Establishing Monoclonal Gammopathy of Undetermined Significance as an Independent Pre-Disease State of Multiple Myeloma Using Raman Spectroscopy, Dynamical Network Biomarker Theory, and Energy Landscape Analysis
Abstract
:1. Introduction
2. Results
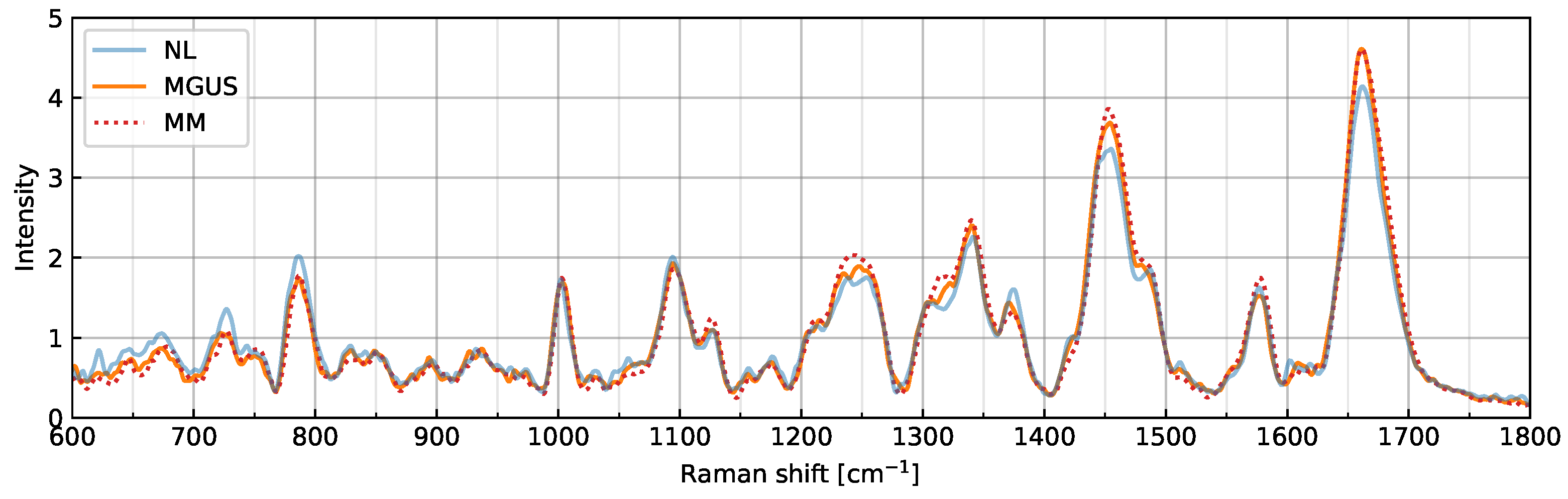
2.1. Overview of Raman Spectra
2.2. The Pre-Disease State Identified through the DNB Analysis
2.3. Frequent Spectral Patterns Revealed through the EL Analysis
3. Discussion
4. Materials and Methods
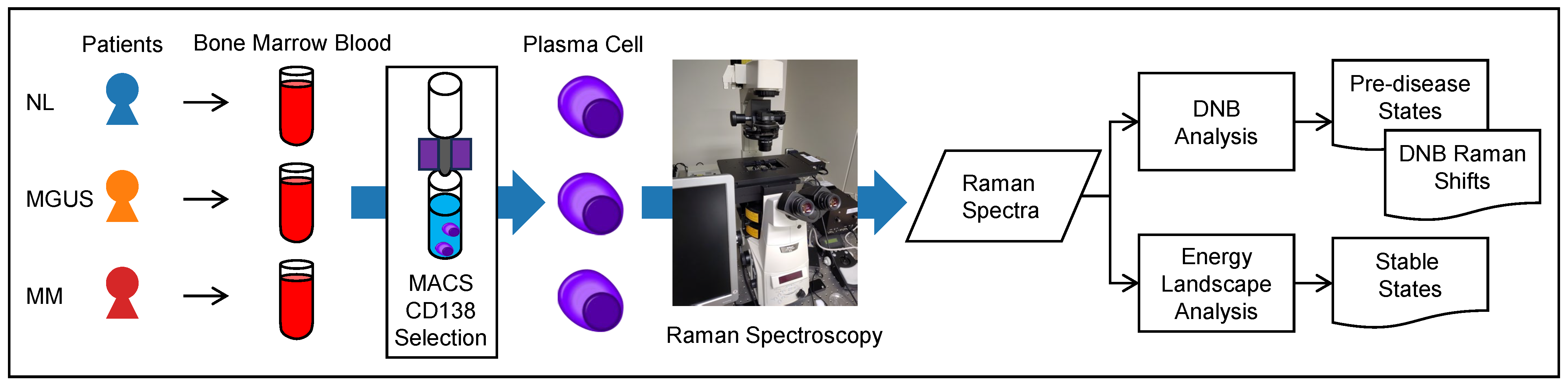
4.1. Overview of our Experimental Design
4.2. Clinical Samples
4.3. Instrumentation and Measurement: Utilizing Raman Spectroscopy
4.4. Datasets
4.5. Preprocessing
4.6. DNB Analysis
4.7. EL Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DNB | Dynamical Network Biomarker |
| EL | Energy Landscape |
| MGUS | Monoclonal Gammopathy of Undetermined Significance |
| MM | Multiple Myeloma |
| NL | Normal |
References
- Abramson, H.N. Immunotherapy of Multiple Myeloma: Current Status as Prologue to the Future. Int. J. Mol. Sci. 2023, 24, 15674. [Google Scholar] [CrossRef]
- Leow, C.C.-Y.; Low, M.S.Y. Targeted Therapies for Multiple Myeloma. J. Pers. Med. 2021, 11, 334. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef]
- Kumar, S.K.; Rajkumar, S.V. The multiple myelomas—Current concepts in cytogenetic classification and therapy. Nat. Rev. Clin. Oncol. 2018, 15, 409–421. [Google Scholar] [CrossRef]
- Kyle, R.A.; Therneau, T.M.; Rajkumar, S.V.; Larson, D.R.; Plevak, M.F.; Offord, J.R.; Dispenzieri, A.; Katzmann, J.A.; Melton, L.J. Prevalence of Monoclonal Gammopathy of Undetermined Significance. N. Engl. J. Med. 2006, 354, 1362–1369. [Google Scholar] [CrossRef]
- Kyle, R.A.; Therneau, T.M.; Rajkumar, S.V.; Offord, J.R.; Larson, D.R.; Plevak, M.F.; Melton, L.J. A Long-Term Study of Prognosis in Monoclonal Gammopathy of Undetermined Significance. N. Engl. J. Med. 2002, 346, 564–569. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Krafft, C.; Schmitt, M.; Schie, I.W.; Cialla-May, D.; Matthäus, C.; Bocklitz, T.; Popp, J. Label-Free Molecular Imaging of Biological Cells and Tissues by Linear and Nonlinear Raman Spectroscopic Approaches. Angew. Chem. 2017, 56, 4392–4430. [Google Scholar] [CrossRef]
- Dodo, K.; Fujita, K.; Sodeoka, M. Raman Spectroscopy for Chemical Biology Research. J. Am. Chem. Soc. 2022, 144, 19651–19667. [Google Scholar] [CrossRef]
- Watanabe, T.M.; Sasaki, K.; Fujita, H. Recent Advances in Raman Spectral Imaging in Cell Diagnosis and Gene Expression Prediction. Genes 2022, 13, 2127. [Google Scholar] [CrossRef]
- Notingher, I.; Jell, G.; Notingher, P.L.; Bisson, I.; Tsigkou, O.; Polak, J.M.; Stevens, M.M.; Hench, L.L. Multivariate analysis of Raman spectra for in vitro non-invasive studies of living cells. J. Mol. Struct. 2005, 744–747, 179–185. [Google Scholar] [CrossRef]
- Chan, J.W.; Taylor, D.S.; Lane, S.M.; Zwerdling, T.; Tuscano, J.; Huser, T. Nondestructive identification of individual leukemia cells by laser trapping Raman spectroscopy. Anal. Chem. 2008, 80, 2180–2187. [Google Scholar] [CrossRef]
- Ishigaki, M.; Hitomi, H.; Ozaki, Y.; Nishiyama, A. Exposing intracellular molecular changes during the differentiation of human-induced pluripotent stem cells into erythropoietin-producing cells using Raman spectroscopy and imaging. Sci. Rep. 2022, 12, 20454. [Google Scholar] [CrossRef]
- Pistiki, A.; Ramoji, A.; Ryabchykov, O.; Thomas-Rüddel, D.; Press, A.T.; Makarewicz, O.; Giamarellos-Bourboulis, E.J.; Bauer, M.; Bocklitz, T.; Popp, J.; et al. Biochemical Analysis of Leukocytes after In Vitro and In Vivo Activation with Bacterial and Fungal Pathogens Using Raman Spectroscopy. Int. J. Mol. Sci. 2021, 22, 10481. [Google Scholar] [CrossRef]
- Chen, L.; Liu, R.; Liu, Z.P.; Li, M.; Aihara, K. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2012, 2, 342. [Google Scholar] [CrossRef]
- Carpenter, S.R.; Brock, W.A. Rising variance: A leading indicator of ecological transition. Ecol. Lett. 2006, 9, 311–318. [Google Scholar] [CrossRef]
- Dakos, V.; Scheffer, M.; Van Nes, E.H.; Brovkin, V.; Petoukhov, V.; Held, H. Slowing down as an early warning signal for abrupt climate change. Proc. Natl. Acad. Sci. USA 2008, 105, 14308–14312. [Google Scholar] [CrossRef]
- Scheffer, M.; Bascompte, J.; Brock, W.A.; Brovkin, V.; Carpenter, S.R.; Dakos, V.; Held, H.; Van Nes, E.H.; Rietkerk, M.; Sugihara, G. Early-warning signals for critical transitions. Nature 2009, 461, 53–59. [Google Scholar] [CrossRef]
- Moon, H.; Lu, T.C. Network Catastrophe: Self-Organized Patterns Reveal both the Instability and the Structure of Complex Networks. Sci. Rep. 2015, 5, 9450. [Google Scholar] [CrossRef]
- Veraart, A.J.; Faassen, E.J.; Dakos, V.; Van Nes, E.H.; Lürling, M.; Scheffer, M. Recovery rates reflect distance to a tipping point in a living system. Nature 2012, 481, 357–359. [Google Scholar] [CrossRef]
- Dakos, V.; Carpenter, S.R.; Brock, W.A.; Ellison, A.M.; Guttal, V.; Ives, A.R.; Kéfi, S.; Livina, V.; Seekell, D.A.; Van Nes, E.H.; et al. Methods for Detecting Early Warnings of Critical Transitions in Time Series Illustrated Using Simulated Ecological Data. PLoS ONE 2012, 7, e41010. [Google Scholar] [CrossRef]
- Olthof, M.; Hasselman, F.; Strunk, G.; Van Rooij, M.; Aas, B.; Helmich, M.A.; Schiepek, G.; Lichtwarck-Aschoff, A. Critical Fluctuations as an Early-Warning Signal for Sudden Gains and Losses in Patients Receiving Psychotherapy for Mood Disorders. Clin. Psychol. Sci. 2020, 8, 25–35. [Google Scholar] [CrossRef]
- Bury, T.M.; Sujith, R.I.; Pavithran, I.; Scheffer, M.; Lenton, T.M.; Anand, M.; Bauch, C.T. Deep learning for early warning signals of tipping points. Proc. Natl. Acad. Sci. USA 2021, 118, e2106140118. [Google Scholar] [CrossRef]
- Liu, R.; Li, M.; Liu, Z.-P.; Wu, J.; Chen, L.; Aihara, K. Identifying critical transitions and their leading biomolecular networks in complex diseases. Sci. Rep. 2012, 2, 813. [Google Scholar] [CrossRef]
- Chen, P.; Liu, R.; Li, Y.; Chen, L. Detecting critical state before phase transition of complex biological systems by hidden Markov model. Bioinformatics 2016, 32, 2143–2150. [Google Scholar] [CrossRef]
- Liu, R.; Yu, X.; Liu, X.; Xu, D.; Aihara, K.; Chen, L. Identifying critical transitions of complex diseases based on a single sample. Bioinformatics 2014, 30, 1579–1586. [Google Scholar] [CrossRef]
- Yang, B.; Li, M.; Tang, W.; Liu, W.; Zhang, S.; Chen, L.; Xia, J. Dynamic network biomarker indicates pulmonary metastasis at the tipping point of hepatocellular carcinoma. Nat. Commun. 2018, 9, 678. [Google Scholar] [CrossRef]
- Jiang, F.; Yang, L.; Jiao, X. Dynamic network biomarker to determine the critical point of breast cancer stage progression. Breast Cancer 2023, 30, 453–465. [Google Scholar] [CrossRef]
- Liu, X.; Liu, R.; Zhao, X.-M.; Chen, L. Detecting early-warning signals of type 1 diabetes and its leading biomolecular networks by dynamical network biomarkers. BMC Med. Genom. 2013, 6, S8. [Google Scholar] [CrossRef]
- Li, M.; Zeng, T.; Liu, R.; Chen, L. Detecting tissue-specific early warning signals for complex diseases based on dynamical network biomarkers: Study of type 2 diabetes by cross-tissue analysis. Brief Bioinform. 2014, 15, 229–243. [Google Scholar] [CrossRef]
- Koizumi, K.; Oku, M.; Hayashi, S.; Inujima, A.; Shibahara, N.; Chen, L.; Igarashi, Y.; Tobe, K.; Saito, S.; Kadowaki, M.; et al. Identifying pre-disease signals before metabolic syndrome in mice by dynamical network biomarkers. Sci. Rep. 2019, 9, 8767. [Google Scholar] [CrossRef]
- Liu, X.; Chang, X.; Leng, S.; Tang, H.; Aihara, K.; Chen, L. Detection for disease tipping points by landscape dynamic network biomarkers. Natl. Sci. Rev. 2019, 6, 775–785. [Google Scholar] [CrossRef]
- Ge, J.; Song, C.; Zhang, C.; Liu, X.; Chen, J.; Dou, K.; Chen, L. Personalized Early-Warning Signals during Progression of Human Coronary Atherosclerosis by Landscape Dynamic Network Biomarker. Genes 2020, 11, 676. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, H.; Ge, J.; Mi, T.; Cui, X.; Tu, F.; Gu, X.; Zeng, T.; Chen, L. Landscape dynamic network biomarker analysis reveals the tipping point of transcriptome reprogramming to prevent skin photodamage. J. Mol. Cell Biol. 2022, 13, 822–833. [Google Scholar] [CrossRef]
- Haruki, T.; Yonezawa, S.; Koizumi, K.; Yoshida, Y.; Watanabe, T.M.; Fujita, H.; Oshima, Y.; Oku, M.; Taketani, A.; Yamazaki, M.; et al. Application of the Dynamical Network Biomarker Theory to Raman Spectra. Biomolecules 2022, 12, 1730. [Google Scholar] [CrossRef]
- Watanabe, T.; Masuda, N.; Megumi, F.; Kanai, R.; Rees, G. Energy landscape and dynamics of brain activity during human bistable perception. Nat. Commun. 2014, 5, 4765. [Google Scholar] [CrossRef]
- Ezaki, T.; Watanabe, T.; Ohzeki, M.; Masuda, N. Energy landscape analysis of neuroimaging data. Philos. Trans. R. Soc. A 2017, 375, 20160287. [Google Scholar] [CrossRef]
- Watanabe, T.; Rees, G. Brain network dynamics in high-functioning individuals with autism. Nat. Commun. 2017, 8, 16048. [Google Scholar] [CrossRef]
- Suzuki, K.; Nakaoka, S.; Fukuda, S.; Masuya, H. Energy landscape analysis elucidates the multistability of ecological communities across environmental gradients. Ecol. Monogr. 2021, 91, e01469. [Google Scholar] [CrossRef]
- Fujita, H.; Ushio, M.; Suzuki, K.; Abe, M.S.; Yamamichi, M.; Iwayama, K.; Canarini, A.; Hayashi, I.; Fukushima, K.; Fukuda, S.; et al. Alternative stable states, nonlinear behavior, and predictability of microbiome dynamics. Microbiome 2023, 11, 63. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Facon, T.; Daviet, A.; Godon, C.; Rapp, M.J.; Harousseau, J.L.; Grosbois, B.; Bataille, R. 14q32 translocations and monosomy 13 observed in monoclonal gammopathy of undetermined significance delineate a multistep process for the oncogenesis of multiple myeloma. Intergroupe Francophone du Myélome. Cancer Res. 1999, 59, 4546–4550. [Google Scholar]
- Harada, H.; Kawano, M.M.; Huang, N.; Harada, Y.; Iwato, K.; Tanabe, O.; Tanaka, H.; Sakai, A.; Asaoku, H.; Kuramoto, A. Phenotypic difference of normal plasma cells from mature myeloma cells. Blood 1993, 81, 2658–2663. [Google Scholar] [CrossRef]
- Tembhare, P.R.; Yuan, C.M.; Venzon, D.; Braylan, R.; Korde, N.; Manasanch, E.; Zuchlinsky, D.; Calvo, K.; Kurlander, R.; Bhutani, M.; et al. Flow cytometric differentiation of abnormal and normal plasma cells in the bone marrow in patients with multiple myeloma and its precursor diseases. Leuk. Res. 2014, 38, 371–376. [Google Scholar] [CrossRef]
- Sato, T.; Tatekoshi, A.; Takada, K.; Iyama, S.; Kamihara, Y.; Jawaid, P.; Rehman, M.U.; Noguchi, K.; Kondo, T.; Kajikawa, S.; et al. DPP8 is a novel therapeutic target for multiple myeloma. Sci. Rep. 2019, 9, 18094. [Google Scholar] [CrossRef]
- Racanelli, V.; Leone, P.; Frassanito, M.A.; Brunetti, C.; Perosa, F.; Ferrone, S.; Dammacco, F. Alterations in the antigen processing-presenting machinery of transformed plasma cells are associated with reduced recognition by CD8+ T cells and characterize the progression of MGUS to multiple myeloma. Blood 2010, 115, 1185–1193. [Google Scholar] [CrossRef]
- Ocqueteau, M.; Orfao, A.; Almeida, J.; Bladé, J.; González, M.; García-Sanz, R.; López-Berges, C.; Moro, M.J.; Hernández, J.; Escribano, L.; et al. Immunophenotypic characterization of plasma cells from monoclonal gammopathy of undetermined significance patients. Implications for the differential diagnosis between MGUS and multiple myeloma. Am. J. Pathol. 1998, 152, 1655–1665. [Google Scholar]
- Lacy, M.Q.; Donovan, K.A.; Heimbach, J.K.; Ahmann, G.J.; Lust, J.A. Comparison of interleukin-1 beta expression by in situ hybridization in monoclonal gammopathy of undetermined significance and multiple myeloma. Blood 1999, 93, 300–305. [Google Scholar] [CrossRef]
- Nakayama-Ichiyama, S.; Yokote, T.; Hirata, Y.; Iwaki, K.; Akioka, T.; Miyoshi, T.; Takayama, A.; Nishiwaki, U.; Masuda, Y.; Nishimura, Y.; et al. Immunohistological diagnosis of plasma cell myeloma based on cytoplasmic kappa/lambda ratio of CD138-positive plasma cells. Leuk. Lymphoma 2012, 53, 2205–2209. [Google Scholar] [CrossRef]
- Pojero, F.; Flores-Montero, J.; Sanoja, L.; Pérez, J.J.; Puig, N.; Paiva, B.; Bottcher, S.; Van Dongen, J.J.; Orfao, A.; EuroFlow group. Utility of CD54, CD229, and CD319 for the identification of plasma cells in patients with clonal plasma cell diseases. Cytom. Part B Clin. Cytom. 2016, 90, 91–100. [Google Scholar] [CrossRef]
- Asaoka, R.; Kiyomatsu, H.; Miura, H.; Jono, A.; Kinoshita, T.; Takao, M.; Katagiri, T.; Oshima, Y. Prognostic potential and pathological validation of a diagnostic application using Raman spectroscopy in the characterization of degenerative changes in the cartilage of the humeral head. J. Biomed. Opt. 2022, 27, 115002. [Google Scholar] [CrossRef]



| NL | MGUS | MM | Total | |||
|---|---|---|---|---|---|---|
| Patients | 25 | 21 | 24 | 70 | ||
| Plasma cells | 396 | 347 | 471 | 1214 | ||
| max. | 21 | 25 | 36 | |||
| median | (cells/patient) | 15 | 15 | 15 | ||
| min. | 9 | 9 | 5 | |||
| Raman spectra | 834 | 711 | 970 | 2515 | ||
| max. | 4 | 4 | 4 | |||
| median | (measurements/cell) | 2 | 2 | 2 | ||
| min. | 2 | 1 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yonezawa, S.; Haruki, T.; Koizumi, K.; Taketani, A.; Oshima, Y.; Oku, M.; Wada, A.; Sato, T.; Masuda, N.; Tahara, J.; et al. Establishing Monoclonal Gammopathy of Undetermined Significance as an Independent Pre-Disease State of Multiple Myeloma Using Raman Spectroscopy, Dynamical Network Biomarker Theory, and Energy Landscape Analysis. Int. J. Mol. Sci. 2024, 25, 1570. https://doi.org/10.3390/ijms25031570
Yonezawa S, Haruki T, Koizumi K, Taketani A, Oshima Y, Oku M, Wada A, Sato T, Masuda N, Tahara J, et al. Establishing Monoclonal Gammopathy of Undetermined Significance as an Independent Pre-Disease State of Multiple Myeloma Using Raman Spectroscopy, Dynamical Network Biomarker Theory, and Energy Landscape Analysis. International Journal of Molecular Sciences. 2024; 25(3):1570. https://doi.org/10.3390/ijms25031570
Chicago/Turabian StyleYonezawa, Shota, Takayuki Haruki, Keiichi Koizumi, Akinori Taketani, Yusuke Oshima, Makito Oku, Akinori Wada, Tsutomu Sato, Naoki Masuda, Jun Tahara, and et al. 2024. "Establishing Monoclonal Gammopathy of Undetermined Significance as an Independent Pre-Disease State of Multiple Myeloma Using Raman Spectroscopy, Dynamical Network Biomarker Theory, and Energy Landscape Analysis" International Journal of Molecular Sciences 25, no. 3: 1570. https://doi.org/10.3390/ijms25031570





