LncRNA LINC02574 Inhibits Influenza A Virus Replication by Positively Regulating the Innate Immune Response
Abstract
:1. Introduction
2. Results
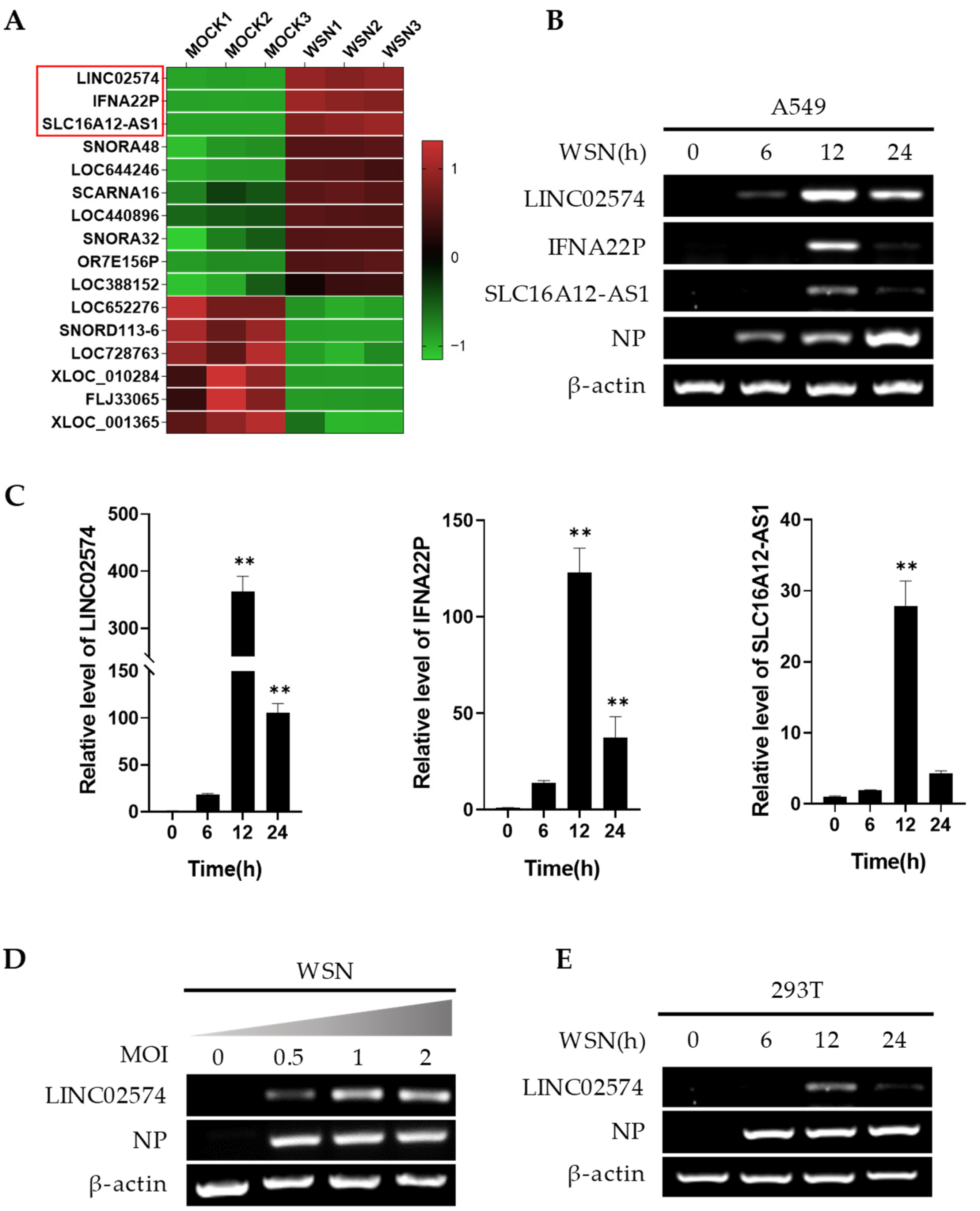
2.1. Influenza Virus Infection Upregulates LINC02574 In Vitro
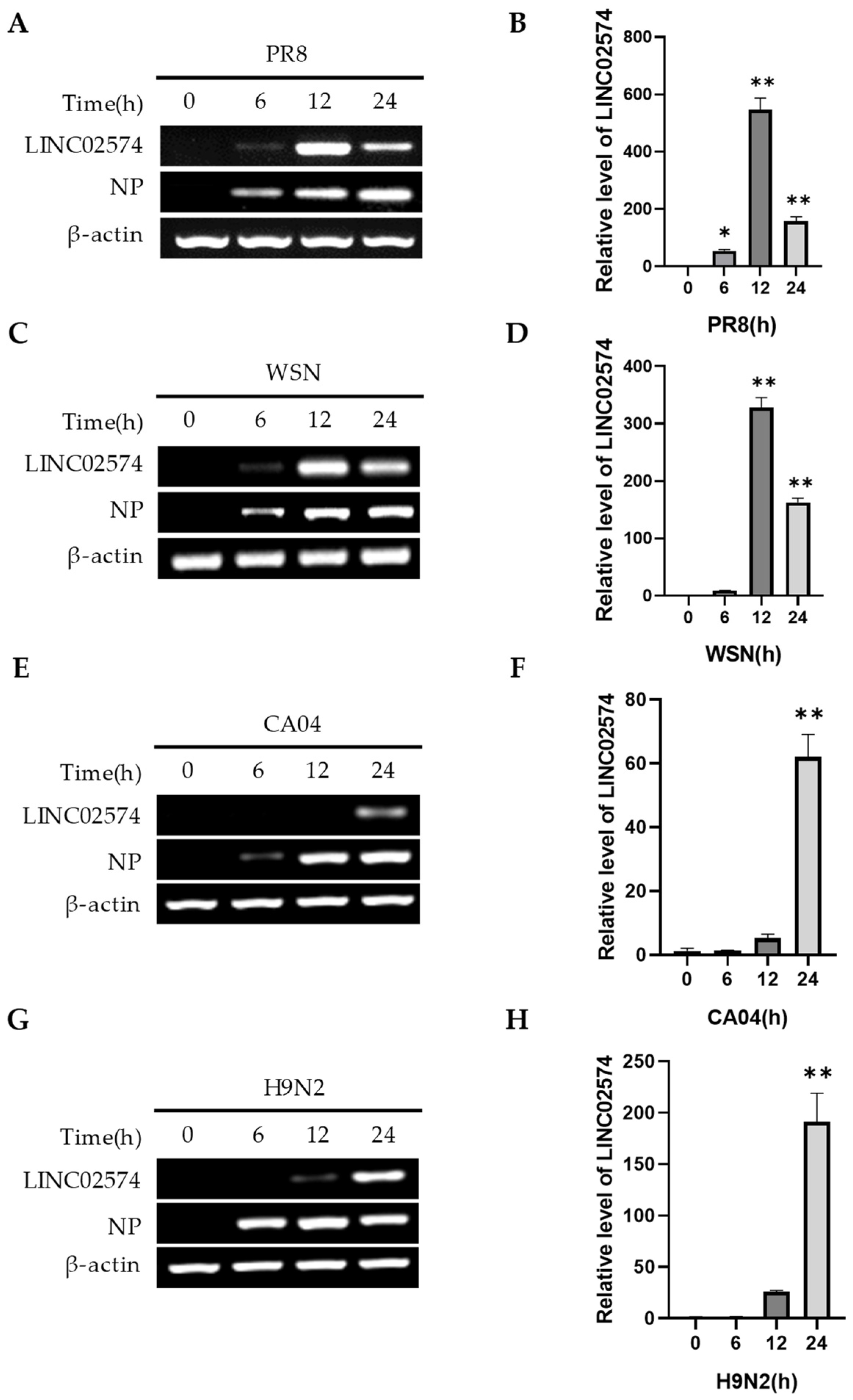
2.2. Multiple Influenza Virus Strains Can Induce LINC02574 Expression
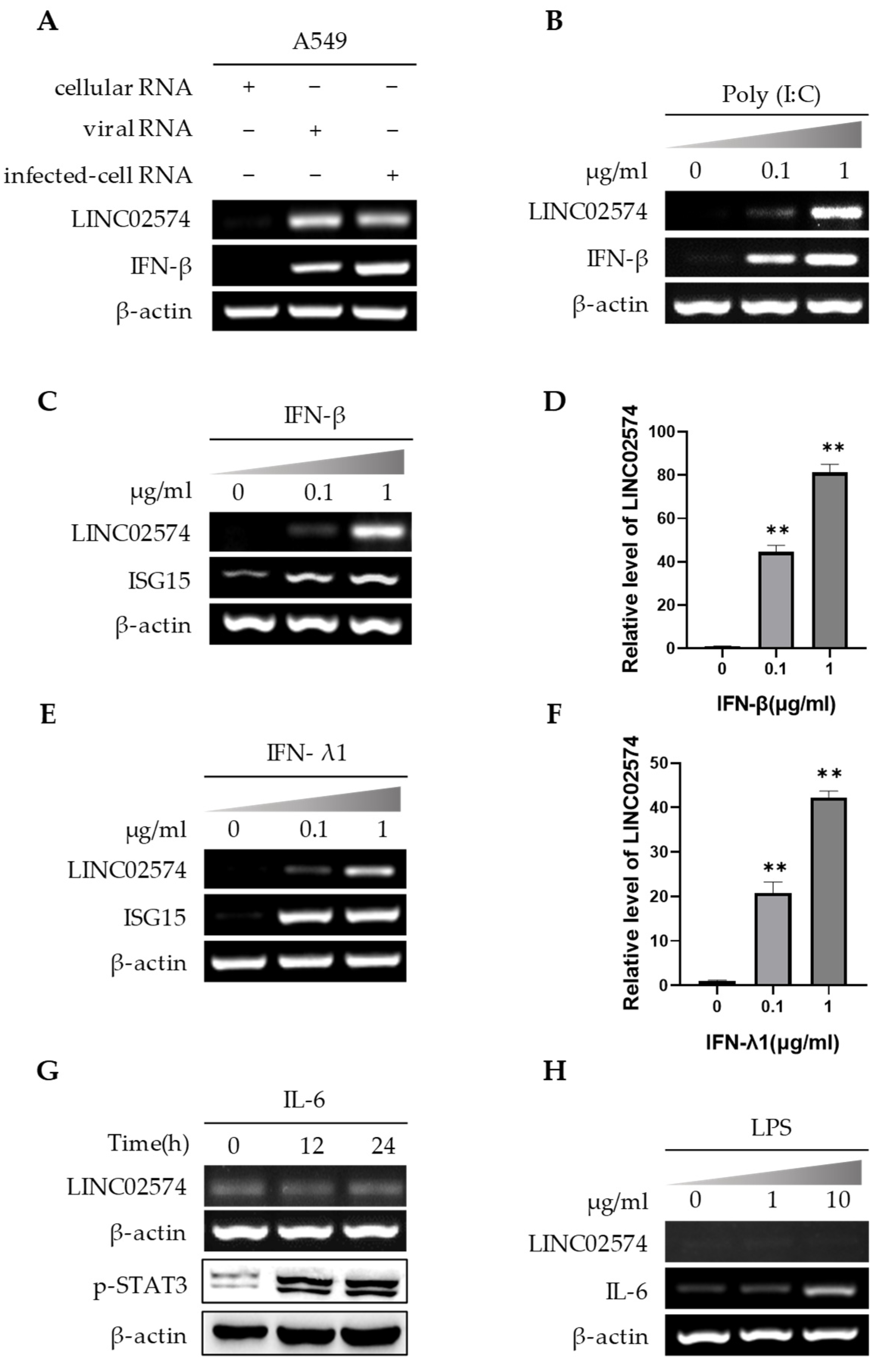
2.3. IFNs Regulate LINC02574 Expression
2.4. Influenza Virus Upregulates LINC02574 through RIG-I/IFN/IFNAR Signaling Pathway
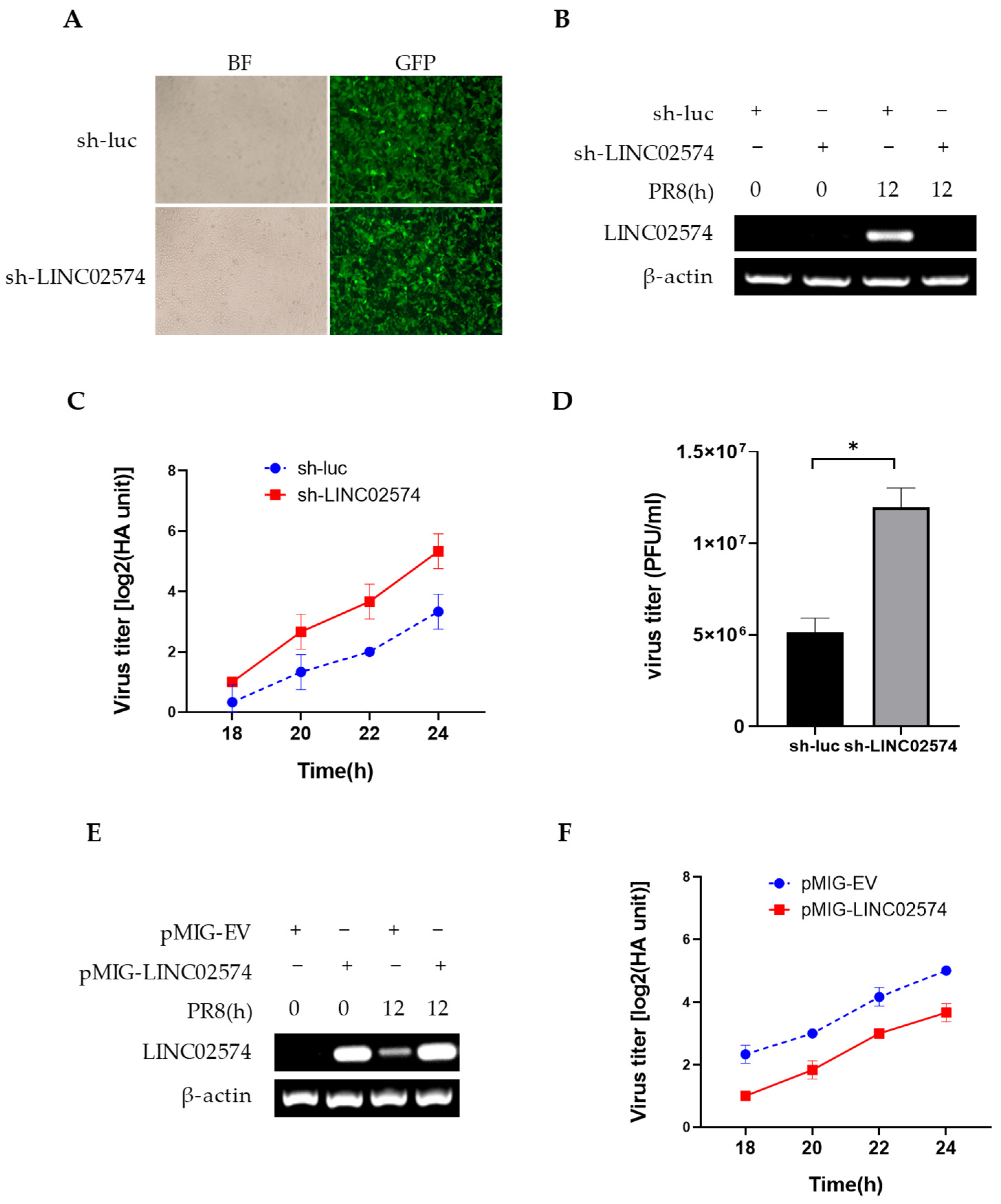
2.5. LINC02574 Inhibits the Replication of Influenza Virus
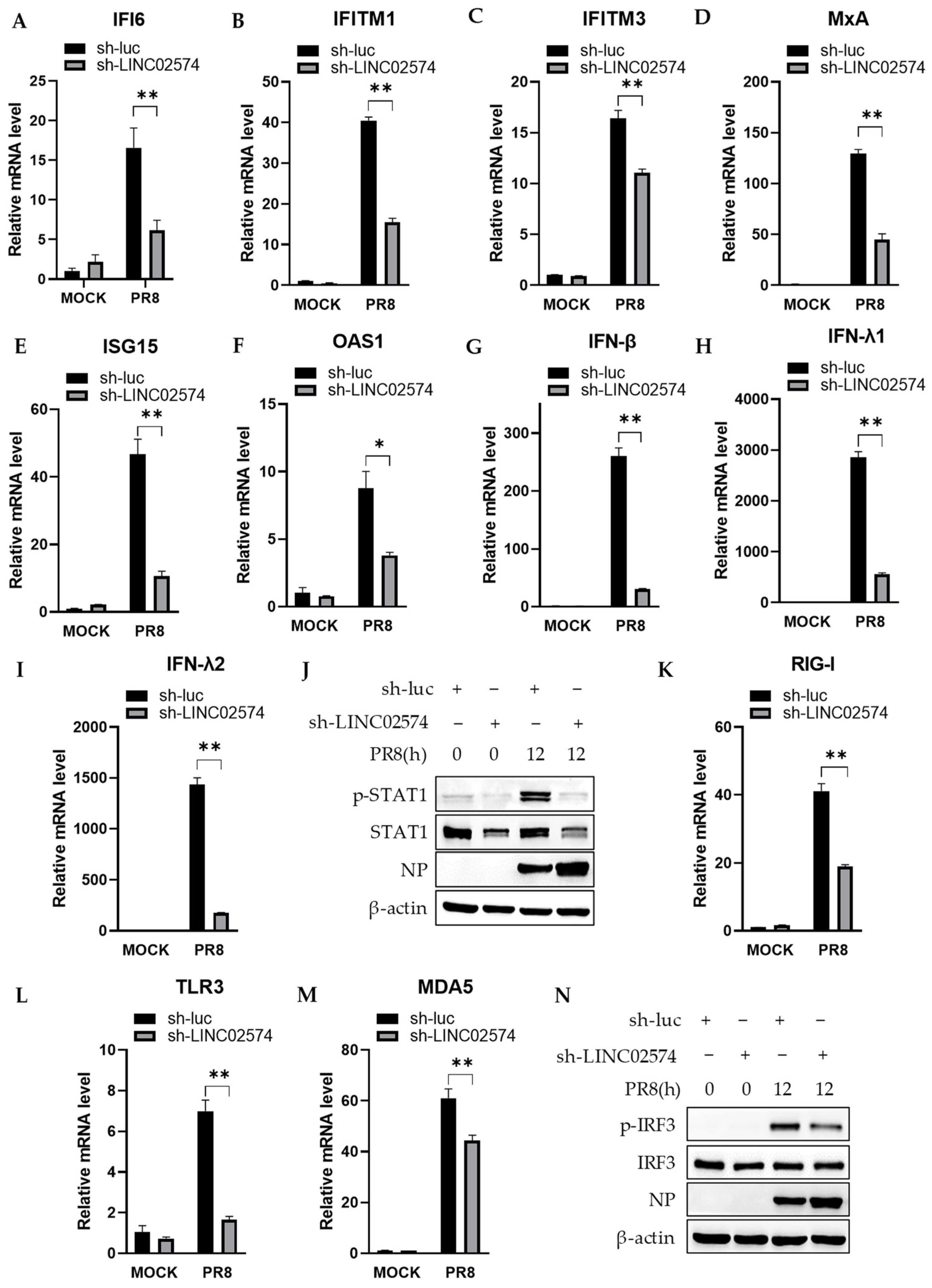
2.6. Knockdown of LINC02574 Attenuates the Innate Immune Response to Influenza Virus Infection
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Viruses and Viral Infection
4.3. Plaque Forming Assay and Hemagglutinin Assay
4.4. RT-PCR and Quantitative Real-Time PCR (qRT-PCR)
4.5. Western Blotting
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Liu, S.; Goraya, M.U.; Maarouf, M.; Huang, S.; Chen, J.L. Host immune response to influenza a virus infection. Front. Immunol. 2018, 9, 320. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin h1n1 influenza virus. Nature 2009, 459, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. The pathology of influenza virus infections. Annu. Rev. Pathol-Mech. 2008, 3, 499–522. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Li, X.; Goraya, M.U.; Wang, S.; Chen, J.-L. Evolution of influenza a virus by mutation and re-assortment. Int. J. Mol. Sci. 2017, 18, 1650. [Google Scholar] [CrossRef] [PubMed]
- Anderson, T.K.; Chang, J.; Arendsee, Z.W.; Venkatesh, D.; Souza, C.K.; Kimble, J.B.; Lewis, N.S.; Davis, C.T.; Vincent, A.L. Swine influenza a viruses and the tangled relationship with humans. Csh. Perspect. Med. 2021, 11, a038737. [Google Scholar] [CrossRef]
- Ciminski, K.; Chase, G.P.; Beer, M.; Schwemmle, M. Influenza a viruses: Understanding human host determinants. Trends. Mol. Med. 2021, 27, 104–112. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M.; Akira, S.; et al. Shared and unique functions of the dexd/h-box helicases rig-i, mda5, and lgp2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-i and melanoma differentiation-associated gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef]
- Unterholzner, L. The interferon response to intracellular DNA: Why so many receptors? Immunobiology 2013, 218, 1312–1321. [Google Scholar] [CrossRef]
- Mansur, D.S.; Smith, G.L.; Ferguson, B.J. Intracellular sensing of viral DNA by the innate immune system. Microbes Infect. 2014, 16, 1002–1012. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Dussurget, O.; Bierne, H.; Cossart, P. The bacterial pathogen listeria monocytogenes and the interferon family: Type i, type ii and type iii interferons. Front. Cell Infect. Microbiol. 2014, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat rna to the mouse x chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Martianov, I.; Ramadass, A.; Barros, A.S.; Chow, N.; Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007, 445, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding rna controls muscle differentiation by functioning as a competing endogenous rna. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Xiao, M.; Chen, Y.; Wang, S.; Liu, S.; Rai, K.R.; Chen, B.; Li, F.; Li, Y.; Maarouf, M.; Chen, J.-L. Long noncoding rna ifitm4p regulates host antiviral responses by acting as a competing endogenous rna. J. Virol. 2021, 95, e0027721. [Google Scholar] [CrossRef]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The stat3-binding long noncoding rna lnc-dc controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef]
- Jain, A.K.; Xi, Y.; McCarthy, R.; Allton, K.; Akdemir, K.C.; Patel, L.R.; Aronow, B.; Lin, C.; Li, W.; Yang, L.; et al. Lncpress1 is a p53-regulated lncrna that safeguards pluripotency by disrupting sirt6-mediated de-acetylation of histone h3k56. Mol. Cell 2016, 64, 967–981. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Luan, W.; Wang, P.; Tao, T.; Zhang, J.; Qian, J.; Liu, N.; You, Y. Long non-coding rna h19 promotes glioma cell invasion by deriving mir-675. PLoS ONE 2014, 9, e86295. [Google Scholar] [CrossRef]
- Ouyang, J.; Zhu, X.M.; Chen, Y.H.; Wei, H.T.; Chen, Q.H.; Chi, X.J.; Qi, B.M.; Zhang, L.F.; Zhao, Y.; Gao, G.F.; et al. Nrav, a long noncoding rna, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014, 16, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Maarouf, M.; Chen, B.; Chen, Y.; Wang, X.; Rai, K.R.; Zhao, Z.; Liu, S.; Li, Y.; Xiao, M.; Chen, J.-L. Identification of lncrna-155 encoded by mir155hg as a novel regulator of innate immunity against influenza a virus infection. Cell Microbiol. 2019, 21, e13036. [Google Scholar] [CrossRef]
- Rai, K.R.; Liao, Y.; Cai, M.; Qiu, H.; Wen, F.; Peng, M.; Wang, S.; Liu, S.; Guo, G.; Chi, X.; et al. Mir155hg plays a bivalent role in regulating innate antiviral immunity by encoding long noncoding rna-155 and microrna-155-5p. mBio 2022, 13, e0251022. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.; Li, J.; Shangguan, Q.; Liu, Q.; Li, X.; Qi, D.; Tong, X.; Liu, W.; Ye, X. Lnc-isg20 inhibits influenza a virus replication by enhancing isg20 expression. J. Virol. 2018, 92, e00539-18. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.C.; Zhang, Q.; Li, Z.; Tiwari, S.K.; Qin, Y.; Yau, E.; Sanchez, A.; Singh, G.; Chang, K.; Kaul, M.; et al. The long noncoding rna heal regulates hiv-1 replication through epigenetic regulation of the hiv-1 promoter. mBio 2019, 10, e2150–e7511. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Kim, H.J.; Lee, T.R. Epidermal long non-coding rnas are regulated by ultraviolet irradiation. Gene 2017, 637, 196–202. [Google Scholar] [CrossRef]
- Li, F.; Chen, Y.; Zhang, Z.; Ouyang, J.; Wang, Y.; Yan, R.; Huang, S.; Gao, G.F.; Guo, G.; Chen, J.-L. Robust expression of vault rnas induced by influenza a virus plays a critical role in suppression of pkr-mediated innate immunity. Nucleic Acids Res. 2015, 43, 10321–10337. [Google Scholar] [CrossRef]
- Mifsud, E.J.; Kuba, M.; Barr, I.G. Innate immune responses to influenza virus infections in the upper respiratory tract. Viruses-Basel 2021, 13, 2090. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; Laurent, G.S.; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding rna is elevated in alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense rna controls uchl1 translation through an embedded sineb2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.L.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. Lincrna-p21 suppresses target mrna translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef] [PubMed]
- van Solingen, C.; Cyr, Y.; Scacalossi, K.R.; de Vries, M.; Barrett, T.J.; de Jong, A.; Gourvest, M.; Zhang, T.; Peled, D.; Kher, R.; et al. Long noncoding rna chromr regulates antiviral immunity in humans. Proc. Natl. Acad. Sci. USA 2022, 119, e2210321119. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Vierbuchen, T.; Ghosh, S.; Chan, J.; Jiang, Z.; Kandasamy, R.K.; Ricci, E.; Fitzgerald, K.A. The long non-coding rna lucat1 is a negative feedback regulator of interferon responses in humans. Nat. Commun. 2020, 11, 6348. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Jiang, M.; Liu, L.; Yang, Z.; Ma, Z.; Liu, S.; Ma, Y.; Zhang, L.; Cao, X. The long noncoding rna lnczc3h7a promotes a trim25-mediated rig-i antiviral innate immune response. Nat. Immunol. 2019, 20, 812–823. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y.; et al. Self-recognition of an inducible host lncrna by rig-i feedback restricts innate immune response. Cell 2018, 173, 906–919.e13. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, S.; Tian, R.; Huang, X.; Deng, R.; Xue, B.; Qin, Y.; Xu, Y.; Wang, J.; Guo, M.; et al. Long noncoding rna itprip-1 positively regulates the innate immune response through promotion of oligomerization and activation of mda5. J. Virol. 2018, 92, e00507–e00518. [Google Scholar] [CrossRef]
- Wang, S.; Luo, X.Q.; Yan, R.X.; Wang, Q.X.; Qi, Q.Y.; Chi, X.J.; Zhang, L.L.; Yu, Z.D.; Cai, B.X.; Chen, J.L.; et al. 3-anhydro-6-hydroxy-ophiobolin a displays high in vitro and in vivo efficacy against influenza a virus infection. Protein Cell 2016, 7, 839–843. [Google Scholar] [CrossRef]
- Wang, S.; Chi, X.J.; Wei, H.T.; Chen, Y.H.; Chen, Z.L.; Huang, S.L.; Chen, J.L. Influenza a virus-induced degradation of eukaryotic translation initiation factor 4b contributes to viral replication by suppressing ifitm3 protein expression. J. Virol. 2014, 88, 8375–8385. [Google Scholar] [CrossRef]
- Liu, S.; Liao, Y.; Chen, B.; Chen, Y.; Yu, Z.; Wei, H.; Zhang, L.; Huang, S.; Rothman, P.B.; Gao, G.F.; et al. Critical role of syk-dependent stat1 activation in innate antiviral immunity. Cell Rep. 2021, 34, 108627. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, N.; Shi, W.; Yin, H.; Chi, X.; Xie, Y.; Hu, J.; Zhang, Y.; Li, H.; Chen, J.L. Co-infection of h9n2 influenza a virus and escherichia coli in a balb/c mouse model aggravates lung injury by synergistic effects. Front. Microbiol. 2021, 12, 670688. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Chi, X.; Hu, J.; Wang, S.; Zhao, S.; Mao, Y.; Peng, B.; Chen, J.-L.; Wang, S. LncRNA LINC02574 Inhibits Influenza A Virus Replication by Positively Regulating the Innate Immune Response. Int. J. Mol. Sci. 2023, 24, 7248. https://doi.org/10.3390/ijms24087248
Zhang Y, Chi X, Hu J, Wang S, Zhao S, Mao Y, Peng B, Chen J-L, Wang S. LncRNA LINC02574 Inhibits Influenza A Virus Replication by Positively Regulating the Innate Immune Response. International Journal of Molecular Sciences. 2023; 24(8):7248. https://doi.org/10.3390/ijms24087248
Chicago/Turabian StyleZhang, Yanwei, Xiaojuan Chi, Jingyun Hu, Shulin Wang, Senhong Zhao, Yanan Mao, Benqun Peng, Ji-Long Chen, and Song Wang. 2023. "LncRNA LINC02574 Inhibits Influenza A Virus Replication by Positively Regulating the Innate Immune Response" International Journal of Molecular Sciences 24, no. 8: 7248. https://doi.org/10.3390/ijms24087248




