Virtual Screening of Peptide Libraries: The Search for Peptide-Based Therapeutics Using Computational Tools
Abstract
:1. Introduction
1.1. Targeting PPIs with Peptides
1.2. Brief Overview of Peptide Therapeutic Potential: Major Drawbacks and Optimization Routes
2. Identifying In Silico Novel Bioactive Peptides: Methodological Aspects
2.1. Design of Virtual Peptide Libraries
2.2. Virtual Screening Approaches in Brief
Limitations of Virtual Screening Approaches
3. Identifying Novel Bioactive Peptides through In Silico Approaches: Applications
3.1. General Overview
3.2. Anticancer Peptides
3.2.1. Introduction
3.2.2. Case Studies Related to Cancer Research

3.3. Antimicrobial/Antiviral Peptides
3.3.1. Introduction
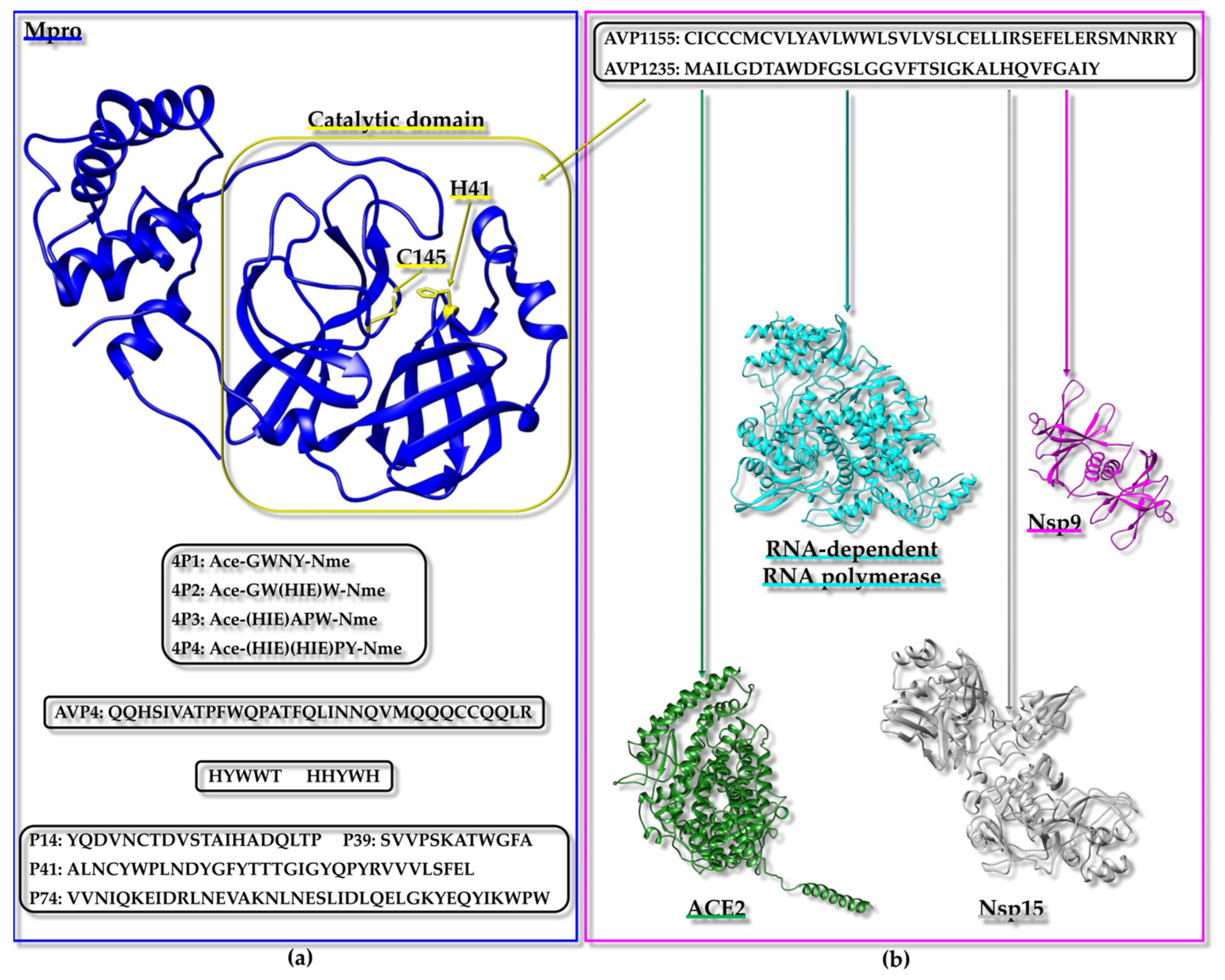
3.3.2. Case Studies: Targeting SARS-CoV-2

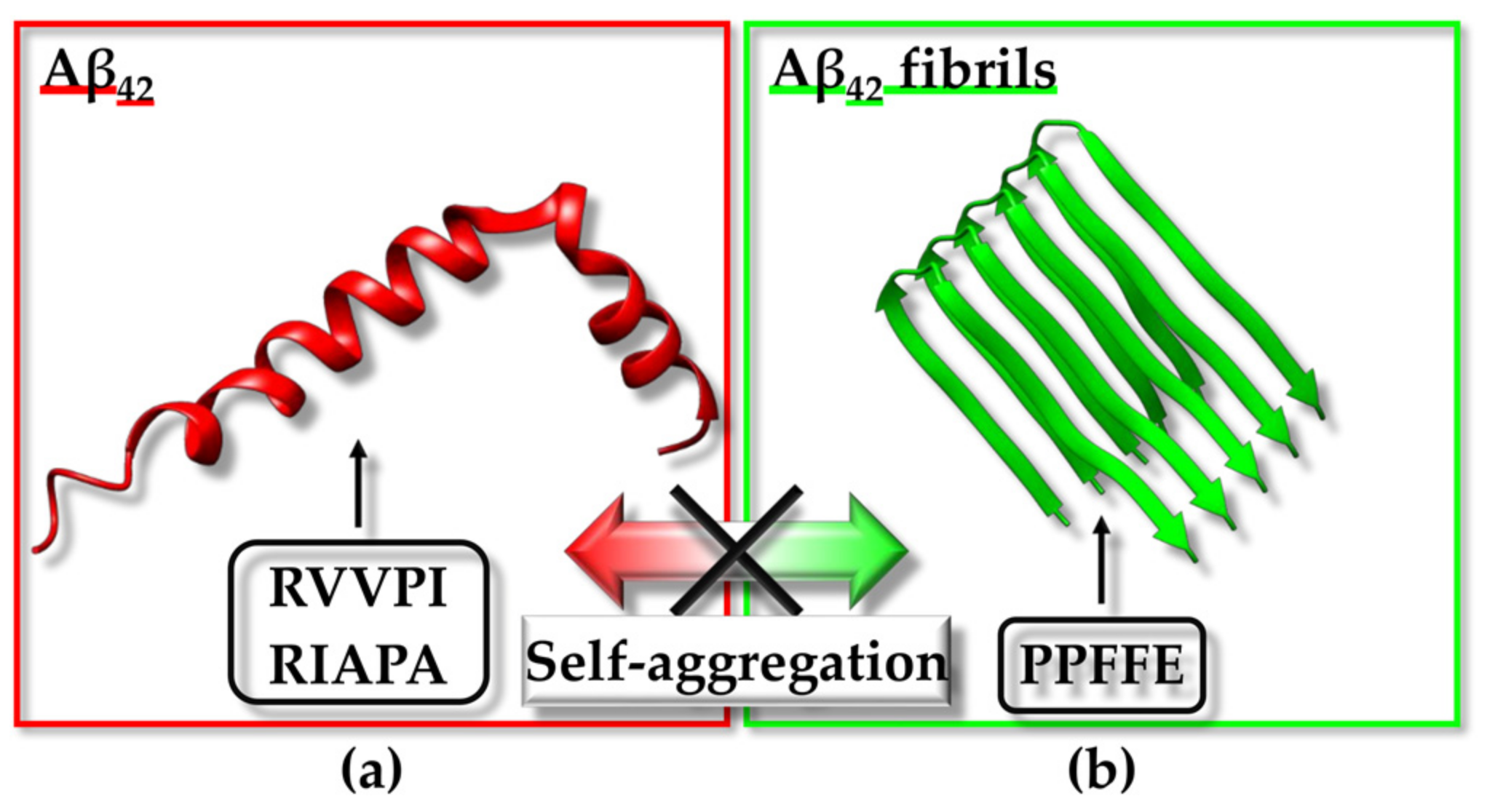
3.4. Inhibiting Fibril Formation: Aβ42 and hIAPP as Targets

4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gasbarri, C.; Rosignoli, S.; Janson, G.; Boi, D.; Paiardini, A. Prediction and Modeling of Protein-Protein Interactions Using “Spotted” Peptides with a Template-Based Approach. Biomolecules 2022, 12, 201. [Google Scholar] [CrossRef] [PubMed]
- Bruzzoni-Giovanelli, H.; Alezra, V.; Wolff, N.; Dong, C.Z.; Tuffery, P.; Rebollo, A. Interfering peptides targeting protein-protein interactions: The next generation of drugs? Drug Discov. Today 2018, 23, 272–285. [Google Scholar] [CrossRef]
- Hashemi, Z.S.; Zarei, M.; Fath, M.K.; Ganji, M.; Farahani, M.S.; Afsharnouri, F.; Pourzardosht, N.; Khalesi, B.; Jahangiri, A.; Rahbar, M.R.; et al. In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein-Protein Interactions. Front. Mol. Biosci. 2021, 8, 669431. [Google Scholar] [CrossRef] [PubMed]
- Anand, U.; Bandyopadhyay, A.; Jha, N.K.; Perez de la Lastra, J.M.; Dey, A. Translational aspect in peptide drug discovery and development: An emerging therapeutic candidate. Biofactors 2023, 49, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Skolnick, J.; Gao, M.; Zhou, H.; Singh, S. AlphaFold 2: Why It Works and Its Implications for Understanding the Relationships of Protein Sequence, Structure, and Function. J. Chem. Inf. Model. 2021, 61, 4827–4831. [Google Scholar] [CrossRef] [PubMed]
- Bertoline, L.M.F.; Lima, A.N.; Krieger, J.E.; Teixeira, S.K. Before and after AlphaFold2: An overview of protein structure prediction. Front. Bioinform. 2023, 3, 1120370. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.L.; Juergens, D.; Bennett, N.R.; Trippe, B.L.; Yim, J.; Eisenach, H.E.; Ahern, W.; Borst, A.J.; Ragotte, R.J.; Milles, L.F.; et al. De novo design of protein structure and function with RFdiffusion. Nature 2023, 620, 1089–1100. [Google Scholar] [CrossRef]
- Swinney, D.C. Phenotypic vs. target-based drug discovery for first-in-class medicines. Clin. Pharmacol. Ther. 2013, 93, 299–301. [Google Scholar] [CrossRef]
- Diller, D.J.; Jarosinski, M.; Sawyer, T.K.; Audie, J. Peptide drug discovery: Innovative technologies and transformational medicines. In Advances in the Discovery and Development of Peptide Therapeutics; Kruger, G., Albericio, F., Eds.; Future Medicine: London, UK, 2015; pp. 8–27. [Google Scholar] [CrossRef]
- Winkler, D.F.; Campbell, W.D. The spot technique: Synthesis and screening of peptide macroarrays on cellulose membranes. Methods Mol. Biol. 2008, 494, 47–70. [Google Scholar] [CrossRef]
- De Vries, S.J.; Rey, J.; Schindler, C.E.M.; Zacharias, M.; Tuffery, P. The pepATTRACT web server for blind, large-scale peptide-protein docking. Nucleic Acids Res. 2017, 45, W361–W364. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Nomine, Y. High-Quality Data of Protein/Peptide Interaction by Isothermal Titration Calorimetry. Methods Mol. Biol. 2019, 1964, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Drescher, D.G.; Selvakumar, D.; Drescher, M.J. Analysis of Protein Interactions by Surface Plasmon Resonance. Adv. Protein Chem. Struct. Biol. 2018, 110, 1–30. [Google Scholar] [CrossRef]
- Ren, B.; Sayed, A.M.M.; Tan, H.L.; Mok, Y.K.; Chen, E.S. Identifying Protein Interactions with Histone Peptides Using Bio-layer Interferometry. Bio Protoc. 2018, 8, e3012. [Google Scholar] [CrossRef]
- Plach, M.G.; Grasser, K.; Schubert, T. MicroScale Thermophoresis as a Tool to Study Protein-peptide Interactions in the Context of Large Eukaryotic Protein Complexes. Bio Protoc. 2017, 7, e2632. [Google Scholar] [CrossRef] [PubMed]
- Pellecchia, M.; Bertini, I.; Cowburn, D.; Dalvit, C.; Giralt, E.; Jahnke, W.; James, T.L.; Homans, S.W.; Kessler, H.; Luchinat, C.; et al. Perspectives on NMR in drug discovery: A technique comes of age. Nat. Rev. Drug Discov. 2008, 7, 738–745. [Google Scholar] [CrossRef]
- Pellecchia, M.; Sem, D.S.; Wuthrich, K. NMR in drug discovery. Nat. Rev. Drug Discov. 2002, 1, 211–219. [Google Scholar] [CrossRef]
- Leone, M.; Barile, E.; Dahl, R.; Pellecchia, M. Design and NMR studies of cyclic peptides targeting the N-terminal domain of the protein tyrosine phosphatase YopH. Chem. Biol. Drug Des. 2011, 77, 12–19. [Google Scholar] [CrossRef]
- Furukawa, A.; Konuma, T.; Yanaka, S.; Sugase, K. Quantitative analysis of protein-ligand interactions by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 96, 47–57. [Google Scholar] [CrossRef]
- Hobbs, B.; Drant, J.; Williamson, M.P. The measurement of binding affinities by NMR chemical shift perturbation. J. Biomol. NMR 2022, 76, 153–163. [Google Scholar] [CrossRef]
- Vincenzi, M.; Mercurio, F.A.; Leone, M. About TFE: Old and New Findings. Curr. Protein Pept. Sci. 2019, 20, 425–451. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, F.A.; Marasco, D.; Di Natale, C.; Pirone, L.; Costantini, S.; Pedone, E.M.; Leone, M. Targeting EphA2-Sam and Its Interactome: Design and Evaluation of Helical Peptides Enriched in Charged Residues. Chembiochem 2016, 17, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Kruger, D.M.; Glas, A.; Bier, D.; Pospiech, N.; Wallraven, K.; Dietrich, L.; Ottmann, C.; Koch, O.; Hennig, S.; Grossmann, T.N. Structure-Based Design of Non-natural Macrocyclic Peptides That Inhibit Protein-Protein Interactions. J. Med. Chem. 2017, 60, 8982–8988. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, F.A.; Pirone, L.; Di Natale, C.; Marasco, D.; Pedone, E.M.; Leone, M. Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2. Bioorg. Chem. 2018, 80, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Masuya, K. New trends in drug discovery and development by constrained peptides. Nihon Yakurigaku Zasshi 2016, 148, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Szabo, I.; Yousef, M.; Soltesz, D.; Bato, C.; Mezo, G.; Banoczi, Z. Redesigning of Cell-Penetrating Peptides to Improve Their Efficacy as a Drug Delivery System. Pharmaceutics 2022, 14, 907. [Google Scholar] [CrossRef]
- Komin, A.; Russell, L.M.; Hristova, K.A.; Searson, P.C. Peptide-based strategies for enhanced cell uptake, transcellular transport, and circulation: Mechanisms and challenges. Adv. Drug Deliv. Rev. 2017, 110–111, 52–64. [Google Scholar] [CrossRef]
- Al Musaimi, O.; Lombardi, L.; Williams, D.R.; Albericio, F. Strategies for Improving Peptide Stability and Delivery. Pharmaceuticals 2022, 15, 1283. [Google Scholar] [CrossRef]
- Fominaya, J.; Bravo, J.; Rebollo, A. Strategies to stabilize cell penetrating peptides for in vivo applications. Ther. Deliv. 2015, 6, 1171–1194. [Google Scholar] [CrossRef]
- Kalafatovic, D.; Giralt, E. Cell-Penetrating Peptides: Design Strategies beyond Primary Structure and Amphipathicity. Molecules 2017, 22, 1929. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Gfeller, D.; Buth, S.A.; Michielin, O.; Leiman, P.G.; Heinis, C. Improving binding affinity and stability of peptide ligands by substituting glycines with D-amino acids. Chembiochem 2013, 14, 1316–1322. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Lamers, C. Overcoming the shortcomings of peptide-based therapeutics. Future Drug Discov. 2022, 4, FDD75. [Google Scholar] [CrossRef]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Hummel, G.; Reineke, U.; Reimer, U. Translating peptides into small molecules. Mol. Biosyst. 2006, 2, 499–508. [Google Scholar] [CrossRef]
- Zane, D.; Feldman, P.L.; Sawyer, T.; Sobol, Z.; Hawes, J. Development and Regulatory Challenges for Peptide Therapeutics. Int. J. Toxicol. 2021, 40, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L., Jr.; Wade, J.D. Current challenges in peptide-based drug discovery. Front. Chem. 2014, 2, 62. [Google Scholar] [CrossRef]
- Cary, D.R.; Ohuchi, M.; Reid, P.C.; Masuya, K. Constrained Peptides in Drug Discovery and Development. J. Synth. Org. Chem. Jpn. 2017, 75, 1171–1178. [Google Scholar] [CrossRef]
- Walensky, L.D.; Bird, G.H. Hydrocarbon-stapled peptides: Principles, practice, and progress. J. Med. Chem. 2014, 57, 6275–6288. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, J.; Fishelson, Z.; Wang, C.; Zhang, S. Cell-Penetrating Peptide-Based Delivery of Macromolecular Drugs: Development, Strategies, and Progress. Biomedicines 2023, 11, 1971. [Google Scholar] [CrossRef]
- Torchilin, V. Intracellular delivery of protein and peptide therapeutics. Drug Discov. Today Technol. 2008, 5, e95–e103. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Sugita, T.; Mukai, Y.; Abe, Y.; Nakagawa, S.; Kamada, H.; Tsunoda, S.; Tsutsumi, Y. The augmentation of intracellular delivery of peptide therapeutics by artificial protein transduction domains. Biomaterials 2009, 30, 3318–3323. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Sylvestre, M.; Prossnitz, A.N.; Yang, L.F.; Pun, S.H. Design of Polymeric Carriers for Intracellular Peptide Delivery in Oncology Applications. Chem. Rev. 2021, 121, 11653–11698. [Google Scholar] [CrossRef] [PubMed]
- Matsson, P.; Doak, B.C.; Over, B.; Kihlberg, J. Cell permeability beyond the rule of 5. Adv. Drug Deliv. Rev. 2016, 101, 42–61. [Google Scholar] [CrossRef]
- Howell, L.A.; Beekman, A.M. In silico peptide-directed ligand design complements experimental peptide-directed binding for protein-protein interaction modulator discovery. RSC Chem. Biol. 2021, 2, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Beekman, A.M.; Cominetti, M.M.D.; Walpole, S.J.; Prabhu, S.; O’Connell, M.A.; Angulo, J.; Searcey, M. Identification of selective protein-protein interaction inhibitors using efficient in silico peptide-directed ligand design. Chem. Sci. 2019, 10, 4502–4508. [Google Scholar] [CrossRef]
- Coscueta, E.R.; Batista, P.; Gomes, J.E.G.; da Silva, R.; Pintado, M.M. Screening of Novel Bioactive Peptides from Goat Casein: In Silico to In Vitro Validation. Int. J. Mol. Sci. 2022, 23, 2439. [Google Scholar] [CrossRef]
- Mahmoodi-Reihani, M.; Abbasitabar, F.; Zare-Shahabadi, V. In Silico Rational Design and Virtual Screening of Bioactive Peptides Based on QSAR Modeling. ACS Omega 2020, 5, 5951–5958. [Google Scholar] [CrossRef]
- Prasasty, V.D.; Istyastono, E.P. Data of small peptides in SMILES and three-dimensional formats for virtual screening campaigns. Data Brief 2019, 27, 104607. [Google Scholar] [CrossRef]
- Joshi, J.; Blankenberg, D. PDAUG: A Galaxy based toolset for peptide library analysis, visualization, and machine learning modeling. BMC Bioinform. 2022, 23, 197. [Google Scholar] [CrossRef]
- Crooks, R.O.; Baxter, D.; Panek, A.S.; Lubben, A.T.; Mason, J.M. Deriving Heterospecific Self-Assembling Protein-Protein Interactions Using a Computational Interactome Screen. J. Mol. Biol. 2016, 428, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Engel, H.; Guischard, F.; Krause, F.; Nandy, J.; Kaas, P.; Hofflin, N.; Kohn, M.; Kilb, N.; Voigt, K.; Wolf, S.; et al. finDr: A web server for in silico D-peptide ligand identification. Synth. Syst. Biotechnol. 2021, 6, 402–413. [Google Scholar] [CrossRef]
- Frederix, P.W.; Ulijn, R.V.; Hunt, N.T.; Tuttle, T. Virtual Screening for Dipeptide Aggregation: Toward Predictive Tools for Peptide Self-Assembly. J. Phys. Chem. Lett. 2011, 2, 2380–2384. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, K.N.; De Maria, L.; Tyrchan, C.; Eriksson, L.A.; Sadowski, J.; Petrovic, D. Virtual Screening Expands the Non-Natural Amino Acid Palette for Peptide Optimization. J. Chem. Inf. Model. 2022, 62, 2999–3007. [Google Scholar] [CrossRef] [PubMed]
- Foight, G.W.; Chen, T.S.; Richman, D.; Keating, A.E. Enriching Peptide Libraries for Binding Affinity and Specificity Through Computationally Directed Library Design. Methods Mol. Biol. 2017, 1561, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Golosov, A.A.; Flyer, A.N.; Amin, J.; Babu, C.; Gampe, C.; Li, J.; Liu, E.; Nakajima, K.; Nettleton, D.; Patel, T.J.; et al. Design of Thioether Cyclic Peptide Scaffolds with Passive Permeability and Oral Exposure. J. Med. Chem. 2021, 64, 2622–2633. [Google Scholar] [CrossRef] [PubMed]
- Saha, I.; Dang, E.K.; Svatunek, D.; Houk, K.N.; Harran, P.G. Computational generation of an annotated gigalibrary of synthesizable, composite peptidic macrocycles. Proc. Natl. Acad. Sci. USA 2020, 117, 24679–24690. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Chaudhary, K.; Kumar, R.; Sharma, A.; Kapoor, P.; Tyagi, A.; Open source drug discovery, c.; Raghava, G.P. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013, 11, 74. [Google Scholar] [CrossRef]
- Park, H.; Park, J.H.; Kim, M.S.; Cho, K.; Shin, J.M. In Silico Screening and Optimization of Cell-Penetrating Peptides Using Deep Learning Methods. Biomolecules 2023, 13, 522. [Google Scholar] [CrossRef]
- Viart, B.; Dias-Lopes, C.; Kozlova, E.; Oliveira, C.F.; Nguyen, C.; Neshich, G.; Chavez-Olortegui, C.; Molina, F.; Felicori, L.F. EPI-peptide designer: A tool for designing peptide ligand libraries based on epitope-paratope interactions. Bioinformatics 2016, 32, 1462–1470. [Google Scholar] [CrossRef]
- Jukic, M.; Kralj, S.; Kolaric, A.; Bren, U. Design of Tetra-Peptide Ligands of Antibody Fc Regions Using In Silico Combinatorial Library Screening. Pharmaceuticals 2023, 16, 1170. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Terada, K.; Noma, T.; Ikebukuro, K.; Sode, K. In silico panning for a non-competitive peptide inhibitor. BMC Bioinform. 2007, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Kalafatovic, D.; Mausa, G.; Todorovski, T.; Giralt, E. Algorithm-supported, mass and sequence diversity-oriented random peptide library design. J. Cheminform. 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Mondal, A.; Perez, A. Towards rational computational peptide design. Front. Bioinform. 2022, 2, 1046493. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.M.; Bandyopadhyay, A. High throughput virtual screening (HTVS) of peptide library: Technological advancement in ligand discovery. Eur. J. Med. Chem. 2022, 243, 114766. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, V.; Moro, S. Bridging Molecular Docking to Molecular Dynamics in Exploring Ligand-Protein Recognition Process: An Overview. Front. Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Weng, G.; Gao, J.; Wang, Z.; Wang, E.; Hu, X.; Yao, X.; Cao, D.; Hou, T. Comprehensive Evaluation of Fourteen Docking Programs on Protein-Peptide Complexes. J. Chem. Theory Comput. 2020, 16, 3959–3969. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Heo, L.; Lee, M.S.; Seok, C. GalaxyPepDock: A protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 2015, 43, W431–W435. [Google Scholar] [CrossRef]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.Y. HPEPDOCK: A web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Rentzsch, R.; Renard, B.Y. Docking small peptides remains a great challenge: An assessment using AutoDock Vina. Brief Bioinform. 2015, 16, 1045–1056. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Geng, C.; Narasimhan, S.; Rodrigues, J.P.; Bonvin, A.M. Information-Driven, Ensemble Flexible Peptide Docking Using HADDOCK. Methods Mol. Biol. 2017, 1561, 109–138. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, G.C.; Bonvin, A.M. Modeling protein-protein complexes using the HADDOCK webserver “modeling protein complexes with HADDOCK”. Methods Mol. Biol. 2014, 1137, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Antunes, D.A.; Moll, M.; Devaurs, D.; Jackson, K.R.; Lizee, G.; Kavraki, L.E. DINC 2.0: A New Protein-Peptide Docking Webserver Using an Incremental Approach. Cancer Res. 2017, 77, e55–e57. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.S.; Karthe, P. Improved docking of peptides and small molecules in iMOLSDOCK. J. Mol. Model. 2022, 29, 12. [Google Scholar] [CrossRef] [PubMed]
- Kurcinski, M.; Badaczewska-Dawid, A.; Kolinski, M.; Kolinski, A.; Kmiecik, S. Flexible docking of peptides to proteins using CABS-dock. Protein Sci. 2020, 29, 211–222. [Google Scholar] [CrossRef]
- Almeida Filho, J.L.d.; Fernandez, J.H. HTP SurflexDock: A web tool for Structure-Based Virtual Screening analysis based on the Ensemble Docking protocol. In Proceedings of the Brazilian e-Science Workshop (BRESCI), Porto Alegre, Brazil, 31 July 2022; Sociedade Brasileira de Computação: Porto Alegre, Brazil, 2022; Volume 16, pp. 81–88. [Google Scholar]
- Ansar, S.; Vetrivel, U. PepVis: An integrated peptide virtual screening pipeline for ensemble and flexible docking protocols. Chem. Biol. Drug Des. 2019, 94, 2041–2050. [Google Scholar] [CrossRef]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sanner, M.F. AutoDock CrankPep: Combining folding and docking to predict protein-peptide complexes. Bioinformatics 2019, 35, 5121–5127. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, D.; Huang, S.Y. Efficient conformational ensemble generation of protein-bound peptides. J. Cheminform. 2017, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Pierce, B.; Weng, Z. A combination of rescoring and refinement significantly improves protein docking performance. Proteins 2008, 72, 270–279. [Google Scholar] [CrossRef]
- Alam, N.; Goldstein, O.; Xia, B.; Porter, K.A.; Kozakov, D.; Schueler-Furman, O. High-resolution global peptide-protein docking using fragments-based PIPER-FlexPepDock. PLoS Comput. Biol. 2017, 13, e1005905. [Google Scholar] [CrossRef]
- Perez, G.M.; Salomon, L.A.; Montero-Cabrera, L.A.; de la Vega, J.M.; Mascini, M. Integrating sampling techniques and inverse virtual screening: Toward the discovery of artificial peptide-based receptors for ligands. Mol. Divers. 2016, 20, 421–438. [Google Scholar] [CrossRef]
- Duffy, F.J.; O’Donovan, D.; Devocelle, M.; Moran, N.; O’Connell, D.J.; Shields, D.C. Virtual screening using combinatorial cyclic peptide libraries reveals protein interfaces readily targetable by cyclic peptides. J. Chem. Inf. Model. 2015, 55, 600–613. [Google Scholar] [CrossRef]
- Santini, B.L.; Zacharias, M. Rapid in silico Design of Potential Cyclic Peptide Binders Targeting Protein-Protein Interfaces. Front. Chem. 2020, 8, 573259. [Google Scholar] [CrossRef]
- Bender, B.J.; Gahbauer, S.; Luttens, A.; Lyu, J.; Webb, C.M.; Stein, R.M.; Fink, E.A.; Balius, T.E.; Carlsson, J.; Irwin, J.J.; et al. A practical guide to large-scale docking. Nat. Protoc. 2021, 16, 4799–4832. [Google Scholar] [CrossRef]
- Paul Daniel Phillips, P.D.; Andersen, T.; McDougal, O.M. Assessing the utility and limitations of high throughput virtual screening. AIMS Mol. Sci. 2016, 3, 238–245. [Google Scholar] [CrossRef]
- Waszkowycz, B.; Clark, D.E.; Gancia, E. Outstanding challenges in protein–ligand docking and structure-based virtual screening. WIREs Comput. Mol. Sci. 2011, 1, 229–259. [Google Scholar] [CrossRef]
- Berry, M.; Fielding, B.; Gamieldien, J. Practical Considerations in Virtual Screening and Molecular Docking. In Emerging Trends in Computational Biology, Bioinformatics, and Systems Biology, 1st ed.; Tran, Q.N., Arabnia, H.R., Eds.; Wiley Inc.: Hoboken, NJ, USA, 2015; pp. 487–502. [Google Scholar] [CrossRef]
- Budipramana, K.; Sangande, F. Molecular docking-based virtual screening: Challenges in hits identification for Anti-SARS-CoV-2 activity. Pharmacia 2022, 69, 1047–1056. [Google Scholar] [CrossRef]
- Gimeno, A.; Ojeda-Montes, M.J.; Tomas-Hernandez, S.; Cereto-Massague, A.; Beltran-Debon, R.; Mulero, M.; Pujadas, G.; Garcia-Vallve, S. The Light and Dark Sides of Virtual Screening: What Is There to Know? Int. J. Mol. Sci. 2019, 20, 1375. [Google Scholar] [CrossRef]
- Macip, G.; Garcia-Segura, P.; Mestres-Truyol, J.; Saldivar-Espinoza, B.; Ojeda-Montes, M.J.; Gimeno, A.; Cereto-Massague, A.; Garcia-Vallve, S.; Pujadas, G. Haste makes waste: A critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition. Med. Res. Rev. 2022, 42, 744–769. [Google Scholar] [CrossRef] [PubMed]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwuja, E.I.; Aja, P.M. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Xie, D.; Du, L.; Lin, H.; Su, E.; Shen, Y.; Xie, J.; Wei, D. In vitro-in silico screening strategy and mechanism of angiotensin I-converting enzyme inhibitory peptides from α-lactalbumin. Food Sci. Technol. 2022, 156, 112984. [Google Scholar] [CrossRef]
- Yu, Z.; Wu, S.; Zhao, W.; Ding, L.; Li, J.; Liu, J. Virtual screening and molecular docking for exploring ACE inhibitory peptides in Larimichthys crocea nebulin protein. Int. Food Res. J. 2019, 26, 1417–1426. [Google Scholar]
- Panyayai, T.; Sangsawad, P.; Pacharawongsakda, E.; Sawatdichaikul, O.; Tongsima, S.; Choowongkomon, K. The potential peptides against angiotensin-I converting enzyme through a virtual tripeptide-constructing library. Comput. Biol. Chem. 2018, 77, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Chaudhary, K.; Singh Chauhan, J.; Nagpal, G.; Kumar, R.; Sharma, M.; Raghava, G.P. An in silico platform for predicting, screening and designing of antihypertensive peptides. Sci. Rep. 2015, 5, 12512. [Google Scholar] [CrossRef]
- Stefanucci, A.; Iobbi, V.; Della Valle, A.; Scioli, G.; Pieretti, S.; Minosi, P.; Mirzaie, S.; Novellino, E.; Mollica, A. In Silico Identification of Tripeptides as Lead Compounds for the Design of KOR Ligands. Molecules 2021, 26, 4767. [Google Scholar] [CrossRef] [PubMed]
- Malyshev, A.V.; Sukhanova, I.A.; Zlobin, A.S.; Gedzun, V.R.; Pavshintsev, V.V.; Vasileva, E.V.; Zalevsky, A.O.; Doronin, I.I.; Mitkin, N.A.; Golovin, A.V.; et al. In silico Screening and Behavioral Validation of a Novel Peptide, LCGA-17, with Anxiolytic-Like Properties. Front. Neurosci. 2021, 15, 705590. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Grouleff, J.J.; Jitkova, Y.; Diaz, D.B.; Griffith, E.C.; Shao, W.; Bogdanchikova, A.F.; Poda, G.; Schimmer, A.D.; Lee, R.E.; et al. De Novo Design of Boron-Based Peptidomimetics as Potent Inhibitors of Human ClpP in the Presence of Human ClpX. J. Med. Chem. 2019, 62, 6377–6390. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Shaw, P.E.; Hirst, J.D. Molecular dynamics simulations and in silico peptide ligand screening of the Elk-1 ETS domain. J. Cheminform. 2011, 3, 49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, H.; Feng, Z.; Wang, S.; Wang, Y.; He, Q.; Li, G.; Lin, W.; Xie, X.Q.; Lin, Z. PD-1-Targeted Discovery of Peptide Inhibitors by Virtual Screening, Molecular Dynamics Simulation, and Surface Plasmon Resonance. Molecules 2019, 24, 3784. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Liu, G.; Chen, T.; Fu, X.; Niu, M.M. Structure-Based Virtual Screening and Biological Evaluation of Peptide Inhibitors for Polo-Box Domain. Molecules 2019, 25, 107. [Google Scholar] [CrossRef] [PubMed]
- Mastouri, M.; Baachaoui, S.; Mosbah, A.; Raouafi, N. In silico screening for oligopeptides useful as capture and reporting probes for interleukin-6 biosensing. RSC Adv. 2022, 12, 13003–13013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wei, X.; Zhang, R.; Mozdziak, P.E.; Si, D.; Ahmad, B.; Cheng, Q.; Tong, Y. Design and Immunological Evaluation of a Hybrid Peptide as a Potent TLR2 Agonist by Structure-Based Virtual Screening. Front. Cell Dev. Biol. 2021, 9, 620370. [Google Scholar] [CrossRef]
- Dama, E.; Cornelie, S.; Camara, M.; Somda, M.B.; Poinsignon, A.; Ilboudo, H.; Elanga Ndille, E.; Jamonneau, V.; Solano, P.; Remoue, F.; et al. In silico identification of a candidate synthetic peptide (Tsgf118-43) to monitor human exposure to tsetse flies in West Africa. PLoS Negl. Trop. Dis. 2013, 7, e2455. [Google Scholar] [CrossRef]
- Aksoydan, B.; Durdagi, S. Virtual drug repurposing study for the CGRPR identifies pentagastrin and leuprorelin as putative candidates. J. Mol. Graph. Model. 2022, 116, 108254. [Google Scholar] [CrossRef] [PubMed]
- Mollica, A.; Zengin, G.; Durdagi, S.; Ekhteiari Salmas, R.; Macedonio, G.; Stefanucci, A.; Dimmito, M.P.; Novellino, E. Combinatorial peptide library screening for discovery of diverse alpha-glucosidase inhibitors using molecular dynamics simulations and binary QSAR models. J. Biomol. Struct. Dyn. 2019, 37, 726–740. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Blythe, M.J.; Flower, D.R. Additive method for the prediction of protein-peptide binding affinity. Application to the MHC class I molecule HLA-A*0201. J. Proteome Res. 2002, 1, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Walshe, V.A.; Jones, N.A.; Gloster, S.E.; Borrow, P.; Flower, D.R. Coupling in silico and in vitro analysis of peptide-MHC binding: A bioinformatic approach enabling prediction of superbinding peptides and anchorless epitopes. J. Immunol. 2004, 172, 7495–7502. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Takeuchi, Y.; Shimizu, K.; Honda, H. In Silico Screening of a Bile Acid Micelle Disruption Peptide for Oral Consumptions from Edible Peptide Database. Foods 2021, 10, 2496. [Google Scholar] [CrossRef] [PubMed]
- Deryusheva, E.; Machulin, A.; Litus, E. Virtual Screening of Human Serum Albumin Mutants to Optimize the Search for its Forms that Increase Affinity to Amyloid-Β Peptide. BIO Web Conf. 2023, 57. [Google Scholar] [CrossRef]
- Martini, S.; Cattivelli, A.; Conte, A.; Tagliazucchi, D. Application of a Combined Peptidomics and In Silico Approach for the Identification of Novel Dipeptidyl Peptidase-IV-Inhibitory Peptides in In Vitro Digested Pinto Bean Protein Extract. Curr. Issues Mol. Biol. 2021, 44, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Sub, L.; Huo, S.; Yua, Z.; Lib, J.; Liuca, J. Virtual screening, molecular docking and identification of umami peptides derived from Oncorhynchus mykiss. Food Sci. Hum. Wellness. 2023, 12, 89–93. [Google Scholar] [CrossRef]
- Mercurio, F.A.; Di Natale, C.; Pirone, L.; Marasco, D.; Calce, E.; Vincenzi, M.; Pedone, E.M.; De Luca, S.; Leone, M. Design and analysis of EphA2-SAM peptide ligands: A multi-disciplinary screening approach. Bioorg. Chem. 2019, 84, 434–443. [Google Scholar] [CrossRef]
- Mercurio, F.A.; Di Natale, C.; Pirone, L.; Vincenzi, M.; Marasco, D.; De Luca, S.; Pedone, E.M.; Leone, M. Exploring the Ability of Cyclic Peptides to Target SAM Domains: A Computational and Experimental Study. ChemBioChem 2020, 21, 702–711. [Google Scholar] [CrossRef]
- Mercurio, F.A.; Vincenzi, M.; Leone, M. Hunting for Novel Routes in Anticancer Drug Discovery: Peptides against Sam-Sam Interactions. Int. J. Mol. Sci. 2022, 23, 10397. [Google Scholar] [CrossRef] [PubMed]
- Leffler, A.E.; Kuryatov, A.; Zebroski, H.A.; Powell, S.R.; Filipenko, P.; Hussein, A.K.; Gorson, J.; Heizmann, A.; Lyskov, S.; Tsien, R.W.; et al. Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models. Proc. Natl. Acad. Sci. USA 2017, 114, E8100–E8109. [Google Scholar] [CrossRef]
- Chiangjong, W.; Chutipongtanate, S.; Hongeng, S. Anticancer peptide: Physicochemical property, functional aspect and trend in clinical application (Review). Int. J. Oncol. 2020, 57, 678–696. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer—World Health Organization. Available online: https://www.iarc.who.int/ (accessed on 25 November 2023).
- Gaspar, D.; Veiga, A.S.; Castanho, M.A. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef]
- Speck-Planche, A.; Cordeiro, M.N.D.S. Speeding Up the Virtual Design and Screening of Therapeutic Peptides. In Multi-Scale Approaches in Drug Discovery: From Empirical Knowledge to In Silico Experiments and Back, 1st ed.; Speck-Planche, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 127–147. [Google Scholar] [CrossRef]
- Mercurio, F.A.; Leone, M. The Sam Domain of EphA2 Receptor and its Relevance to Cancer: A Novel Challenge for Drug Discovery? Curr. Med. Chem. 2016, 23, 4718–4734. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Kapoor, P.; Kumar, R.; Chaudhary, K.; Gautam, A.; Raghava, G.P. In silico models for designing and discovering novel anticancer peptides. Sci. Rep. 2013, 3, 2984. [Google Scholar] [CrossRef] [PubMed]
- Chai, T.T.; Koh, J.A.; Wong, C.C.; Sabri, M.Z.; Wong, F.C. Computational Screening for the Anticancer Potential of Seed-Derived Antioxidant Peptides: A Cheminformatic Approach. Molecules 2021, 26, 7396. [Google Scholar] [CrossRef]
- Das, D.; Jaiswal, M.; Khan, F.N.; Ahamad, S.; Kumar, S. PlantPepDB: A manually curated plant peptide database. Sci. Rep. 2020, 10, 2194. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P.S. AntiCP 2.0: An updated model for predicting anticancer peptides. Brief Bioinform. 2021, 22, bbaa153. [Google Scholar] [CrossRef]
- Shaw, S.A.; Vokits, B.P.; Dilger, A.K.; Viet, A.; Clark, C.G.; Abell, L.M.; Locke, G.A.; Duke, G.; Kopcho, L.M.; Dongre, A.; et al. Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase. Bioorg. Med. Chem. 2020, 28, 115723. [Google Scholar] [CrossRef]
- Cao, H.; Pauff, J.M.; Hille, R. X-ray crystal structure of a xanthine oxidase complex with the flavonoid inhibitor quercetin. J. Nat. Prod. 2014, 77, 1693–1699. [Google Scholar] [CrossRef]
- Lo, S.C.; Li, X.; Henzl, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef] [PubMed]
- Ogura, K.; Nobuhisa, I.; Yuzawa, S.; Takeya, R.; Torikai, S.; Saikawa, K.; Sumimoto, H.; Inagaki, F. NMR solution structure of the tandem Src homology 3 domains of p47phox complexed with a p22phox-derived proline-rich peptide. J. Biol. Chem. 2006, 281, 3660–3668. [Google Scholar] [CrossRef]
- Lamiable, A.; Thevenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tuffery, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Kochnev, Y.; Hellemann, E.; Cassidy, K.C.; Durrant, J.D. Webina: An open-source library and web app that runs AutoDock Vina entirely in the web browser. Bioinformatics 2020, 36, 4513–4515. [Google Scholar] [CrossRef]
- Manavalan, B.; Subramaniyam, S.; Shin, T.H.; Kim, M.O.; Lee, G. Machine-learning-based prediction of cell-penetrating peptides and their uptake efficiency with improved accuracy. J. Proteome Res. 2018, 17, 2715–2726. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Patiyal, S.; Dhall, A.; Sharma, N.; Raghava, G.P.S. B3Pred: A Random-Forest-Based Method for Predicting and Designing Blood-Brain Barrier Penetrating Peptides. Pharmaceutics 2021, 13, 1237. [Google Scholar] [CrossRef] [PubMed]
- Mathur, D.; Singh, S.; Mehta, A.; Agrawal, P.; Raghava, G.P.S. In silico approaches for predicting the half-life of natural and modified peptides in blood. PLoS ONE 2018, 13, e0196829. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef]
- Wood, C.W.; Ibarra, A.A.; Bartlett, G.J.; Wilson, A.J.; Woolfson, D.N.; Sessions, R.B. BAlaS: Fast, interactive and accessible computational alanine-scanning using BudeAlaScan. Bioinformatics 2020, 36, 2917–2919. [Google Scholar] [CrossRef]
- Ferguson, K.M.; Berger, M.B.; Mendrola, J.M.; Cho, H.S.; Leahy, D.J.; Lemmon, M.A. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 2003, 11, 507–517. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated Docking Using a Lamarckian Genetic Algorithm and and Empirical Binding Free Energy Function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Song, S.; Liu, D.; Peng, J.; Deng, H.; Guo, Y.; Xu, L.X.; Miller, A.D.; Xu, Y. Novel peptide ligand directs liposomes toward EGF-R high-expressing cancer cells in vitro and in vivo. FASEB J. 2009, 23, 1396–1404. [Google Scholar] [CrossRef]
- Sogabe, S.; Kawakita, Y.; Igaki, S.; Iwata, H.; Miki, H.; Cary, D.R.; Takagi, T.; Takagi, S.; Ohta, Y.; Ishikawa, T. Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors. ACS Med. Chem. Lett. 2013, 4, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Li, Z.; Wan, H.; Shi, Y.; Ouyang, P. Personal experience with four kinds of chemical structure drawing software: Review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J. Chem. Inf. Comput. Sci. 2004, 44, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.S.; Jadhav, G.; Pradeep, S.; Dharmashekar, C.; Jain, A.S.; Devegowda, D.; Shivamallu, C.; Kollur, S.P.; Prasad, S.K. In Silico Examination of Peptides Containing Selenium and Ebselen Backbone to Assess Their Tumoricidal Potential. Int. J. Health Allied Sci. 2022, 11, 9. [Google Scholar] [CrossRef]
- Schwartz, J.C.; Zhang, X.; Fedorov, A.A.; Nathenson, S.G.; Almo, S.C. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature 2001, 410, 604–608. [Google Scholar] [CrossRef]
- Darnell, S.J.; Page, D.; Mitchell, J.C. An automated decision-tree approach to predicting protein interaction hot spots. Proteins 2007, 68, 813–823. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Raveh, B.; London, N.; Zimmerman, L.; Schueler-Furman, O. Rosetta FlexPepDock ab-initio: Simultaneous folding, docking and refinement of peptides onto their receptors. PLoS ONE 2011, 6, e18934. [Google Scholar] [CrossRef]
- Chaudhury, S.; Lyskov, S.; Gray, J.J. PyRosetta: A script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 2010, 26, 689–691. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 2004, 55, 383–394. [Google Scholar] [CrossRef]
- Fu, H.; Gumbart, J.C.; Chen, H.; Shao, X.; Cai, W.; Chipot, C. BFEE: A User-Friendly Graphical Interface Facilitating Absolute Binding Free-Energy Calculations. J. Chem. Inf. Model. 2018, 58, 556–560. [Google Scholar] [CrossRef]
- Thakkar, R.; Upreti, D.; Ishiguro, S.; Tamura, M.; Comer, J. Computational design of a cyclic peptide that inhibits the CTLA4 immune checkpoint. RSC Med. Chem. 2023, 14, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Henin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Kuncewicz, K.; Battin, C.; Sieradzan, A.; Karczynska, A.; Orlikowska, M.; Wardowska, A.; Pikula, M.; Steinberger, P.; Rodziewicz-Motowidlo, S.; Spodzieja, M. Fragments of gD Protein as Inhibitors of BTLA/HVEM Complex Formation-Design, Synthesis, and Cellular Studies. Int. J. Mol. Sci. 2020, 21, 8876. [Google Scholar] [CrossRef]
- Lerksuthirat, T.; On-Yam, P.; Chitphuk, S.; Stitchantrakul, W.; Newburg, D.S.; Morrow, A.L.; Hongeng, S.; Chiangjong, W.; Chutipongtanate, S. ALA-A2 Is a Novel Anticancer Peptide Inspired by Alpha-Lactalbumin: A Discovery from a Computational Peptide Library, In Silico Anticancer Peptide Screening and In Vitro Experimental Validation. Glob. Chall. 2023, 7, 2200213. [Google Scholar] [CrossRef] [PubMed]
- Gan, B.H.; Gaynord, J.; Rowe, S.M.; Deingruber, T.; Spring, D.R. The multifaceted nature of antimicrobial peptides: Current synthetic chemistry approaches and future directions. Chem. Soc. Rev. 2021, 50, 7820–7880. [Google Scholar] [CrossRef]
- Lee, Y.J.; Shirkey, J.D.; Park, J.; Bisht, K.; Cowan, A.J. An Overview of Antiviral Peptides and Rational Biodesign Considerations. Biodes. Res. 2022, 2022, 9898241. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, S.; Zhang, C. Antimicrobial peptides with antiviral and anticancer properties and their modification and nanodelivery systems. Curr. Res. Biotechnol. 2023, 5, 100121. [Google Scholar] [CrossRef]
- Izadpanah, A.; Gallo, R.L. Antimicrobial peptides. J. Am. Acad. Dermatol. 2005, 52, 381–390, quiz 391–382. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, D933–D937. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37–51. [Google Scholar] [CrossRef]
- Huang, J.; Xu, Y.; Xue, Y.; Huang, Y.; Li, X.; Chen, X.; Xu, Y.; Zhang, D.; Zhang, P.; Zhao, J.; et al. Identification of potent antimicrobial peptides via a machine-learning pipeline that mines the entire space of peptide sequences. Nat. Biomed. Eng. 2023, 7, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Veltri, D.; Kamath, U.; Shehu, A. Improving Recognition of Antimicrobial Peptides and Target Selectivity through Machine Learning and Genetic Programming. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 300–313. [Google Scholar] [CrossRef]
- Kleandrova, V.V.; Ruso, J.M.; Speck-Planche, A.; Dias Soeiro Cordeiro, M.N. Enabling the Discovery and Virtual Screening of Potent and Safe Antimicrobial Peptides. Simultaneous Prediction of Antibacterial Activity and Cytotoxicity. ACS Comb. Sci. 2016, 18, 490–498. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard—World Health Organization. Available online: https://covid19.who.int/ (accessed on 2 December 2023).
- Vincenzi, M.; Mercurio, F.A.; Leone, M. Looking for SARS-CoV-2 Therapeutics Through Computational Approaches. Curr. Med. Chem. 2023, 30, 3158–3214. [Google Scholar] [CrossRef]
- Vincenzi, M.; Leone, M. The Fight against Human Viruses: How NMR Can Help? Curr. Med. Chem. 2021, 28, 4380–4453. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef]
- Jamison, D.A., Jr.; Anand Narayanan, S.; Trovao, N.S.; Guarnieri, J.W.; Topper, M.J.; Moraes-Vieira, P.M.; Zaksas, V.; Singh, K.K.; Wurtele, E.S.; Beheshti, A. A comprehensive SARS-CoV-2 and COVID-19 review, Part 1: Intracellular overdrive for SARS-CoV-2 infection. Eur. J. Hum. Genet. 2022, 30, 889–898. [Google Scholar] [CrossRef]
- Narayanan, S.A.; Jamison, D.A., Jr.; Guarnieri, J.W.; Zaksas, V.; Topper, M.; Koutnik, A.P.; Park, J.; Clark, K.B.; Enguita, F.J.; Leitao, A.L.; et al. A comprehensive SARS-CoV-2 and COVID-19 review, Part 2: Host extracellular to systemic effects of SARS-CoV-2 infection. Eur. J. Hum. Genet. 2024, 32, 10–20. [Google Scholar] [CrossRef]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef]
- Satpathy, R. In silico Tools and Techniques for Screening and Development of Peptide-Based Spike Protein Inhibitors against Novel Coronavirus (SARS-CoV-2). Jordan J. Biol. Sci. 2022, 15, 729–738. [Google Scholar] [CrossRef]
- Badhe, Y.; Gupta, R.; Rai, B. In silico design of peptides with binding to the receptor binding domain (RBD) of the SARS-CoV-2 and their utility in bio-sensor development for SARS-CoV-2 detection. RSC Adv. 2021, 11, 3816–3826. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, L.; Li, L.; Lu, F.; Liu, F. Bionics design of affinity peptide inhibitors for SARS-CoV-2 RBD to block SARS-CoV-2 RBD-ACE2 interactions. Heliyon 2023, 9, e12890. [Google Scholar] [CrossRef]
- Hu, C.; Guo, T.; Zou, Y.; Gao, J.; Gao, Y.; Niu, M.; Xia, Y.; Shen, X.; Li, J. Discovery of dual S-RBD/NRP1-targeting peptides: Structure-based virtual screening, synthesis, biological evaluation, and molecular dynamics simulation studies. J. Enzyme Inhib. Med. Chem. 2023, 38, 2212327. [Google Scholar] [CrossRef]
- Panda, S.K.; Sen Gupta, P.S.; Biswal, S.; Ray, A.K.; Rana, M.K. ACE-2-Derived Biomimetic Peptides for the Inhibition of Spike Protein of SARS-CoV-2. J. Proteome Res. 2021, 20, 1296–1303. [Google Scholar] [CrossRef]
- Xu, Z.; Zou, Y.; Gao, X.; Niu, M.M.; Li, J.; Xue, L.; Jiang, S. Dual-targeting cyclic peptides of receptor-binding domain (RBD) and main protease (Mpro) as potential drug leads for the treatment of SARS-CoV-2 infection. Front. Pharmacol. 2022, 13, 1041331. [Google Scholar] [CrossRef] [PubMed]
- Hernandez Gonzalez, J.E.; Eberle, R.J.; Willbold, D.; Coronado, M.A. A Computer-Aided Approach for the Discovery of D-Peptides as Inhibitors of SARS-CoV-2 Main Protease. Front. Mol. Biosci. 2021, 8, 816166. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Acosta, K.; Rosales-Fuerte, I.A.; Perez-Sanchez, J.E.; Nunez-Rivera, A.; Juarez, J.; Cadena-Nava, R.D. Design and selection of peptides to block the SARS-CoV-2 receptor binding domain by molecular docking. Beilstein J. Nanotechnol. 2022, 13, 699–711. [Google Scholar] [CrossRef]
- Harnkit, N.; Khongsonthi, T.; Masuwan, N.; Prasartkul, P.; Noikaew, T.; Chumnanpuen, P. Virtual Screening for SARS-CoV-2 Main Protease Inhibitory Peptides from the Putative Hydrolyzed Peptidome of Rice Bran. Antibiotics 2022, 11, 1318. [Google Scholar] [CrossRef]
- Porto, W.F. Virtual screening of peptides with high affinity for SARS-CoV-2 main protease. Comput. Biol. Med. 2021, 133, 104363. [Google Scholar] [CrossRef]
- Waqas, M.; Haider, A.; Rehman, A.; Qasim, M.; Umar, A.; Sufyan, M.; Akram, H.N.; Mir, A.; Razzaq, R.; Rasool, D.; et al. Immunoinformatics and Molecular Docking Studies Predicted Potential Multiepitope-Based Peptide Vaccine and Novel Compounds against Novel SARS-CoV-2 through Virtual Screening. Biomed Res. Int. 2021, 2021, 1596834. [Google Scholar] [CrossRef]
- Qureshi, A.; Thakur, N.; Tandon, H.; Kumar, M. AVPdb: A database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 2014, 42, D1147–D1153. [Google Scholar] [CrossRef] [PubMed]
- Khater, I.; Nassar, A. Potential antiviral peptides targeting the SARS-CoV-2 spike protein. BMC Pharmacol. Toxicol. 2022, 23, 91. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Wu, L.; Chen, Q.; Liu, K.; Wang, J.; Han, P.; Zhang, Y.; Hu, Y.; Meng, Y.; Pan, X.; Qiao, C.; et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020, 6, 68. [Google Scholar] [CrossRef]
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Pearce, R.; Zhang, Y. De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging 2020, 12, 11263–11276. [Google Scholar] [CrossRef]
- Huang, X.; Pearce, R.; Zhang, Y. EvoEF2: Accurate and fast energy function for computational protein design. Bioinformatics 2020, 36, 1135–1142. [Google Scholar] [CrossRef]
- Pearce, R.; Huang, X.; Setiawan, D.; Zhang, Y. EvoDesign: Designing Protein-Protein Binding Interactions Using Evolutionary Interface Profiles in Conjunction with an Optimized Physical Energy Function. J. Mol. Biol. 2019, 431, 2467–2476. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Molecular Operating Environment (MOE), 2022.02; Chemical Computing Group ULC: Montreal, QC, Canada, 2023.
- Wang, Z.; Wang, G. APD: The Antimicrobial Peptide Database. Nucleic Acids Res. 2004, 32, D590–D592. [Google Scholar] [CrossRef]
- MacCarthy, E.A.; Zhang, C.; Zhang, Y.; Kc, D.B. GPU-I-TASSER: A GPU accelerated I-TASSER protein structure prediction tool. Bioinformatics 2022, 38, 1754–1755. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Sidney, J.; Sette, A.; Peters, B. TepiTool: A Pipeline for Computational Prediction of T Cell Epitope Candidates. Curr. Protoc. Immunol. 2016, 114, 18.19.11–18.19.24. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, R.; Agarwal, D.K.; Ranaweera, C.B.; Ishiguro, S.; Conda-Sheridan, M.; Gaudreault, N.N.; Richt, J.A.; Tamura, M.; Comer, J. De novo design of a stapled peptide targeting SARS-CoV-2 spike protein receptor-binding domain. RSC Med. Chem. 2023, 14, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Johansen-Leete, J.; Ullrich, S.; Fry, S.E.; Frkic, R.; Bedding, M.J.; Aggarwal, A.; Ashhurst, A.S.; Ekanayake, K.B.; Mahawaththa, M.C.; Sasi, V.M.; et al. Antiviral cyclic peptides targeting the main protease of SARS-CoV-2. Chem. Sci. 2022, 13, 3826–3836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. Amber 2020; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Mahmud, S.; Paul, G.K.; Biswas, S.; Afrose, S.; Mita, M.A.; Hasan, M.R.; Shimu, M.S.S.; Hossain, A.; Promi, M.M.; Ema, F.K.; et al. Prospective Role of Peptide-Based Antiviral Therapy Against the Main Protease of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 628585. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Kim, Y.; Jedrzejczak, R.; Maltseva, N.I.; Wilamowski, M.; Endres, M.; Godzik, A.; Michalska, K.; Joachimiak, A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020, 29, 1596–1605. [Google Scholar] [CrossRef]
- Tan, K.; Kim, Y.; Jedrzejczak, R.; Maltseva, N.; Endres, M.; Michalska, K.; Joachimiak, A. The Crystal Structure of Nsp9 Replicase Protein of COVID-19 (2020)—Center for Structural Genomics of Infectious Diseases (CSGID). Available online: https://www.rcsb.org/structure/6w4b (accessed on 12 December 2023).
- Singh, S.; Chauhan, P.; Sharma, V.; Rao, A.; Kumbhar, B.V.; Prajapati, V.K. Identification of multi-targeting natural antiviral peptides to impede SARS-CoV-2 infection. Struct. Chem. 2022, 34, 1743–1758. [Google Scholar] [CrossRef]
- Thakur, N.; Qureshi, A.; Kumar, M. AVPpred: Collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 2012, 40, W199–W204. [Google Scholar] [CrossRef] [PubMed]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. Meta-iAVP: A Sequence-Based Meta-Predictor for Improving the Prediction of Antiviral Peptides Using Effective Feature Representation. Int. J. Mol. Sci. 2019, 20, 5743. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.R.; Kuo, T.R.; Wu, L.C.; Lee, T.Y.; Horng, J.T. Characterization and identification of antimicrobial peptides with different functional activities. Brief Bioinform. 2019, 21, 1098–1114. [Google Scholar] [CrossRef] [PubMed]
- Timmons, P.B.; Hewage, C.M. ENNAVIA is a novel method which employs neural networks for antiviral and anti-coronavirus activity prediction for therapeutic peptides. Brief Bioinform. 2021, 22, bbab258. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Thakur, V.; Raghava, G.P. COPid: Composition based protein identification. Silico Biol. 2008, 8, 121–128. [Google Scholar]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Open Source Drug Discovery, C.; Raghava, G.P. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhong, F.; Lin, C.; Hu, X.; Zhang, Y.; Xiong, B.; Yin, X.; Fu, J.; He, W.; Duan, J.; et al. Structure of SARS-CoV-2 main protease in the apo state. Sci. China Life Sci. 2021, 64, 656–659. [Google Scholar] [CrossRef]
- Dai, W.; Zhang, B.; Jiang, X.M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef]
- Mashiach, E.; Schneidman-Duhovny, D.; Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008, 36, W229–W232. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhu, Y.; Liu, X.; Jin, Z.; Duan, Y.; Zhang, Q.; Wu, C.; Feng, L.; Du, X.; Zhao, J.; et al. Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2022, 119, e2117142119. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Goyal, B. Identification of new pentapeptides as potential inhibitors of amyloid-beta(42) aggregation using virtual screening and molecular dynamics simulations. J. Mol. Graph. Model. 2023, 124, 108558. [Google Scholar] [CrossRef] [PubMed]
- Shuaib, S.; Narang, S.S.; Goyal, D.; Goyal, B. Computational design and evaluation of beta-sheet breaker peptides for destabilizing Alzheimer’s amyloid-beta(42) protofibrils. J. Cell. Biochem. 2019, 120, 17935–17950. [Google Scholar] [CrossRef]
- Kaur, A.; Goyal, B. In silico design and identification of new peptides for mitigating hIAPP aggregation in type 2 diabetes. J. Biomol. Struct. Dyn. 2023, 1–16. [Google Scholar] [CrossRef]
- Crescenzi, O.; Tomaselli, S.; Guerrini, R.; Salvadori, S.; D’Ursi, A.M.; Temussi, P.A.; Picone, D. Solution structure of the Alzheimer amyloid beta-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur. J. Biochem. 2002, 269, 5642–5648. [Google Scholar] [CrossRef]
- Barrow, C.J.; Yasuda, A.; Kenny, P.T.; Zagorski, M.G. Solution conformations and aggregational properties of synthetic amyloid beta-peptides of Alzheimer’s disease. Analysis of circular dichroism spectra. J. Mol. Biol. 1992, 225, 1075–1093. [Google Scholar] [CrossRef]
- Luhrs, T.; Ritter, C.; Adrian, M.; Riek-Loher, D.; Bohrmann, B.; Dobeli, H.; Schubert, D.; Riek, R. 3D structure of Alzheimer’s amyloid-beta(1–42) fibrils. Proc. Natl. Acad. Sci. USA 2005, 102, 17342–17347. [Google Scholar] [CrossRef]
- Nanga, R.P.; Brender, J.R.; Vivekanandan, S.; Ramamoorthy, A. Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment. Biochim. Biophys. Acta 2011, 1808, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Boyer, D.R.; Sawaya, M.R.; Abskharon, R.; Saelices, L.; Nguyen, B.A.; Lu, J.; Murray, K.A.; Kandeel, F.; Eisenberg, D.S. Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores. Nat. Struct. Mol. Biol. 2021, 28, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ni, D.; Liu, Y.; Lu, S. Rational Design of Peptide-Based Inhibitors Disrupting Protein-Protein Interactions. Front. Chem. 2021, 9, 682675. [Google Scholar] [CrossRef] [PubMed]
- Mo, Q.; Xu, Z.; Yan, H.; Chen, P.; Lu, Y. VSTH: A user-friendly web server for structure-based virtual screening on Tianhe-2. Bioinformatics 2023, 39, btac740. [Google Scholar] [CrossRef] [PubMed]
- Jofily, P.; Pascutti, P.G.; Torres, P.H.M. Improving Blind Docking in DOCK6 through an Automated Preliminary Fragment Probing Strategy. Molecules 2021, 26, 1224. [Google Scholar] [CrossRef]
- Andrusier, N.; Mashiach, E.; Nussinov, R.; Wolfson, H.J. Principles of flexible protein-protein docking. Proteins 2008, 73, 271–289. [Google Scholar] [CrossRef]
- Zhu, T.; Cao, S.; Su, P.C.; Patel, R.; Shah, D.; Chokshi, H.B.; Szukala, R.; Johnson, M.E.; Hevener, K.E. Hit identification and optimization in virtual screening: Practical recommendations based on a critical literature analysis. J. Med. Chem. 2013, 56, 6560–6572. [Google Scholar] [CrossRef]
- Corbeil, C.R.; Williams, C.I.; Labute, P. Variability in docking success rates due to dataset preparation. J. Comput. Aided Mol. Des. 2012, 26, 775–786. [Google Scholar] [CrossRef]
| ACP | Protein Target | Computational Tools | Exp. | Ref. |
|---|---|---|---|---|
| LYSPH PSYLNTPLL | MPO (PDB ID: 6WYD [136]); XO (PDB ID: 3NVY [137]); Keap1 (PDB ID: 2FLU [138]); p47phox (PDB ID: 1WLP [139]) | PlantPepDB [134]; AntiCP 2.0 [135]; PEP-FOLD3 [140]; Autodock Vina [72]/Webina 1.0.3 [141]; HPEPDOCK [71]; MLCPP [142]; B3Pred [143]; PlifePred [144]; BIOPEP-UWM [145]; BUDE Alanine [146]; GROMACS 2020 [87] | N | [133] |
| LARLLT (D4) | Extracellular domain of EGFR (PDB ID: 1NQL [147]) | PSCAN 2.2.2 Autodock 3 [148] | Y | [149] |
| Ebselen-WE | Kinase domain of EGFR (PDB ID: 3W2S [150]) | CASTp [151]; Chemsketch [152]; Autodock Vina [72] | N | [153] |
| Cyclo- (EIDTVLTPTGWVAKRYS) | CTLA-4 (PDB ID: 1I85 [154]) | KFC [155]; Phyre2 [156]; FlexPepDock [157]; PyRosetta [158]; NAMD 2.13 [159]; MM-GBSA calculations [160]; BFEE in VMD [161] | Y | [162] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vincenzi, M.; Mercurio, F.A.; Leone, M. Virtual Screening of Peptide Libraries: The Search for Peptide-Based Therapeutics Using Computational Tools. Int. J. Mol. Sci. 2024, 25, 1798. https://doi.org/10.3390/ijms25031798
Vincenzi M, Mercurio FA, Leone M. Virtual Screening of Peptide Libraries: The Search for Peptide-Based Therapeutics Using Computational Tools. International Journal of Molecular Sciences. 2024; 25(3):1798. https://doi.org/10.3390/ijms25031798
Chicago/Turabian StyleVincenzi, Marian, Flavia Anna Mercurio, and Marilisa Leone. 2024. "Virtual Screening of Peptide Libraries: The Search for Peptide-Based Therapeutics Using Computational Tools" International Journal of Molecular Sciences 25, no. 3: 1798. https://doi.org/10.3390/ijms25031798






