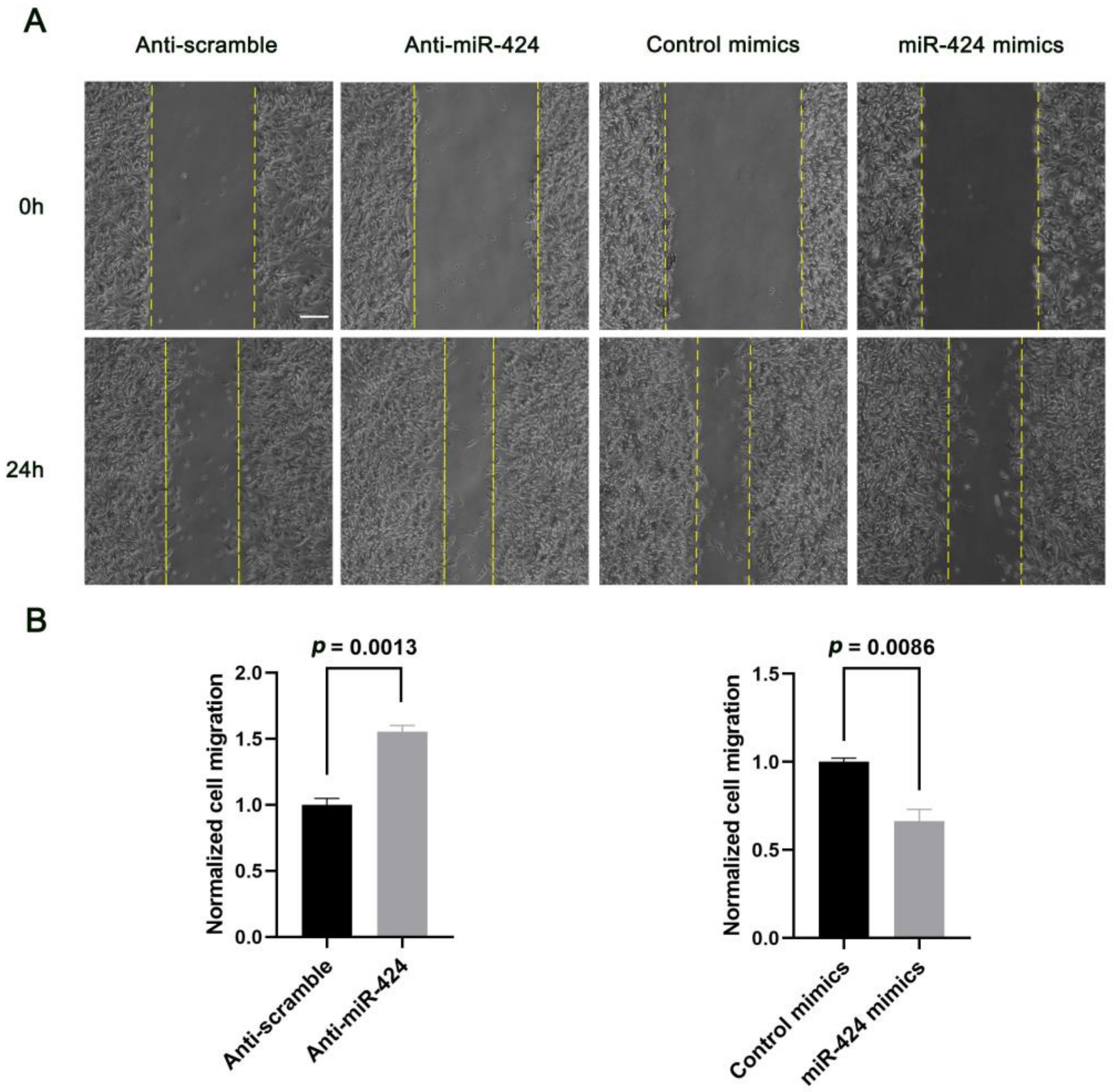
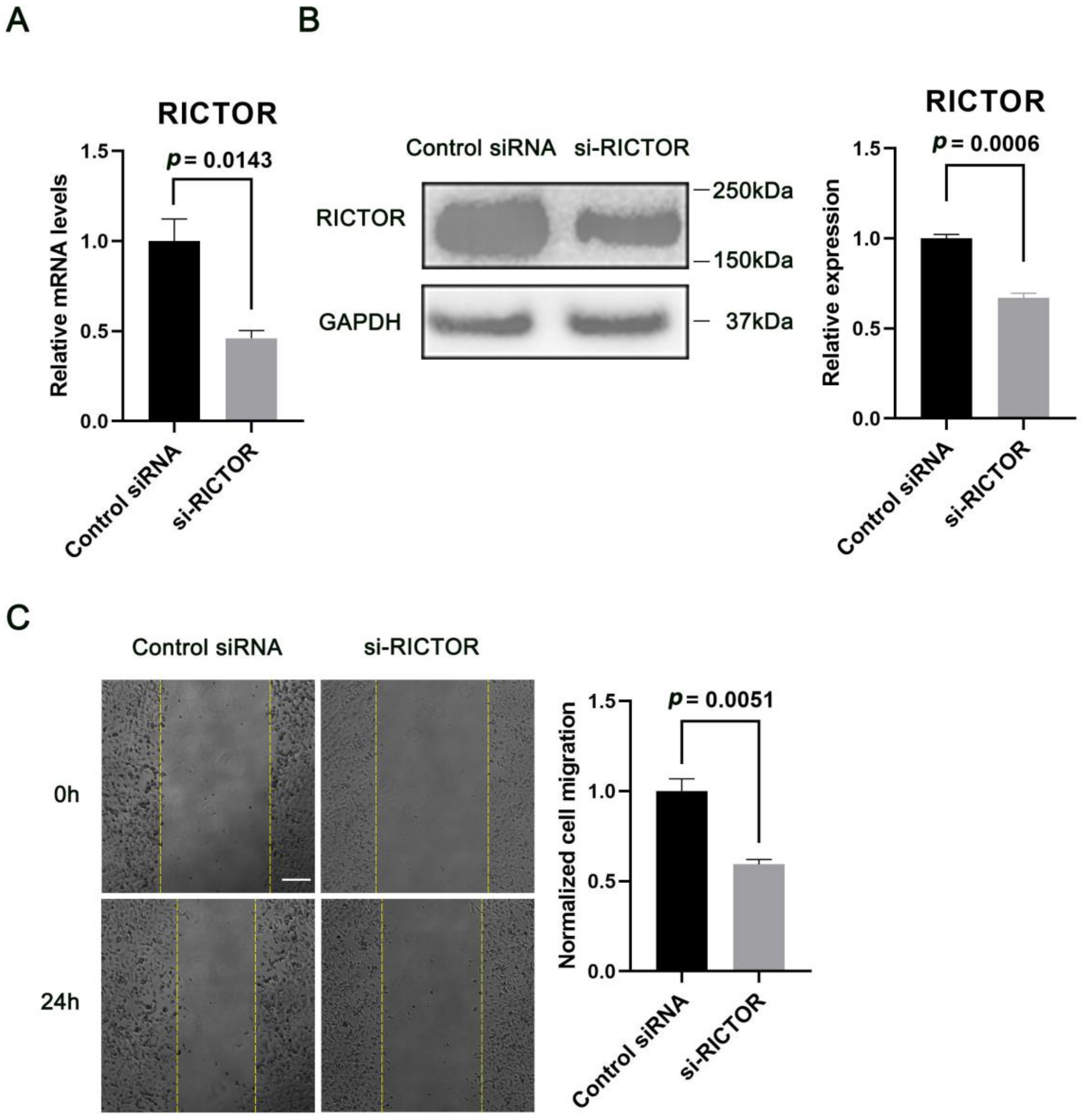
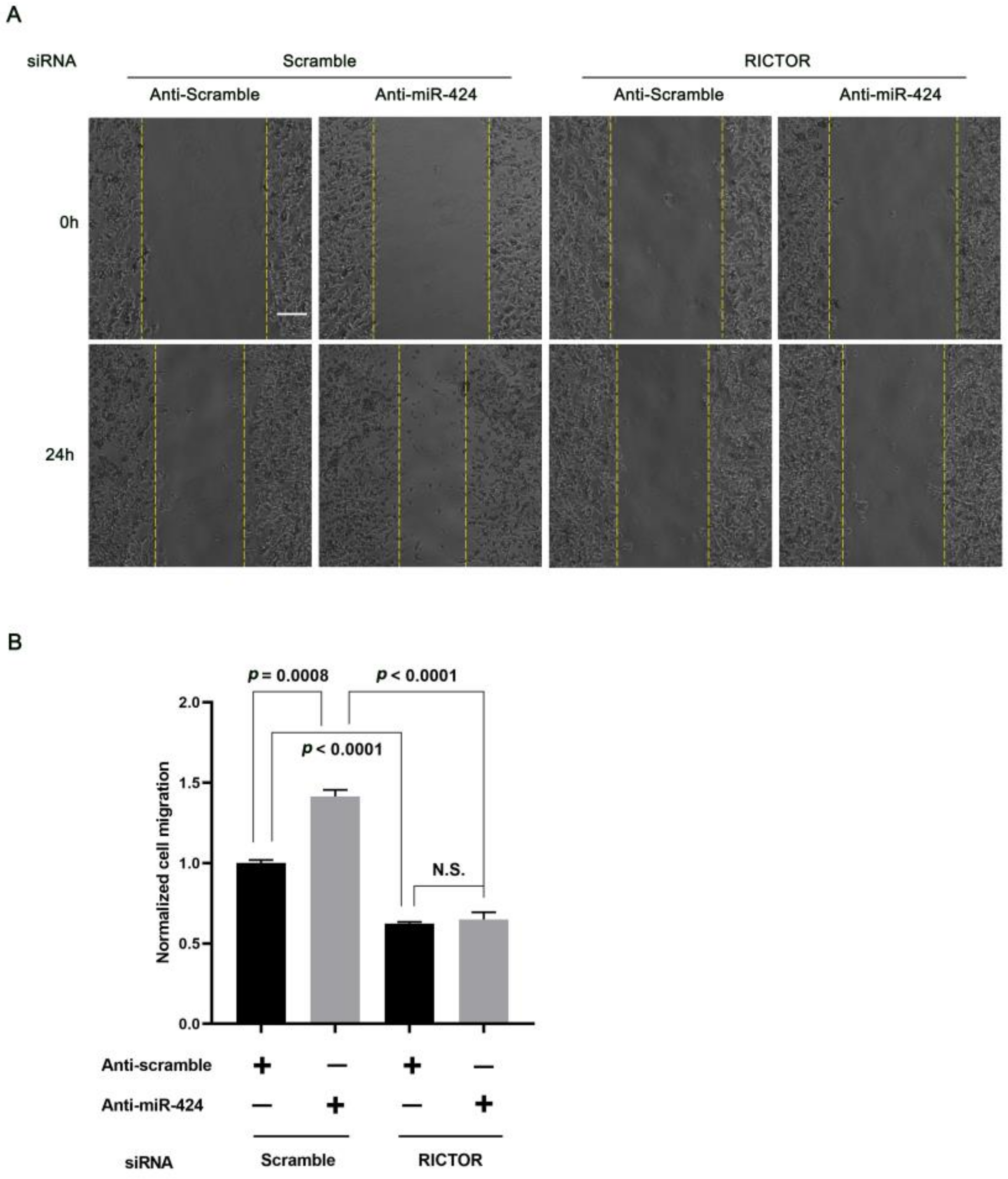
2.3. MiR-424 Modulates Human Neural Cell Migration in a RICTOR-Dependent Manner
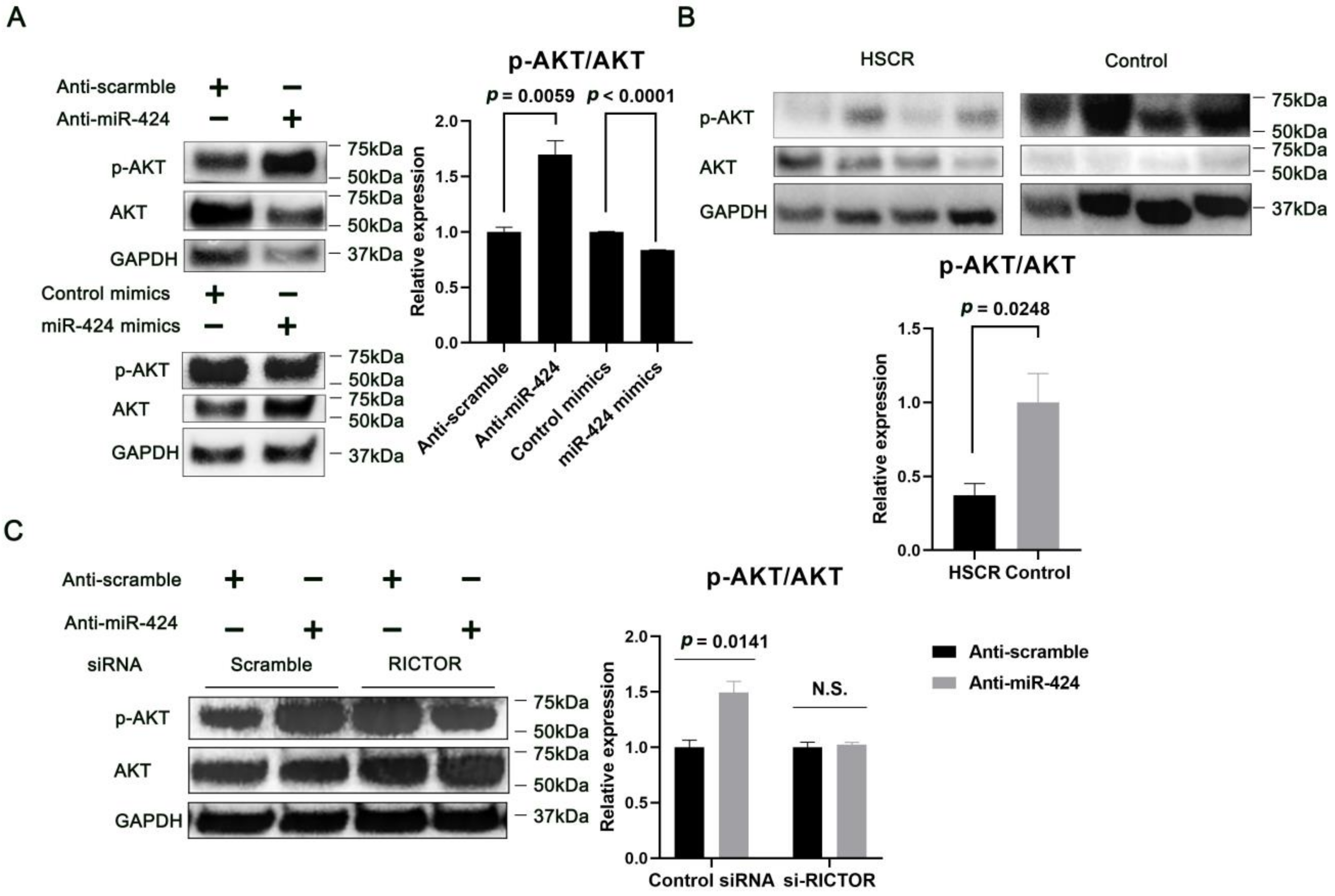
2.4. MiR-424 Regulates AKT Phosphorylation through RICTOR-Dependent Mechanism
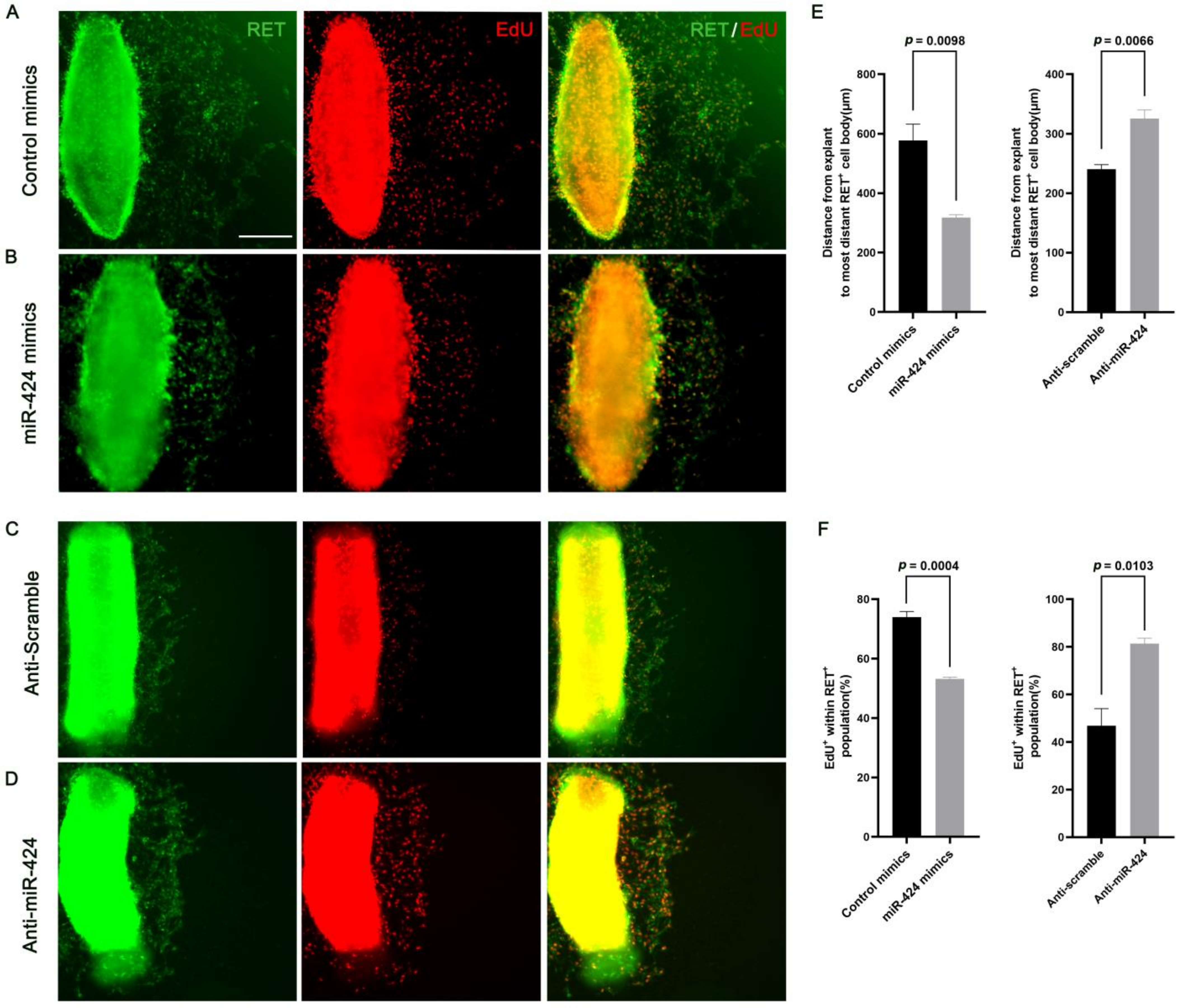
2.5. MiR-424 Regulates the Development of Enteric Neural Crest Cell Ex Vivo
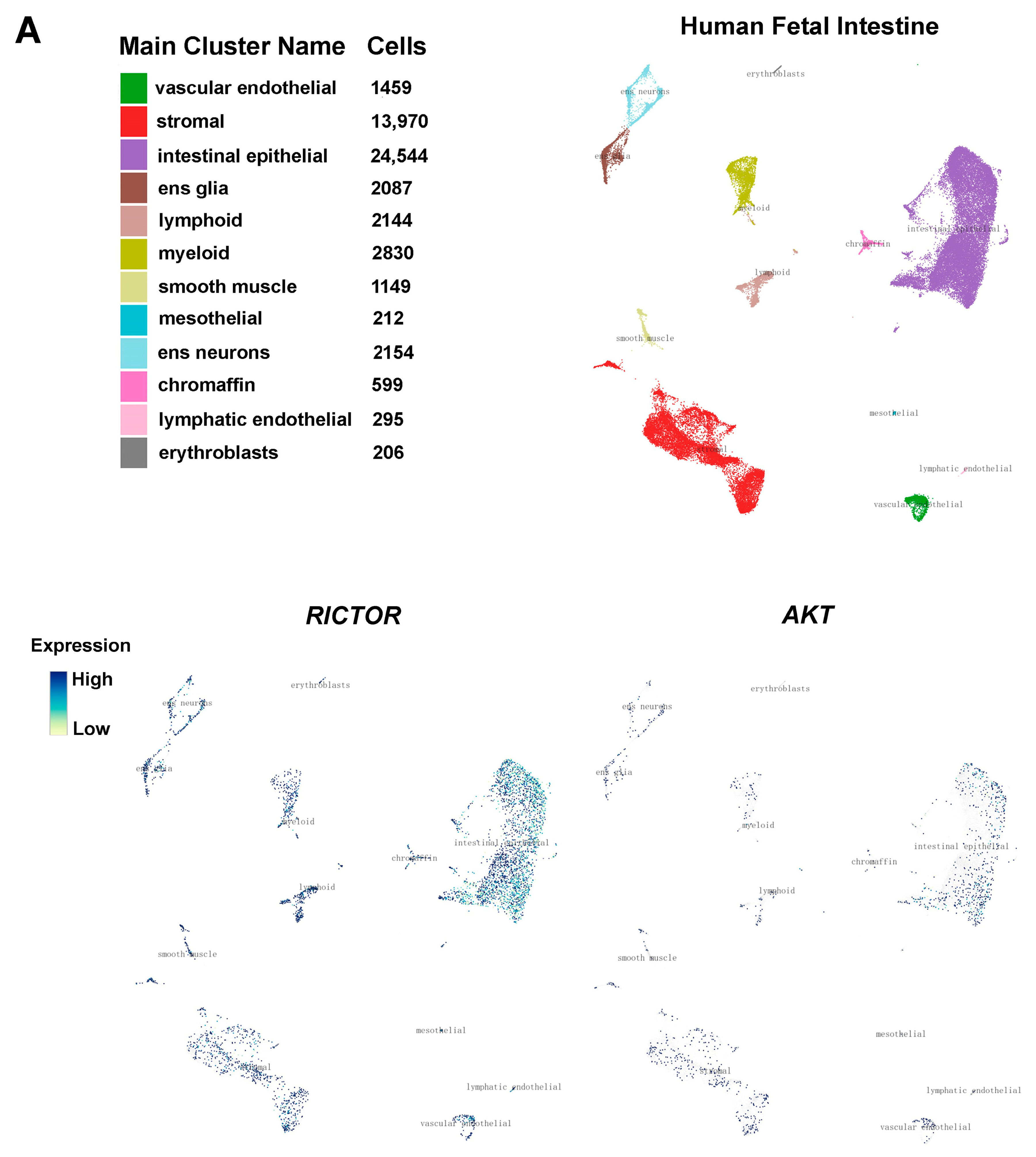
2.6. Interrogation of the Cell Type Specificity of RICTOR and AKT Expression in ENS Development
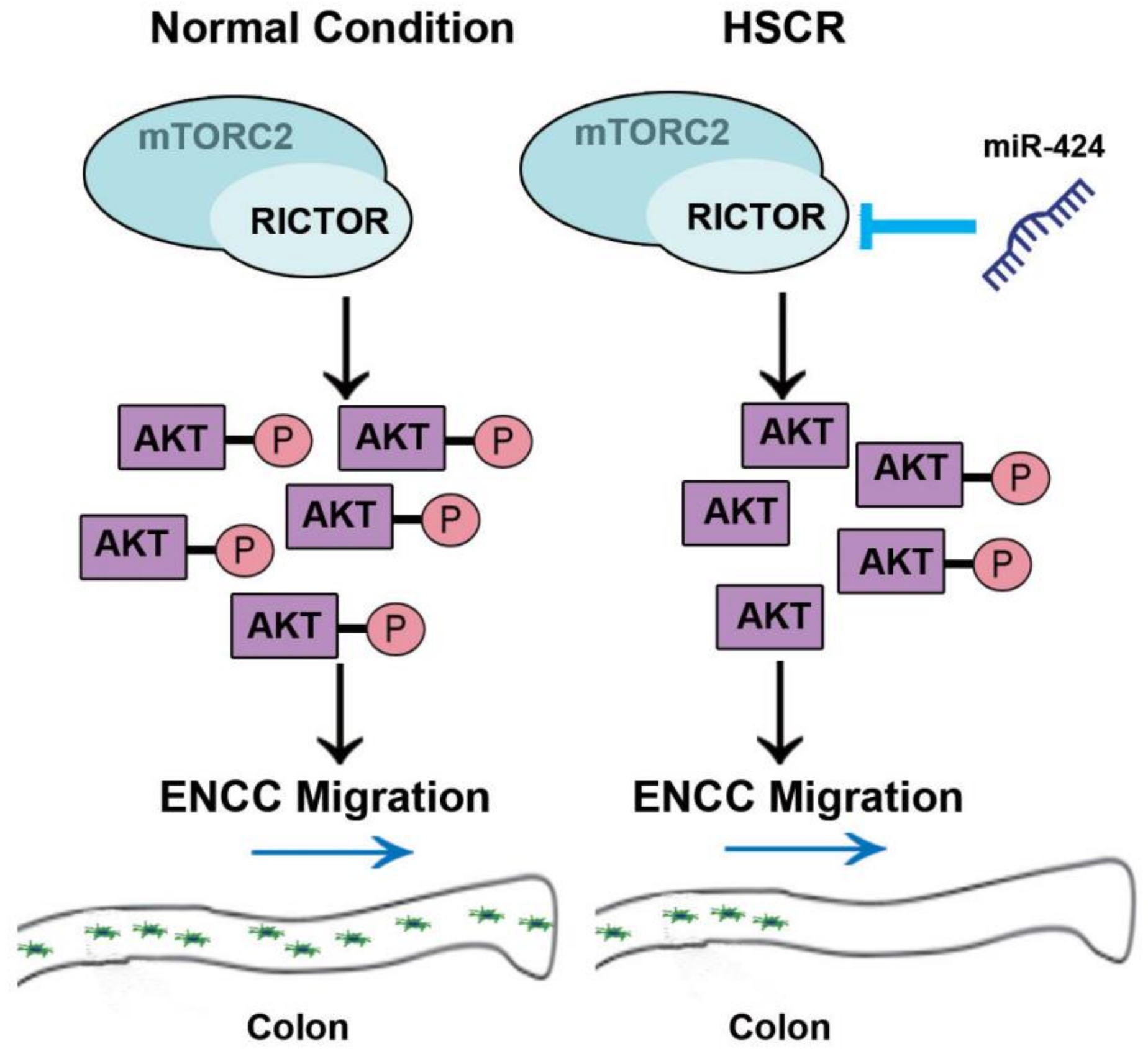
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Cell Cultures and Transfections
4.3. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
4.4. Western Blot Analysis
4.5. Validation of miR-424 Targets by Luciferase Reporter Assay
4.6. Wound Healing Assay
4.7. Mouse Strain and Primary ENCC Culture
4.8. Immunohistochemistry
4.9. Single-Cell RNA Sequencing Data Sets and Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Avetisyan, M.; Schill, E.M.; Heuckeroth, R.O. Building a second brain in the bowel. J. Clin. Investig. 2015, 125, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Graham, K.D.; Lopez, S.H.; Sengupta, R.; Shenoy, A.; Schneider, S.; Wright, C.M.; Feldman, M.; Furth, E.; Valdivieso, F.; Lemke, A.; et al. Robust, 3-Dimensional Visualization of Human Colon Enteric Nervous System without Tissue Sectioning. Gastroenterology 2020, 158, 2221–2235.e5. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.S.; Li, P.; Lai, F.P.; Fu, A.X.; Lau, S.T.; So, M.T.; Lui, K.N.; Li, Z.; Zhuang, X.; Yu, M.; et al. Identification of Genes Associated with Hirschsprung Disease, Based on Whole-Genome Sequence Analysis, and Potential Effects on Enteric Nervous System Development. Gastroenterology 2018, 155, 1908–1922.e5. [Google Scholar] [CrossRef]
- Emison, E.S.; Garcia-Barcelo, M.; Grice, E.A.; Lantieri, F.; Amiel, J.; Burzynski, G.; Fernandez, R.M.; Hao, L.; Kashuk, C.; West, K.; et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am. J. Hum. Genet. 2010, 87, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Tilghman, J.M.; Ling, A.Y.; Turner, T.N.; Sosa, M.X.; Krumm, N.; Chatterjee, S.; Kapoor, A.; Coe, B.P.; Nguyen, K.H.; Gupta, N.; et al. Molecular Genetic Anatomy and Risk Profile of Hirschsprung’s Disease. N. Engl. J. Med. 2019, 380, 1421–1432. [Google Scholar] [CrossRef]
- Jiang, Q.; Arnold, S.; Heanue, T.; Kilambi, K.P.; Doan, B.; Kapoor, A.; Ling, A.Y.; Sosa, M.X.; Guy, M.; Jiang, Q.; et al. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability. Am. J. Hum. Genet. 2015, 96, 581–596. [Google Scholar] [CrossRef]
- Nie, X.; Zheng, J.; Ricupero, C.L.; He, L.; Jiao, K.; Mao, J.J. mTOR acts as a pivotal signaling hub for neural crest cells during craniofacial development. PLoS Genet. 2018, 14, e1007491. [Google Scholar] [CrossRef]
- Carson, R.P.; Fu, C.; Winzenburger, P.; Ess, K.C. Deletion of Rictor in neural progenitor cells reveals contributions of mTORC2 signaling to tuberous sclerosis complex. Hum. Mol. Genet. 2013, 22, 140–152. [Google Scholar] [CrossRef]
- Hagan, G.N.; Lin, Y.; Magnuson, M.A.; Avruch, J.; Czech, M.P. A Rictor-Myo1c complex participates in dynamic cortical actin events in 3T3-L1 adipocytes. Mol. Cell Biol. 2008, 28, 4215–4226. [Google Scholar] [CrossRef]
- McDonald, P.C.; Oloumi, A.; Mills, J.; Dobreva, I.; Maidan, M.; Gray, V.; Wederell, E.D.; Bally, M.B.; Foster, L.J.; Dedhar, S. Rictor and integrin-linked kinase interact and regulate Akt phosphorylation and cancer cell survival. Cancer Res. 2008, 68, 1618–1624. [Google Scholar] [CrossRef]
- Dada, S.; Demartines, N.; Dormond, O. mTORC2 regulates PGE2-mediated endothelial cell survival and migration. Biochem. Biophys. Res. Commun. 2008, 372, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.K.; Chen, C.H.; Cho, H.; Boulbes, D.R.; Spooner, E.; Sarbassov, D.D. Rictor regulates cell migration by suppressing RhoGDI2. Oncogene 2013, 32, 2521–2526. [Google Scholar] [CrossRef]
- Olde Loohuis, N.F.; Kos, A.; Martens, G.J.; Van Bokhoven, H.; Nadif Kasri, N.; Aschrafi, A. MicroRNA networks direct neuronal development and plasticity. Cell Mol. Life Sci. 2012, 69, 89–102. [Google Scholar] [CrossRef]
- Wang, Y.; Veremeyko, T.; Wong, A.H.; El Fatimy, R.; Wei, Z.; Cai, W.; Krichevsky, A.M. Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer’s disease. Neurobiol. Aging 2017, 51, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug. Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Li, Y.; Li, H.; Zhou, L.; Wang, B.; Zhi, Z.; Tang, W. Molecular function predictions and diagnostic value analysis of plasma exosomal miRNAs in Hirschsprung’s disease. Epigenomics 2020, 12, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, Q.; Chakravarti, A.; Cai, H.; Xu, Z.; Wu, W.; Gu, B.; Li, L.; Cai, W. MicroRNA-4516-mediated regulation of MAPK10 relies on 3’ UTR cis-acting variants and contributes to the altered risk of Hirschsprung disease. J. Med. Genet. 2020, 57, 634–642. [Google Scholar] [CrossRef]
- Merlet, E.; Atassi, F.; Motiani, R.K.; Mougenot, N.; Jacquet, A.; Nadaud, S.; Capiod, T.; Trebak, M.; Lompre, A.M.; Marchand, A. miR-424/322 regulates vascular smooth muscle cell phenotype and neointimal formation in the rat. Cardiovasc. Res. 2013, 98, 458–468. [Google Scholar] [CrossRef]
- Gheidari, F.; Arefian, E.; Adegani, F.J.; Kalhori, M.R.; Seyedjafari, E.; Kabiri, M.; Teimoori-Toolabi, L.; Soleimani, M. miR-424 induces apoptosis in glioblastoma cells and targets AKT1 and RAF1 oncogenes from the ERBB signaling pathway. Eur. J. Pharmacol. 2021, 906, 174273. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiao, P.; Zhong, Y.; Ji, H.; Zhang, Y.; Song, H.; Du, H.; Ding, X.; Wu, H. The Endoplasmic Reticulum-Stressed Head and Neck Squamous Cell Carcinoma Cells Induced Exosomal miR-424-5p Inhibits Angiogenesis and Migration of Humanumbilical Vein Endothelial Cells through LAMC1-Mediated Wnt/beta-Catenin Signaling Pathway. Cell Transplant. 2022, 31, 9636897221083549. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, N.; Hosseinpourfeizi, M.A.; Khojasteh, S.M.B.; Baradaran, B.; Safaralizadeh, R. Tumor suppressive activity of miR-424-5p in breast cancer cells through targeting PD-L1 and modulating PTEN/PI3K/AKT/mTOR signaling pathway. Life Sci. 2020, 259, 118239. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, T.; Zhao, Z.; Wang, Z.; Liu, S.; Yang, X. SPTBN2 regulated by miR-424-5p promotes endometrial cancer progression via CLDN4/PI3K/AKT axis. Cell Death Discov. 2021, 7, 382. [Google Scholar] [CrossRef]
- Senoo, H.; Kamimura, Y.; Kimura, R.; Nakajima, A.; Sawai, S.; Sesaki, H.; Iijima, M. Phosphorylated Rho-GDP directly activates mTORC2 kinase towards AKT through dimerization with Ras-GTP to regulate cell migration. Nat. Cell Biol. 2019, 21, 867–878. [Google Scholar] [CrossRef]
- Peng, B.; Ortega, J.; Gu, L.; Chang, Z.; Li, G.M. Phosphorylation of proliferating cell nuclear antigen promotes cancer progression by activating the ATM/Akt/GSK3beta/Snail signaling pathway. J. Biol. Chem. 2019, 294, 7037–7045. [Google Scholar] [CrossRef]
- Mathavan, K.; Khedgikar, V.; Bartolo, V.; Alfandari, D. The ectodomain of cadherin-11 binds to erbB2 and stimulates Akt phosphorylation to promote cranial neural crest cell migration. PLoS ONE 2017, 12, e0188963. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Yu, Y.C.; Chen, S.P.; Liang, H.C.; Lin, C.W.; Fang, K. The collective nuclear migration of p53 and phosphorylated S473 of Akt during ellipticine-mediated apoptosis in human lung epithelial cancer cells. Mol. Cell Biochem. 2015, 407, 123–133. [Google Scholar] [CrossRef]
- Fu, M.; Sato, Y.; Lyons-Warren, A.; Zhang, B.; Kane, M.A.; Napoli, J.L.; Heuckeroth, R.O. Vitamin A facilitates enteric nervous system precursor migration by reducing Pten accumulation. Development 2010, 137, 631–640. [Google Scholar] [CrossRef]
- Simpson, M.J.; Zhang, D.C.; Mariani, M.; Landman, K.A.; Newgreen, D.F. Cell proliferation drives neural crest cell invasion of the intestine. Dev. Biol. 2007, 302, 553–568. [Google Scholar] [CrossRef]
- Cao, J.; O’Day, D.R.; Pliner, H.A.; Kingsley, P.D.; Deng, M.; Daza, R.M.; Zager, M.A.; Aldinger, K.A.; Blecher-Gonen, R.; Zhang, F.; et al. A human cell atlas of fetal gene expression. Science 2020, 370, eaba7721. [Google Scholar] [CrossRef]
- Morarach, K.; Mikhailova, A.; Knoflach, V.; Memic, F.; Kumar, R.; Li, W.; Ernfors, P.; Marklund, U. Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nat. Neurosci. 2021, 24, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, A.; Ravan, H.; Mehrabani, M. Multiplex monitoring of Alzheimer associated miRNAs based on the modular logic circuit operation and doping of catalytic hairpin assembly. Biosens. Bioelectron. 2020, 170, 112710. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, Y.; Kojima, Y.; Lighthouse, J.K.; Hu, X.; Aldred, M.A.; McLean, D.L.; Park, H.; Comhair, S.A.; Greif, D.M.; et al. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat. Med. 2013, 19, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Volkers, M.; Konstandin, M.H.; Doroudgar, S.; Toko, H.; Quijada, P.; Din, S.; Joyo, A.; Ornelas, L.; Samse, K.; Thuerauf, D.J.; et al. Mechanistic target of rapamycin complex 2 protects the heart from ischemic damage. Circulation 2013, 128, 2132–2144. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yue, P.; Kim, Y.A.; Fu, H.; Khuri, F.R.; Sun, S.Y. Enhancing mammalian target of rapamycin (mTOR)-targeted cancer therapy by preventing mTOR/raptor inhibition-initiated, mTOR/rictor-independent Akt activation. Cancer Res 2008, 68, 7409–7418. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, S.; Paterson, C.; Yang, F.; Hood, V.L.; Law, A.J. PKBbeta/AKT2 deficiency impacts brain mTOR signaling, prefrontal cortical physiology, hippocampal plasticity and select murine behaviors. Mol. Psychiatry 2021, 26, 411–428. [Google Scholar] [CrossRef]
- Iaconelli, J.; Lalonde, J.; Watmuff, B.; Liu, B.; Mazitschek, R.; Haggarty, S.J.; Karmacharya, R. Lysine Deacetylation by HDAC6 Regulates the Kinase Activity of AKT in Human Neural Progenitor Cells. ACS Chem. Biol. 2017, 12, 2139–2148. [Google Scholar] [CrossRef]
- Chadha, R.; Alganem, K.; McCullumsmith, R.E.; Meador-Woodruff, J.H. mTOR kinase activity disrupts a phosphorylation signaling network in schizophrenia brain. Mol. Psychiatry 2021, 26, 6868–6879. [Google Scholar] [CrossRef]
- Roychaudhuri, R.; Snyder, S.H. Mammalian D-cysteine: A novel regulator of neural progenitor cell proliferation Mammalian D-cysteine: A novel regulator of neural progenitor cell proliferation Endogenous D-cysteine, the stereoisomer with rapid spontaneous in vitro racemization rate, has major neural roles. Bioessays 2022, 44, e2200002. [Google Scholar]
- Rozehnal, V.; Nakai, D.; Hoepner, U.; Fischer, T.; Kamiyama, E.; Takahashi, M.; Yasuda, S.; Mueller, J. Human small intestinal and colonic tissue mounted in the Ussing chamber as a tool for characterizing the intestinal absorption of drugs. Eur. J. Pharm. Sci. 2012, 46, 367–373. [Google Scholar] [CrossRef]
- Randall, K.J.; Turton, J.; Foster, J.R. Explant culture of gastrointestinal tissue: A review of methods and applications. Cell Biol. Toxicol. 2011, 27, 267–284. [Google Scholar] [CrossRef]
- Marsman, W.A.; Buskens, C.J.; Wesseling, J.G.; Offerhaus, G.J.; Bergman, J.J.; Tytgat, G.N.; van Lanschot, J.J.; Bosma, P.J. Gene therapy for esophageal carcinoma: The use of an explant model to test adenoviral vectors ex vivo. Cancer Gene Ther. 2004, 11, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Creus-Muncunill, J.; Rue, L.; Alcala-Vida, R.; Badillos-Rodriguez, R.; Romani-Aumedes, J.; Marco, S.; Alberch, J.; Perez-Otano, I.; Malagelada, C.; Perez-Navarro, E. Increased Levels of Rictor Prevent Mutant Huntingtin-Induced Neuronal Degeneration. Mol. Neurobiol. 2018, 55, 7728–7742. [Google Scholar] [CrossRef] [PubMed]
- Dahl, K.D.; Almeida, A.R.; Hathaway, H.A.; Bourne, J.; Brown, T.L.; Finseth, L.T.; Wood, T.L.; Macklin, W.B. mTORC2 loss in oligodendrocyte progenitor cells results in regional hypomyelination in the central nervous system. J. Neurosci. 2022, 43, 540–558. [Google Scholar] [CrossRef] [PubMed]
- Obermayr, F.; Hotta, R.; Enomoto, H.; Young, H.M. Development and developmental disorders of the enteric nervous system. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D.; Gershon, M.D.; Rothman, T.P. Time of origin of neurons in the murine enteric nervous system: Sequence in relation to phenotype. J. Comp. Neurol. 1991, 314, 789–798. [Google Scholar] [CrossRef]
- Popescu, D.M.; Botting, R.A.; Stephenson, E.; Green, K.; Webb, S.; Jardine, L.; Calderbank, E.F.; Polanski, K.; Goh, I.; Efremova, M.; et al. Decoding human fetal liver haematopoiesis. Nature 2019, 574, 365–371. [Google Scholar] [CrossRef]
- Parikh, K.; Antanaviciute, A.; Fawkner-Corbett, D.; Jagielowicz, M.; Aulicino, A.; Lagerholm, C.; Davis, S.; Kinchen, J.; Chen, H.H.; Alham, N.K.; et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019, 567, 49–55. [Google Scholar] [CrossRef]
- Martin, J.C.; Chang, C.; Boschetti, G.; Ungaro, R.; Giri, M.; Grout, J.A.; Gettler, K.; Chuang, L.S.; Nayar, S.; Greenstein, A.J.; et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 2019, 178, 1493–1508.e1420. [Google Scholar] [CrossRef]
- Srinivasan, S.; Anitha, M.; Mwangi, S.; Heuckeroth, R.O. Enteric neuroblasts require the phosphatidylinositol 3-kinase/Akt/Forkhead pathway for GDNF-stimulated survival. Mol. Cell Neurosci. 2005, 29, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Lindow, M.; Kauppinen, S. Discovering the first microRNA-targeted drug. J. Cell Biol. 2012, 199, 407–412. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Yan, Y.; Gu, B.; Cai, W.; Wang, Y. Up-Regulation of microRNA-424 Causes an Imbalance in AKT Phosphorylation and Impairs Enteric Neural Crest Cell Migration in Hirschsprung Disease. Int. J. Mol. Sci. 2023, 24, 6700. https://doi.org/10.3390/ijms24076700
Xu Z, Yan Y, Gu B, Cai W, Wang Y. Up-Regulation of microRNA-424 Causes an Imbalance in AKT Phosphorylation and Impairs Enteric Neural Crest Cell Migration in Hirschsprung Disease. International Journal of Molecular Sciences. 2023; 24(7):6700. https://doi.org/10.3390/ijms24076700
Chicago/Turabian StyleXu, Ze, Yingnan Yan, Beilin Gu, Wei Cai, and Yang Wang. 2023. "Up-Regulation of microRNA-424 Causes an Imbalance in AKT Phosphorylation and Impairs Enteric Neural Crest Cell Migration in Hirschsprung Disease" International Journal of Molecular Sciences 24, no. 7: 6700. https://doi.org/10.3390/ijms24076700
APA StyleXu, Z., Yan, Y., Gu, B., Cai, W., & Wang, Y. (2023). Up-Regulation of microRNA-424 Causes an Imbalance in AKT Phosphorylation and Impairs Enteric Neural Crest Cell Migration in Hirschsprung Disease. International Journal of Molecular Sciences, 24(7), 6700. https://doi.org/10.3390/ijms24076700





