Molecular Classifications in Gastric Cancer: A Call for Interdisciplinary Collaboration
Abstract
:1. Introduction
2. Gastric Cancer Characterization, Prognosis, and Management in the Molecular Era
2.1. The Molecular Era: Recent Advances in Molecular Techniques
2.2. Main Molecular Alterations in Gastric Cancer
2.3. Current Treatment of Gastric Cancer
2.4. Gastric Cancer: Therapeutic Advances and Challenges
3. Molecular Classifications in Gastric Cancer
3.1. The Cancer Genome Atlas (2014) [42]
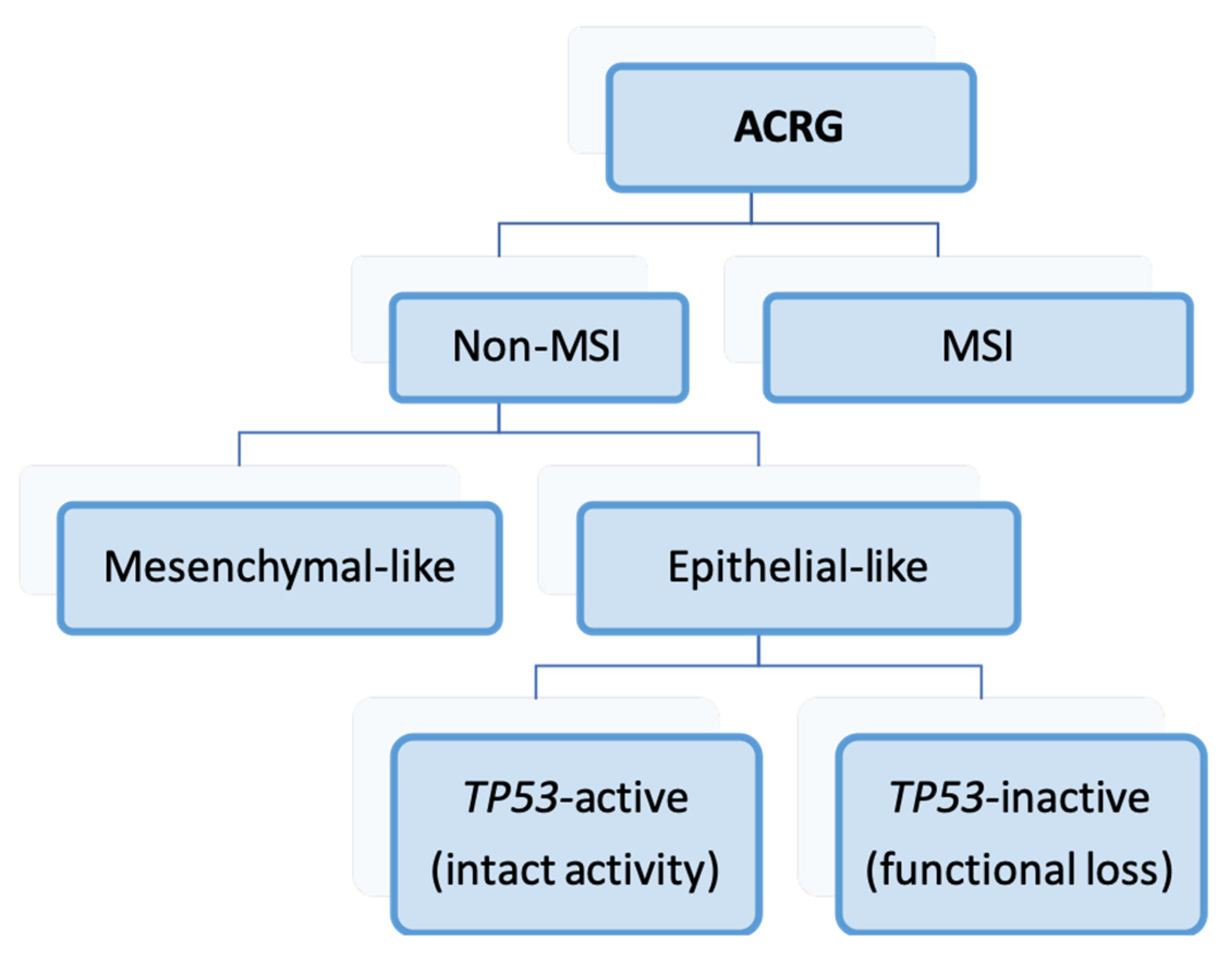
3.2. Asian Cancer Research Group (2015) [43]
3.3. Intrinsic Subtypes (2011) [155]
Relationship with Clinicopathological, Prognostic, and Therapeutic Variables
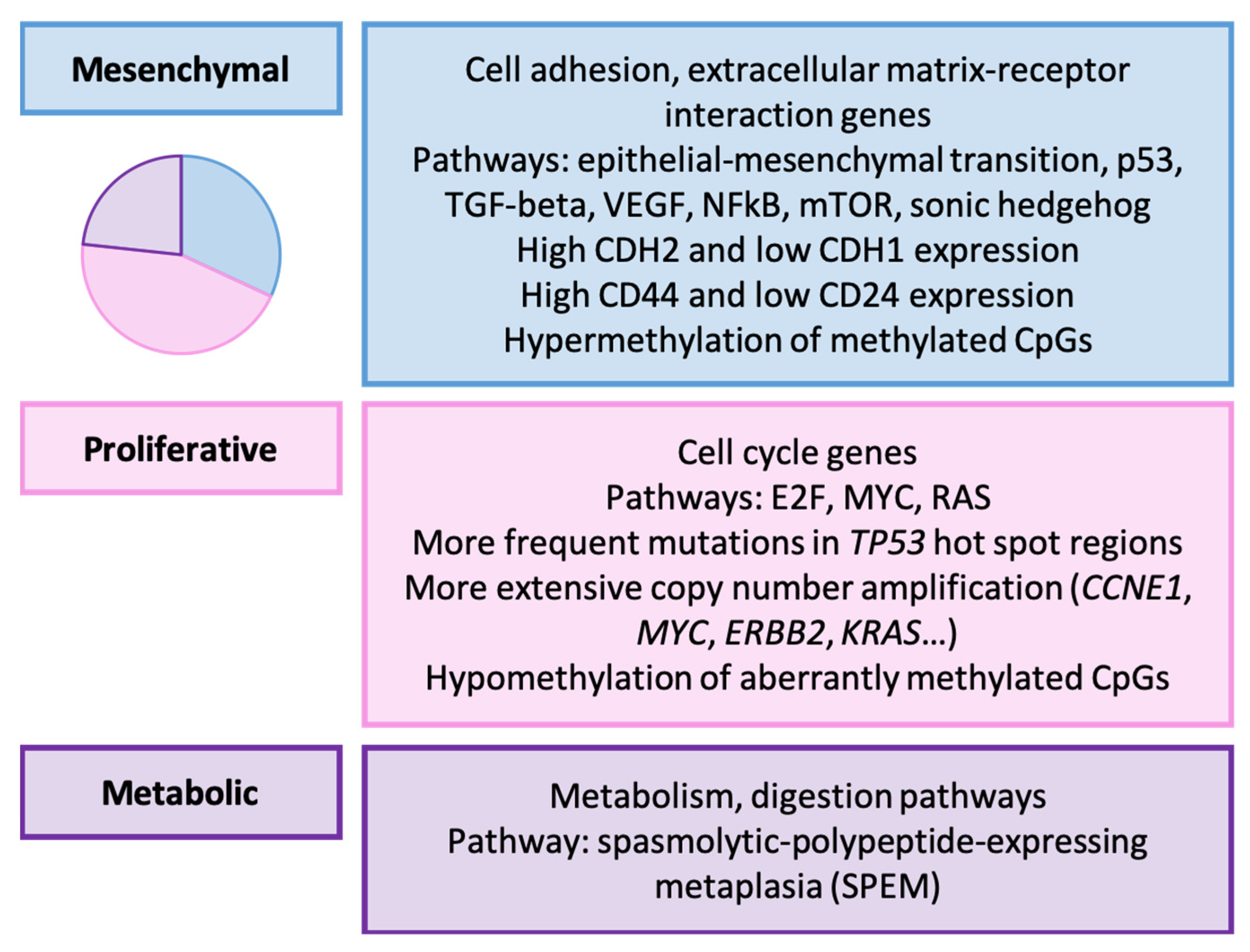
3.4. Lei’s Classification (2013) [156]
Relationship with Clinicopathological, Prognostic, and Therapeutic Variables
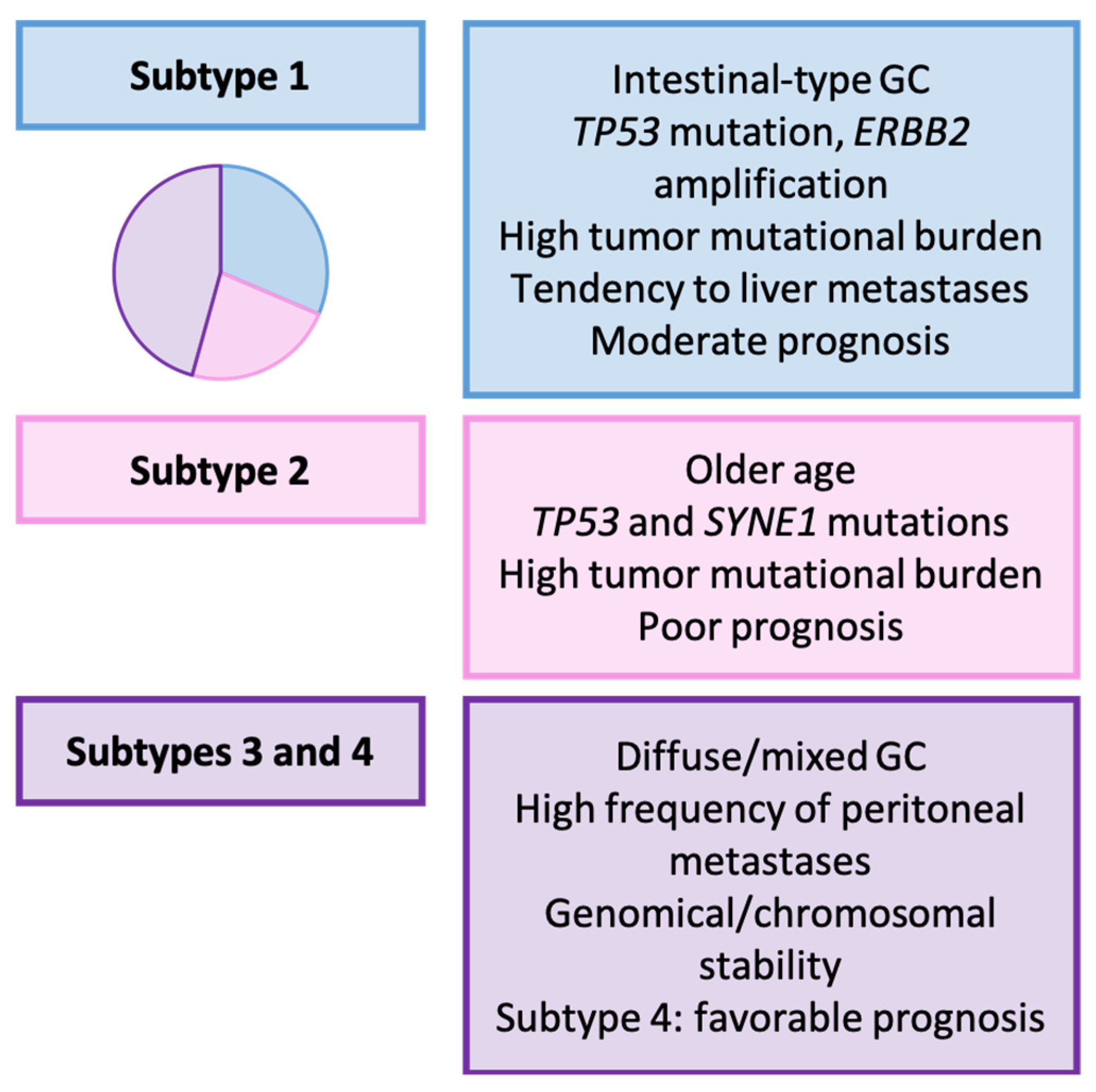
3.5. Wang’s Classification (2021) [157]
Relationship with Clinicopathological, Prognostic, and Therapeutic Variables
3.6. Shah’s Classification (2011) [158]
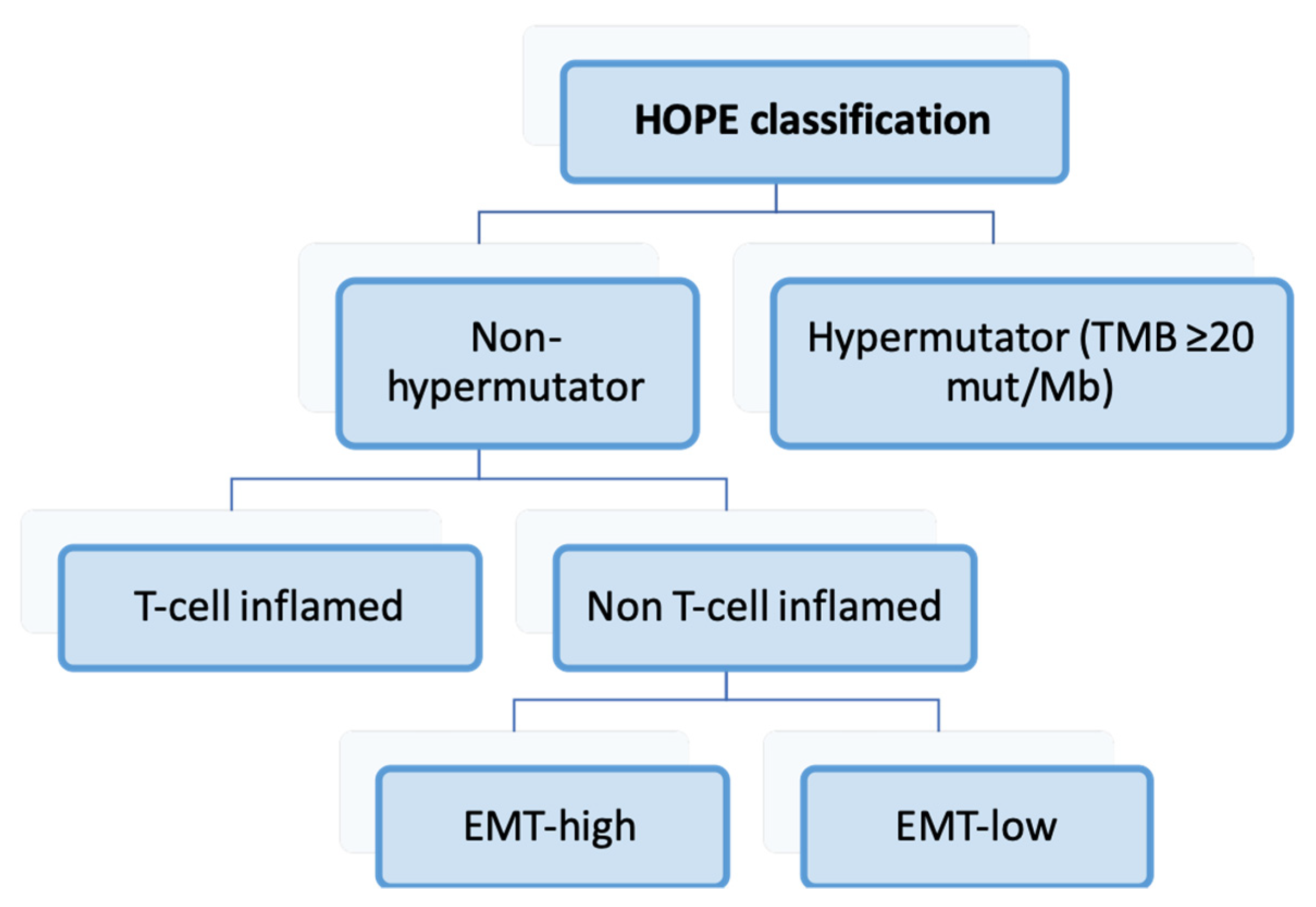
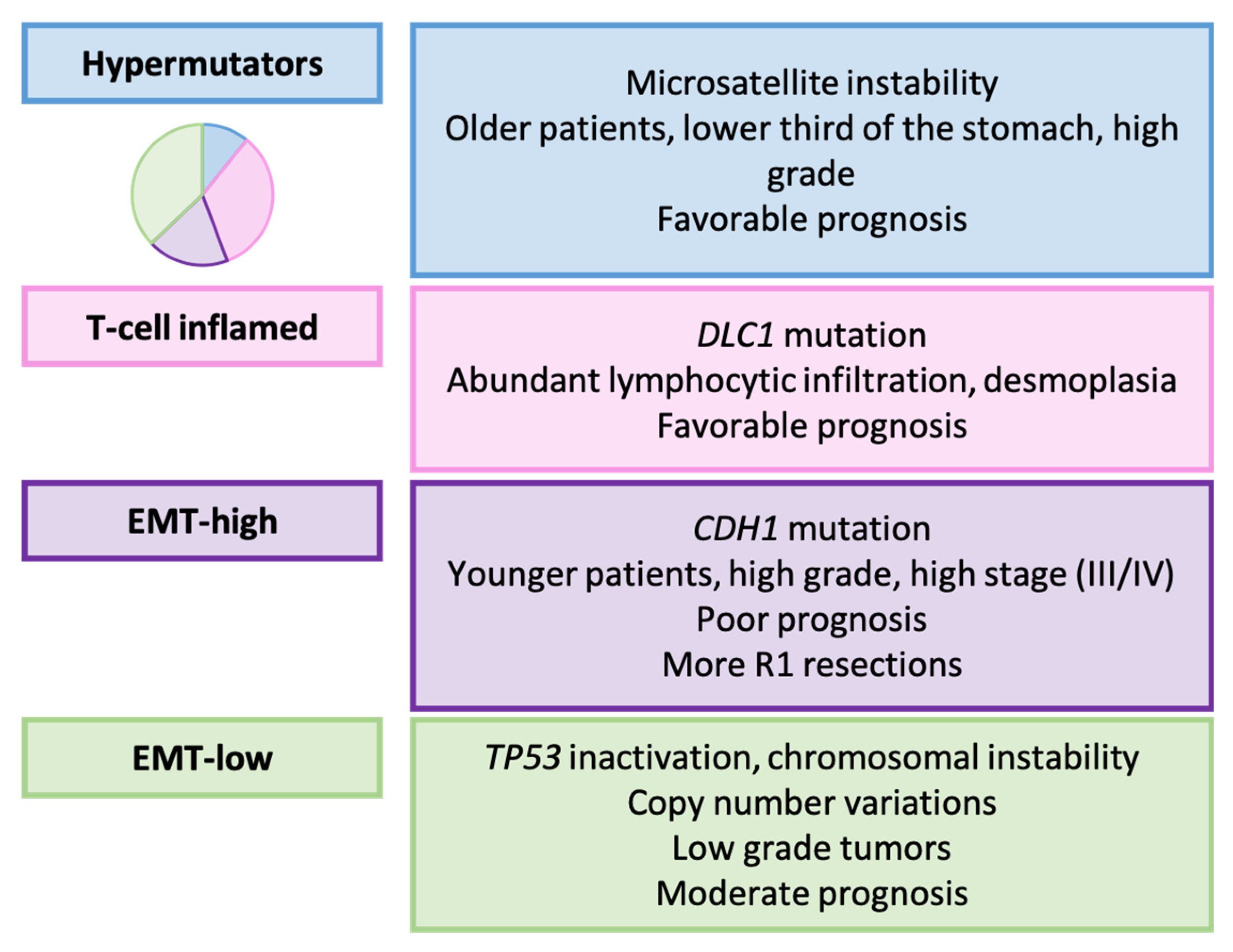
3.7. HOPE Classification (2022) [161]
Relationship with Clinicopathological, Prognostic, and Therapeutic Variables
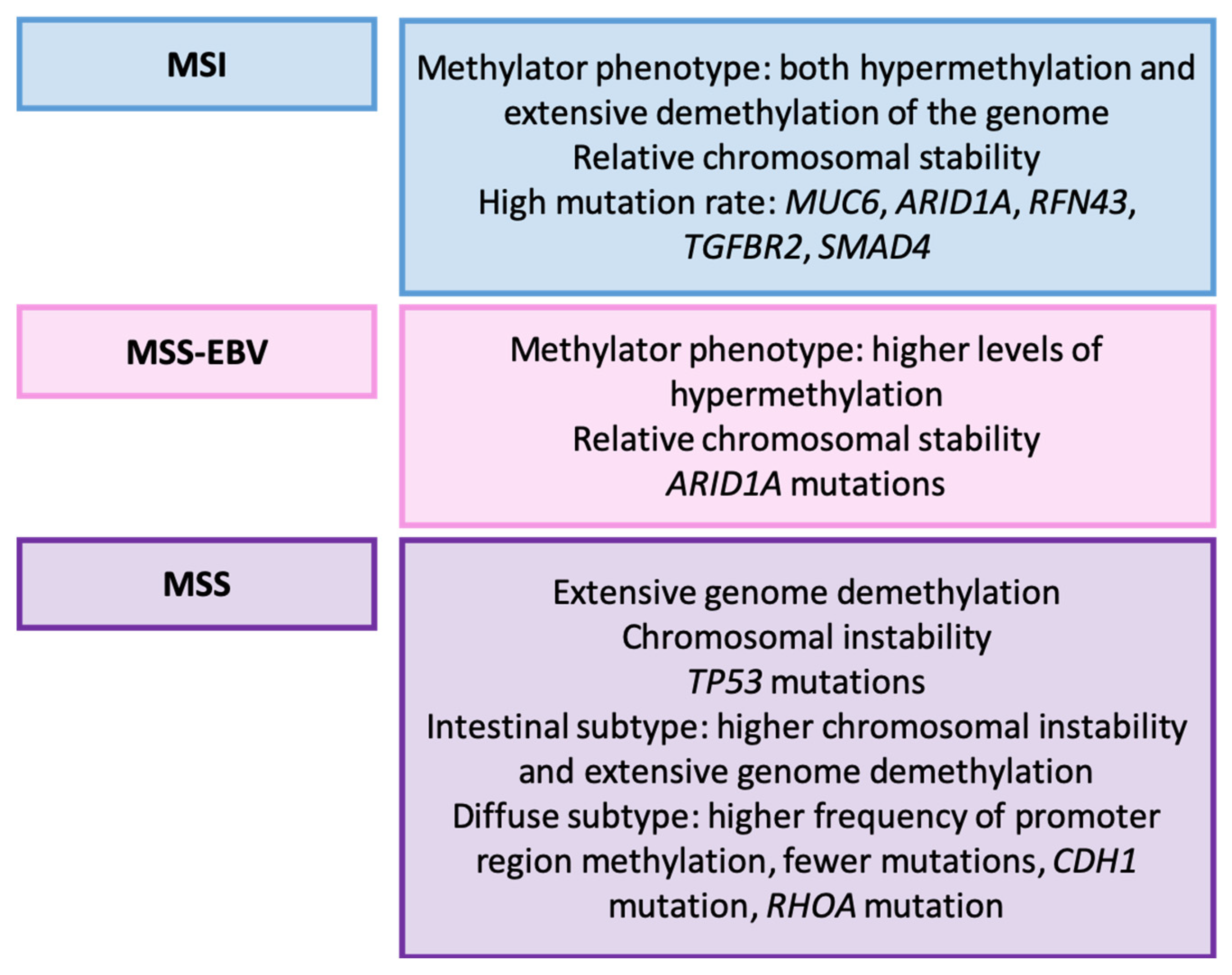
3.8. Wang’s Classification (2014) [162]
Relationship with Clinicopathological, Prognostic, and Therapeutic Variables
3.9. Cheong’s Classification (2022) [164]
3.10. Zhou et al. (2023): Functional Status-Based Classification [165]
3.11. Ye et al. (2022): Metabolism-Based Classification [166]
3.12. Li et al. (2021): Metabolism-Based Classification [167]
3.13. Lin et al. (2021): Immune-Based Classification [168]
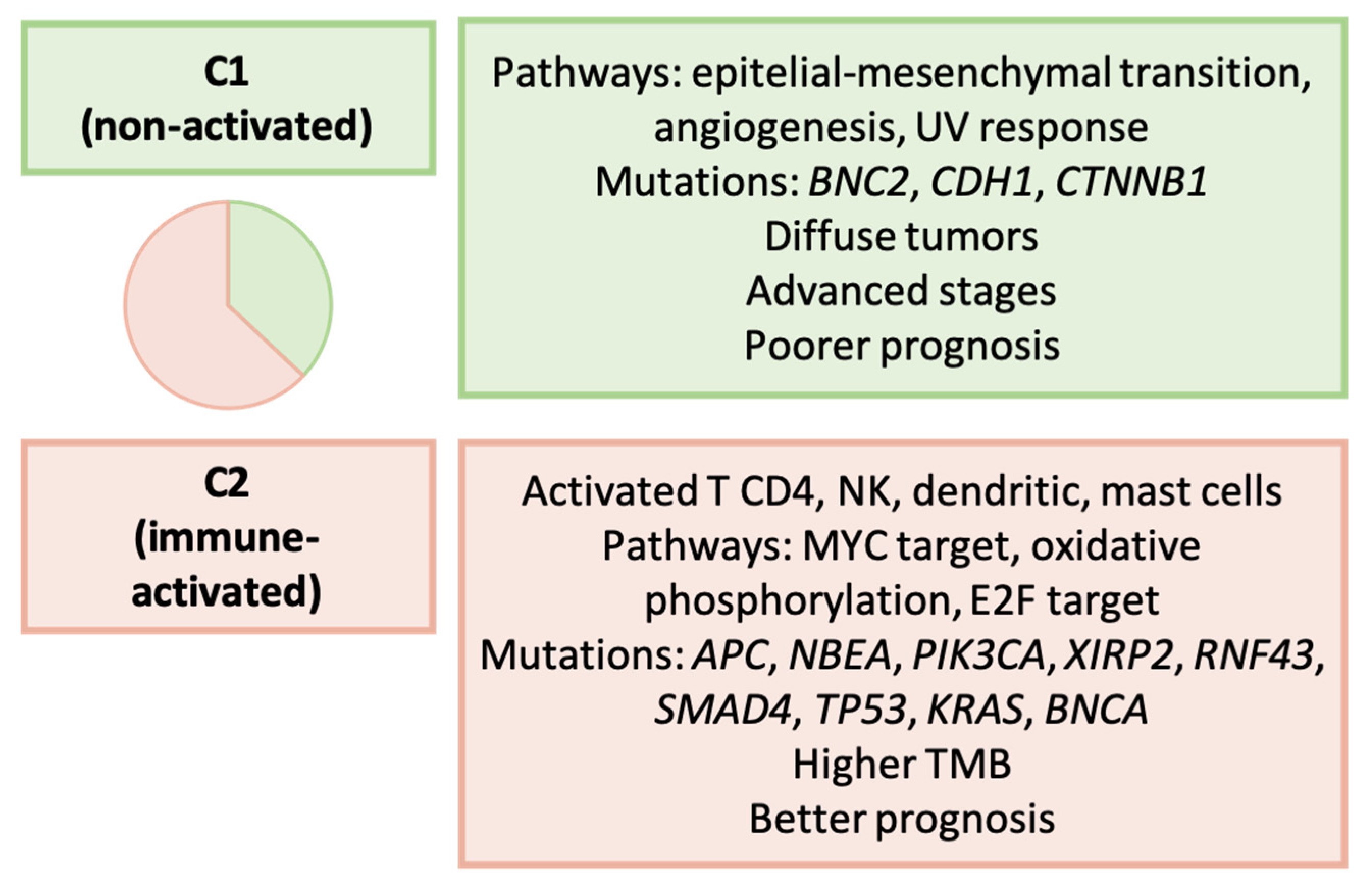
3.14. Wu et al. (2022): Immune-Based Classification [169]
3.15. Li et al. (2016): Tumor Mutational Burden [170]
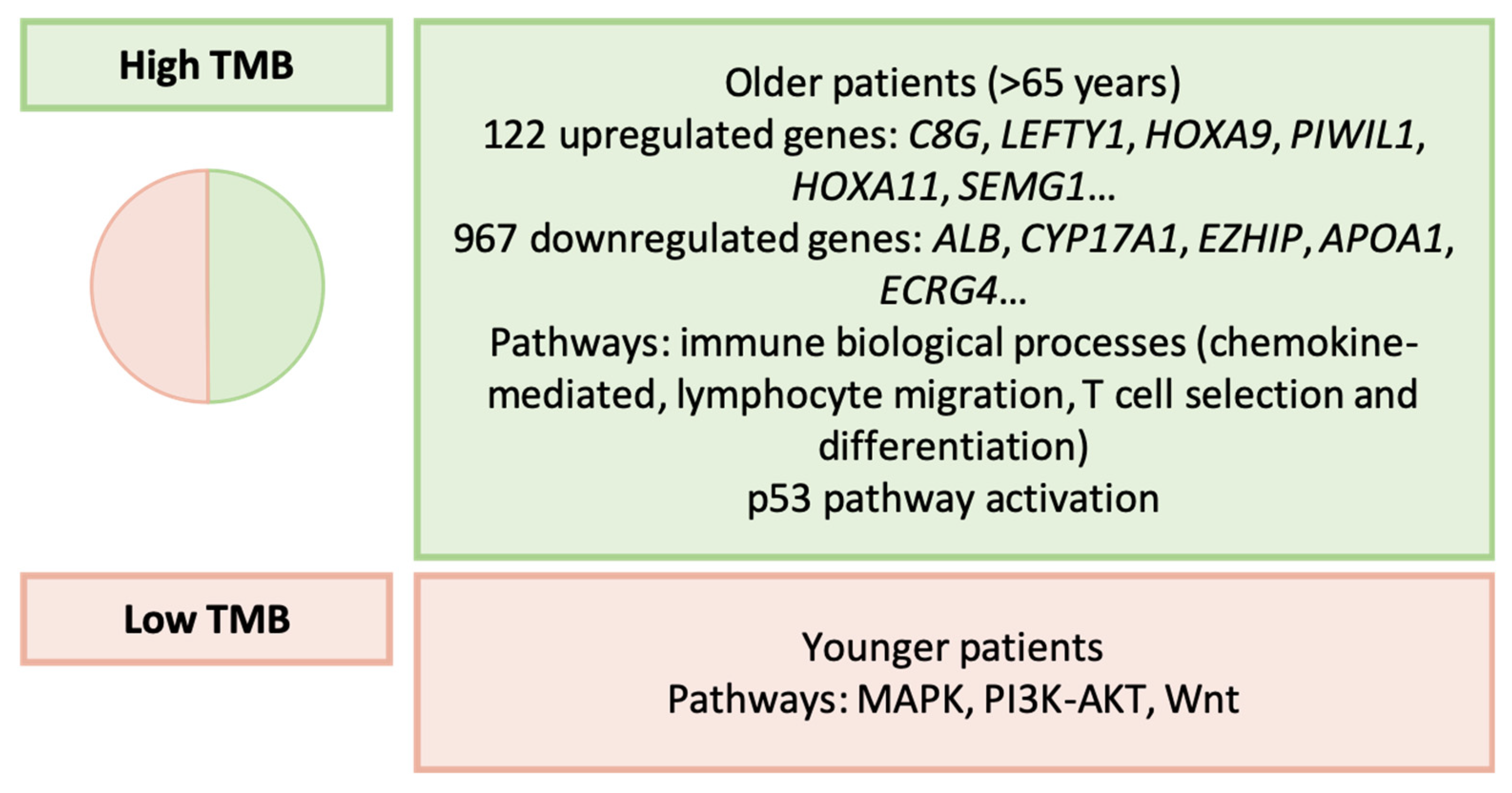
3.16. Wei et al. (2022): Tumor Mutational Burden [171]
3.17. Weng et al. (2023): Epigenetic-Based Classification [172]
3.18. Li et al. (2022): Proteomic-Based Classification [173]
3.19. Tanaka et al. (2021): Ascites-Disseminated GC [175]
4. Clinical Impact of Molecular Classifications
4.1. Application of Molecular Classifications through Surrogate Markers
4.2. Equivalencies between Classifications, Prognostic, and Therapeutic Value
5. Maximizing Impact through Interdisciplinary Collaboration
6. Future Challenges
- Technological Advancements and Improvement of Novel Molecular Techniques:Novel molecular techniques beyond NGS or microarrays hold significant promise in GC. For instance, third-generation or SCS techniques can be valuable for characterizing GC in small samples, such as pre-surgical biopsies or liquid biopsy specimens, and for assessing intratumoral heterogeneity. These methods offer significant potential for analyzing the molecular profile of pre-neoadjuvant GC, monitoring patients, and identifying resistance mechanisms. Unfortunately, their broad availability or integration into clinical routine still requires significant progress.
- Validation of Molecular Classifications:Despite the publication of numerous molecular classifications in GC, those beyond TCGA and the ACRG have not been extensively validated. In addition, it is crucial to confirm previous results in geographically distinct cohorts due to the regional differences observed in the molecular, clinicopathological, and treatment features of GC. In this context, the promotion of open and collaborative GC databases could be beneficial.
- Identification of Surrogate Markers:Identifying optimal surrogate markers for applying TCGA and the ACRG classifications is essential, given the complex approaches used in these studies. Furthermore, exploring suitable surrogate markers for molecular classifications beyond those published by TCGA and the ACRG is also recommended, as this would address a significant gap in the GC literature.
- Consensus on Molecular Classifications:Given the heterogeneity of GC, achieving a consensus molecular classification is necessary for standardizing comparisons between studies in different cohorts. If complete consensus proves challenging, harmonizing the most consistent molecular types across classifications, such as MSI or high TMB tumors, those related to EMT, or those with TP53 alterations, is advisable.
- Incorporation of Clinical and Histological Features:A notable correlation exists between molecular types and certain clinicopathological factors in GC, particularly the Laurén type, and, to a lesser extent, tumor location and other features. Integrating molecular alterations with histopathological findings that carry prognostic or therapeutic significance, akin to the methodologies applied in endometrial cancer, could prove to be a more effective strategy than presuming that a novel molecular classification will completely replace the value of histological features in GC. Achieving such an integrative classification relies on collaboration among different scientific disciplines and a holistic approach, involving all stakeholders in the context of GC diagnosis and treatment.
- Interdisciplinary Collaboration:Fostering an integrative approach necessitates collaboration among diverse professionals, encompassing biologists, clinicians, and pathologists. Essential to this is the training of clinicians and pathologists in molecular pathology, coupled with a deep understanding of clinical practice realities by basic researchers. Open forums for interdisciplinary discussions and knowledge exchanges remain vital for the successful translation of basic research into clinical practice.
- Improved Patient Stratification in Clinical Trials:Enhancing patient stratification in clinical trials could yield valuable insights into new biomarkers. For instance, studies have shown that categorizing patients according to the Laurén type enables the identification of distinct subgroups with diverse treatment responses [8]. The classification of patients in clinical trials based on molecular alterations or prominent histological factors, such as the Laurén type, would facilitate the individualized search for targeted therapies within more homogeneous groups, ultimately enhancing the precision and effectiveness of personalized approaches.
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Ilic, M.; Ilic, I. Epidemiology of stomach cancer. World J. Gastroenterol. 2022, 28, 1187–1203. [Google Scholar] [CrossRef] [PubMed]
- Sexton, R.E.; Al Hallak, M.N.; Uddin, M.H.; Diab, M.; Azmi, A.S. Gastric Cancer Heterogeneity and Clinical Outcomes. Technol. Cancer Res. Treat. 2020, 19, 1533033820935477. [Google Scholar] [CrossRef] [PubMed]
- Gullo, I.; Carneiro, F.; Oliveira, C.; Almeida, G.M. Heterogeneity in Gastric Cancer: From Pure Morphology to Molecular Classifications. Pathobiology 2018, 85, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.E.; Strong, V.E. Gastric Cancer Etiology and Management in Asia and the West. Annu. Rev. Med. 2019, 70, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Rahman, R.; Asombang, A.W.; Ibdah, J.A. Characteristics of gastric cancer in Asia. World J. Gastroenterol. 2014, 20, 4483–4490. [Google Scholar] [CrossRef] [PubMed]
- Yashima, K.; Shabana, M.; Kurumi, H.; Kawaguchi, K.; Isomoto, H. Gastric Cancer Screening in Japan: A Narrative Review. J. Clin. Med. 2022, 11, 4337. [Google Scholar] [CrossRef]
- Díaz del Arco, C.; Ortega Medina, L.; Estrada Muñoz, L.; García Gómez de las Heras, S.; Fernández Aceñero, M.J. Is there still a place for conventional histopathology in the age of molecular medicine? Laurén classification, inflammatory infiltration and other current topics in gastric cancer diagnosis and prognosis. Histol. Histopathol. 2021, 36, 587–613. [Google Scholar]
- Ye, X.S.; Yu, C.; Aggarwal, A.; Reinhard, C. Genomic alterations and molecular subtypes of gastric cancers in Asians. Chin. J. Cancer 2016, 35, 42. [Google Scholar] [CrossRef]
- Kim, J.; Sun, C.L.; Mailey, B.; Prendergast, C.; Artinyan, A.; Bhatia, S.; Pigazzi, A.; Ellenhorn, J.D.I. Race and ethnicity correlate with survival in patients with gastric adenocarcinoma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2010, 21, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Park, J.Y.; Camargo, M.C.; Lunet, N.; Forman, D.; Soerjomataram, I. Is gastric cancer becoming a rare disease? A global assessment of predicted incidence trends to 2035. Gut 2020, 69, 823–829. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Sano, T.; Kodera, Y. Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer 2011, 14, 101–112. [Google Scholar]
- The Paris endoscopic classification of superficial neoplastic lesions: Esophagus, stomach, and colon: November 30 to December 1, 2002. Gastrointest. Endosc. 2003, 58, S3–S43. [CrossRef]
- Song, X.H.; Zhang, W.H.; Kai-Liu; Chen, X.L.; Zhao, L.Y.; Chen, X.Z.; Kun-Yang; Zhou, Z.G.; Hu, J.K. Prognostic impact of Borrmann classification on advanced gastric cancer: A retrospective cohort from a single institution in western China. World J. Surg. Oncol. 2020, 18, 204. [Google Scholar] [CrossRef]
- Laurén, P. The two histological main types of gastric carcinoma: Diffuse and so-alled intestinal-type arcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Yang, H.; Yang, W.J.; Hu, B. Gastric epithelial histology and precancerous conditions. World J. Gastrointest. Oncol. 2022, 14, 396–412. [Google Scholar] [CrossRef] [PubMed]
- Correa, P.; Piazuelo, M.B. The gastric precancerous cascade. J. Dig. Dis. 2012, 13, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Kushima, R. The updated WHO classification of digestive system tumours-gastric adenocarcinoma and dysplasia. Pathologe 2022, 43, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Agnes, A.; Estrella, J.S.; Badgwell, B. The significance of a nineteenth century definition in the era of genomics: Linitis plastica. World J. Surg. Oncol. 2017, 15, 123. [Google Scholar] [CrossRef] [PubMed]
- Tegels, J.J.W.; De Maat, M.F.G.; Hulsewé, K.W.E.; Hoofwijk, A.G.M.; Stoot, J.H. Improving the outcomes in gastric cancer surgery. World J. Gastroenterol. 2014, 20, 13692–13704. [Google Scholar] [CrossRef]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Wagner, A.D.; Syn, N.L.X.; Moehler, M.; Grothe, W.; Yong, W.P.; Tai, B.C.; Ho, J.; Unverzagt, S. Chemotherapy for advanced gastric cancer. Cochrane Database Syst. Rev. 2017, 8, CD004064. [Google Scholar] [CrossRef] [PubMed]
- Hironaka, S. Anti-angiogenic therapies for gastric cancer. Asia Pac. J. Clin. Oncol. 2019, 15, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhu, X.; Wei, X.; Tang, C.; Zhang, W. HER2-targeted therapies in gastric cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188549. [Google Scholar] [CrossRef] [PubMed]
- Salati, M.; Orsi, G.; Smyth, E.; Beretta, G.; De Vita, F.; Di Bartolomeo, M.; Fanotto, V.; Lonardi, S.; Morano, F.; Pietrantonio, F.; et al. Gastric cancer: Translating novels concepts into clinical practice. Cancer Treat. Rev. 2019, 79, 101889. [Google Scholar] [CrossRef] [PubMed]
- Boyiadzis, M.M.; Kirkwood, J.M.; Marshall, J.L.; Pritchard, C.C.; Azad, N.S.; Gulley, J.L. Significance and implications of FDA approval of pembrolizumab for biomarker-defined disease. J. Immunother. Cancer 2018, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.D.; Mathias-Machado, M.C.; Jácome, A.; Gil, M.; Fogacci, J.; Sodré, B.; Passarini, T.; Chaves, A.; Diniz, P.H.; Lino, F.; et al. PD-L1 testing in advanced gastric cancer-what physicians who treat this disease must know-a literature review. J. Gastrointest. Oncol. 2023, 14, 1560–1575. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Kawazoe, A.; Lordick, F.; Janjigian, Y.Y.; Shitara, K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: An emerging paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 473–487. [Google Scholar] [CrossRef]
- Agnarelli, A.; Vella, V.; Samuels, M.; Papanastasopoulos, P.; Giamas, G. Incorporating Immunotherapy in the Management of Gastric Cancer: Molecular and Clinical Implications. Cancers 2022, 14, 4378. [Google Scholar] [CrossRef]
- Del Prete, C.; Muthiah, A.; Almhanna, K. Does tumor profile in gastric and gastroesophageal (GE) junction cancer justify off-label use of targeted therapy?—A narrative review. Ann. Transl. Med. 2020, 8, 1110. [Google Scholar] [CrossRef]
- Sarhadi, V.K.; Armengol, G. Molecular Biomarkers in Cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef]
- Saller, J.J.; Boyle, T.A. Molecular Pathology of Lung Cancer. Cold Spring Harb. Perspect. Med. 2022, 12, a037812. [Google Scholar] [CrossRef]
- Zhang, X. Molecular Classification of Breast Cancer: Relevance and Challenges. Arch. Pathol. Lab. Med. 2023, 147, 46–51. [Google Scholar] [CrossRef]
- Guo, J.; Yu, W.; Su, H.; Pang, X. Genomic landscape of gastric cancer: Molecular classification and potential targets. Sci. China. Life Sci. 2017, 60, 126–137. [Google Scholar] [CrossRef]
- Rostami-Nejad, M.; Rezaei-Tavirani, M.; Mansouri, V.; Akbari, Z.; Abdi, S. Impact of proteomics investigations on gastric cancer treatment and diagnosis. Gastroenterol. Hepatol. Bed Bench 2019, 12, S1. [Google Scholar]
- Yuan, Q.; Deng, D.; Pan, C.; Ren, J.; Wei, T.; Wu, Z.; Zhang, B.; Li, S.; Yin, P.; Shang, D. Integration of transcriptomics, proteomics, and metabolomics data to reveal HER2-associated metabolic heterogeneity in gastric cancer with response to immunotherapy and neoadjuvant chemotherapy. Front. Immunol. 2022, 13, 951137. [Google Scholar] [CrossRef] [PubMed]
- Pužar Dominkuš, P.; Hudler, P. Mutational Signatures in Gastric Cancer and Their Clinical Implications. Cancers 2023, 15, 3788. [Google Scholar] [CrossRef] [PubMed]
- Carino, A.; Graziosi, L.; Marchianò, S.; Biagioli, M.; Marino, E.; Sepe, V.; Zampella, A.; Distrutti, E.; Donini, A.; Fiorucci, S. Analysis of Gastric Cancer Transcriptome Allows the Identification of Histotype Specific Molecular Signatures with Prognostic Potential. Front. Oncol. 2021, 11, 663771. [Google Scholar] [CrossRef] [PubMed]
- Kadam, W.; Wei, B.; Li, F. Metabolomics of Gastric Cancer. Adv. Exp. Med. Biol. 2021, 1280, 291–301. [Google Scholar] [PubMed]
- Tang, S.Y.; Zhou, P.J.; Meng, Y.; Zeng, F.R.; Deng, G.T. Gastric cancer: An epigenetic view. World J. Gastrointest. Oncol. 2022, 14, 90–109. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Min, H.Y.; Lee, H.Y. Molecular targeted therapy for anticancer treatment. Exp. Mol. Med. 2022, 54, 1670–1694. [Google Scholar] [CrossRef]
- Bumgarner, R. Overview of DNA microarrays: Types, applications, and their future. Curr. Protoc. Mol. Biol. 2013, 101, 22.1.1–22.1.11. [Google Scholar] [CrossRef]
- Sealfon, S.C.; Chu, T.T. RNA and DNA microarrays. Methods Mol. Biol. 2011, 671, 3–34. [Google Scholar]
- Sutandy, F.X.R.; Qian, J.; Chen, C.S.; Zhu, H. Overview of protein microarrays. Curr. Protoc. Protein Sci. 2013, 72, 27.1.1–27.1.16. [Google Scholar] [CrossRef]
- Kao, K.J.; Chang, K.M.; Hsu, H.C.; Huang, A.T. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer 2011, 11, 143. [Google Scholar] [CrossRef]
- Yao, K.; Tong, C.Y.; Cheng, C. A framework to predict the applicability of Oncotype DX, MammaPrint, and E2F4 gene signatures for improving breast cancer prognostic prediction. Sci. Rep. 2022, 12, 2211. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G.; Di Rienzo, T.; Ojetti, V. Microarray analysis in gastric cancer: A review. World J. Gastroenterol. 2014, 20, 11972–11976. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Wang, Y.C.; Huang, Q.J. Microarray and next-generation sequencing to analyse gastric cancer. Asian Pac. J. Cancer Prev. 2014, 15, 8033–8039. [Google Scholar] [CrossRef]
- Verma, R.; Sharma, P.C. Next generation sequencing-based emerging trends in molecular biology of gastric cancer. Am. J. Cancer Res. 2018, 8, 207–225. [Google Scholar] [PubMed]
- Businello, G.; Galuppini, F.; Fassan, M. The impact of recent next generation sequencing and the need for a new classification in gastric cancer. Best Pract. Res. Clin. Gastroenterol. 2021, 50–51, 101730. [Google Scholar] [CrossRef]
- Ku, G.Y. Next generation sequencing in gastric or gastroesophageal adenocarcinoma. Transl. Gastroenterol. Hepatol. 2020, 5, 56. [Google Scholar] [CrossRef]
- Puliga, E.; Corso, S.; Pietrantonio, F.; Giordano, S. Microsatellite instability in Gastric Cancer: Between lights and shadows. Cancer Treat. Rev. 2021, 95, 102175. [Google Scholar] [CrossRef]
- Duan, Y.; Xu, D. Microsatellite instability and immunotherapy in gastric cancer: A narrative review. Precis. Cancer Med. 2023, 6. [Google Scholar] [CrossRef]
- Ke, L.; Li, S.; Huang, D. The predictive value of tumor mutation burden on survival of gastric cancer patients treated with immune checkpoint inhibitors: A systematic review and meta-analysis. Int. Immunopharmacol. 2023, 124, 110986. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, Q.; Zhang, Z.; Peng, S.; Liu, J.; Pang, J.; Jia, Z.; Xi, H.; Li, J.; Chen, L.; et al. Identification of rare thalassemia variants using third-generation sequencing. Front. Genet. 2023, 13, 1076035. [Google Scholar] [CrossRef] [PubMed]
- Ambardar, S.; Gupta, R.; Trakroo, D.; Lal, R.; Vakhlu, J. High Throughput Sequencing: An Overview of Sequencing Chemistry. Indian J. Microbiol. 2016, 56, 394–404. [Google Scholar] [CrossRef]
- Athanasopoulou, K.; Boti, M.A.; Adamopoulos, P.G.; Skourou, P.C.; Scorilas, A. Third-Generation Sequencing: The Spearhead towards the Radical Transformation of Modern Genomics. Life 2021, 12, 30. [Google Scholar] [CrossRef]
- Xiao, T.; Zhou, W. The third generation sequencing: The advanced approach to genetic diseases. Transl. Pediatr. 2020, 9, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Liu, Z.; Zhang, Y.; Lu, P.; Wen, L.; Tang, F. High-throughput and high-sensitivity full-length single-cell RNA-seq analysis on third-generation sequencing platform. Cell Discov. 2023, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Deng, E.; Wang, J.; Zhou, W.; Ao, J.; Liu, R.; Su, D.; Fan, X. Single-cell third-generation sequencing-based multi-omics uncovers gene expression changes governed by ecDNA and structural variants in cancer cells. Clin. Transl. Med. 2023, 13, e1351. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Tang, F. Recent advances in single-cell sequencing technologies. Precis. Clin. Med. 2022, 5, pbac002. [Google Scholar] [CrossRef] [PubMed]
- Anaparthy, N.; Ho, Y.J.; Martelotto, L.; Hammell, M.; Hicks, J. Single-Cell Applications of Next-Generation Sequencing. Cold Spring Harb. Perspect. Med. 2019, 9, a026898. [Google Scholar] [CrossRef] [PubMed]
- Melnekoff, D.T.; Laganà, A. Single-Cell Sequencing Technologies in Precision Oncology. Adv. Exp. Med. Biol. 2022, 1361, 269–282. [Google Scholar] [PubMed]
- Zhang, Y.; Wang, D.; Peng, M.; Tang, L.; Ouyang, J.; Xiong, F.; Guo, C.; Tang, Y.; Zhou, Y.; Liao, Q.; et al. Single-cell RNA sequencing in cancer research. J. Exp. Clin. Cancer Res. 2021, 40, 81. [Google Scholar] [CrossRef]
- Xu, J.; Liao, K.; Yang, X.; Wu, C.; Wu, W.; Han, S. Using single-cell sequencing technology to detect circulating tumor cells in solid tumors. Mol. Cancer 2021, 20, 104. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Z.; Li, Y.; Du, Y.; Chen, G. Application of single-cell sequencing to the research of tumor microenvironment. Front. Immunol. 2023, 14, 1285540. [Google Scholar] [CrossRef]
- Pfisterer, U.; Bräunig, J.; Brattås, P.; Heidenblad, M.; Karlsson, G.; Fioretos, T. Single-cell sequencing in translational cancer research and challenges to meet clinical diagnostic needs. Genes. Chromosomes Cancer 2021, 60, 504–524. [Google Scholar] [CrossRef]
- Deng, G.; Zhang, X.; Chen, Y.; Liang, S.; Liu, S.; Yu, Z.; Lü, M. Single-cell transcriptome sequencing reveals heterogeneity of gastric cancer: Progress and prospects. Front. Oncol. 2023, 13, 1074268. [Google Scholar] [CrossRef]
- Bian, S.; Wang, Y.; Zhou, Y.; Wang, W.; Guo, L.; Wen, L.; Fu, W.; Zhou, X.; Tang, F. Integrative single-cell multiomics analyses dissect molecular signatures of intratumoral heterogeneities and differentiation states of human gastric cancer. Natl. Sci. Rev. 2023, 10, nwad094. [Google Scholar] [CrossRef]
- Wang, R.; Dang, M.; Harada, K.; Han, G.; Wang, F.; Pool Pizzi, M.; Zhao, M.; Tatlonghari, G.; Zhang, S.; Hao, D.; et al. Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma. Nat. Med. 2021, 27, 141–151. [Google Scholar] [CrossRef]
- Katona, B.W.; Rustgi, A.K. Gastric Cancer Genomics: Advances and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Hao, Q.; Vadgama, J.; Wu, Y. Wild-Type TP53 Predicts Poor Prognosis in Patients with Gastric Cancer. J. Cancer Sci. Clin. Ther. 2021, 5, 134–153. [Google Scholar] [CrossRef] [PubMed]
- Donehower, L.A.; Soussi, T.; Korkut, A.; Liu, Y.; Schultz, A.; Cardenas, M.; Li, X.; Babur, O.; Hsu, T.K.; Lichtarge, O.; et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep. 2019, 28, 1370–1384.e5. [Google Scholar] [CrossRef] [PubMed]
- Graziano, F.; Fischer, N.W.; Bagaloni, I.; Di Bartolomeo, M.; Lonardi, S.; Vincenzi, B.; Perrone, G.; Fornaro, L.; Ongaro, E.; Aprile, G.; et al. TP53 Mutation Analysis in Gastric Cancer and Clinical Outcomes of Patients with Metastatic Disease Treated with Ramucirumab/Paclitaxel or Standard Chemotherapy. Cancers 2020, 12, 2049. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhou, Y.; Huang, L.; Ou, W.; Wu, J.; Li, S.; Xu, J.; Feng, J.; Liu, B. TP53 mutation is associated with a poor clinical outcome for non-small cell lung cancer: Evidence from a meta-analysis. Mol. Clin. Oncol. 2016, 5, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Zheng, X.; Shariati, M.; Damodaran, S.; Wathoo, C.; Brusco, L.; Demirhan, M.E.; Tapia, C.; Eterovic, A.K.; Basho, R.K.; et al. Survival Outcomes by TP53 Mutation Status in Metastatic Breast Cancer. JCO Precis. Oncol. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.C.; Lowe, S.W. Mutant p53: It’s not all one and the same. Cell Death Differ. 2022, 29, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, M.; Kaya, V.; Demirpence, O.; Gunduz, S.; Bozcuk, H. Prognostic significance of p53 in gastric cancer: A meta- analysis. Asian Pac. J. Cancer Prev. 2015, 16, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Fenoglio-Preiser, C.M.; Wang, J.; Stemmermann, G.N.; Noffsinger, A. TP53 and gastric carcinoma: A review. Hum. Mutat. 2003, 21, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chu, K.M. E-cadherin and gastric cancer: Cause, consequence, and applications. Biomed Res. Int. 2014, 2014, 637308. [Google Scholar] [CrossRef]
- Garziera, M.; Canzonieri, V.; Cannizzaro, R.; Geremia, S.; Caggiari, L.; De Zorzi, M.; Maiero, S.; Orzes, E.; Perin, T.; Zanussi, S.; et al. Identification and characterization of CDH1 germline variants in sporadic gastric cancer patients and in individuals at risk of gastric cancer. PLoS ONE 2013, 8, e77305. [Google Scholar] [CrossRef]
- Cho, S.Y.; Park, J.W.; Liu, Y.; Park, Y.S.; Kim, J.H.; Yang, H.; Um, H.; Ko, W.R.; Lee, B.I.; Kwon, S.Y.; et al. Sporadic Early-Onset Diffuse Gastric Cancers Have High Frequency of Somatic CDH1 Alterations, but Low Frequency of Somatic RHOA Mutations Compared with Late-Onset Cancers. Gastroenterology 2017, 153, 536–549. [Google Scholar] [CrossRef]
- Machado, J.C.; Oliveira, C.; Carvalho, R.; Soares, P.; Berx, G.; Caldas, C.; Seruca, R.; Carneiro, F.; Sobrinho-Simöes, M. E-cadherin gene (CDH1) promoter methylation as the second hit in sporadic diffuse gastric carcinoma. Oncogene 2001, 20, 1525–1528. [Google Scholar] [CrossRef]
- Kim, Y.S.; Jeong, H.; Choi, J.W.; Oh, H.; Lee, J.H. Unique characteristics of ARID1A mutation and protein level in gastric and colorectal cancer: A meta-analysis. Saudi J. Gastroenterol. 2017, 23, 268–274. [Google Scholar]
- Buckley, K.H.; Niccum, B.A.; Maxwell, K.N.; Katona, B.W. Gastric Cancer Risk and Pathogenesis in BRCA1 and BRCA2 Carriers. Cancers 2022, 14, 5953. [Google Scholar] [CrossRef]
- Kim, J.W.; Lee, H.S.; Nam, K.H.; Ahn, S.; Kim, J.W.; Ahn, S.H.; Park, D.J.; Kim, H.H.; Lee, K.W. PIK3CA mutations are associated with increased tumor aggressiveness and Akt activation in gastric cancer. Oncotarget 2017, 8, 90948–90958. [Google Scholar] [CrossRef]
- Morishita, A.; Gong, J.; Masaki, T. Targeting receptor tyrosine kinases in gastric cancer. World J. Gastroenterol. 2014, 20, 4536–4545. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Nagamura, Y.; Miyazaki, M. Receptor Tyrosine Kinases Amplified in Diffuse-Type Gastric Carcinoma: Potential Targeted Therapies and Novel Downstream Effectors. Cancers 2022, 14, 3750. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, J.H.; Jang, H.J. Pathologic and prognostic impacts of FGFR2 amplification in gastric cancer: A meta-analysis and systemic review. J. Cancer 2019, 10, 2560–2567. [Google Scholar] [CrossRef]
- Park, C.K.; Park, J.S.; Kim, H.S.; Rha, S.Y.; Hyung, W.J.; Cheong, J.H.; Noh, S.H.; Lee, S.K.; Lee, Y.C.; Huh, Y.m.; et al. Receptor tyrosine kinase amplified gastric cancer: Clinicopathologic characteristics and proposed screening algorithm. Oncotarget 2016, 7, 72099–72112. [Google Scholar] [CrossRef]
- Roviello, G.; Catalano, M.; Iannone, L.F.; Marano, L.; Brugia, M.; Rossi, G.; Aprile, G.; Antonuzzo, L. Current status and future perspectives in HER2 positive advanced gastric cancer. Clin. Transl. Oncol. 2022, 24, 981–996. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Xu, Y.H. Trastuzumab combined chemotherapy for the treatment of HER2-positive advanced gastric cancer: A systematic review and meta-analysis of randomized controlled trial. Medicine 2022, 101, E29992. [Google Scholar] [CrossRef]
- Rüschoff, J.; Hanna, W.; Bilous, M.; Hofmann, M.; Osamura, R.Y.; Penault-Llorca, F.; Van De Vijver, M.; Viale, G. HER2 testing in gastric cancer: A practical approach. Mod. Pathol. 2012, 25, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Lago, N.M.; Villar, M.V.; Ponte, R.V.; Nallib, I.A.; Carrera Alvarez, J.J.; Antúnez López, J.R.; López, R.L.; Padin Iruegas, M.E. Impact of HER2 status in resected gastric or gastroesophageal junction adenocarcinoma in a Western population. Ecancermedicalscience 2020, 14, 1020. [Google Scholar]
- Wang, H.B.; Liao, X.F.; Zhang, J. Clinicopathological factors associated with HER2-positive gastric cancer: A meta-analysis. Medicine 2017, 96, e8437. [Google Scholar] [CrossRef]
- Kim, G.H.; Jung, H.Y. Endoscopic Resection of Gastric Cancer. Gastrointest. Endosc. Clin. N. Am. 2021, 31, 563–579. [Google Scholar] [CrossRef]
- Lordick, F.; Carneiro, F.; Cascinu, S.; Fleitas, T.; Haustermans, K.; Piessen, G.; Vogel, A.; Smyth, E.C. Gastric cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2022, 33, 1005–1020. [Google Scholar] [CrossRef]
- Cunningham, D.; Allum, W.H.; Stenning, S.P.; Thompson, J.N.; Van de Velde, C.J.H.; Nicolson, M.; Scarffe, J.H.; Lofts, F.J.; Falk, S.J.; Iveson, T.J.; et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N. Engl. J. Med. 2006, 355, 11–20. [Google Scholar] [CrossRef]
- Al-Batran, S.E.; Homann, N.; Pauligk, C.; Goetze, T.O.; Meiler, J.; Kasper, S.; Kopp, H.G.; Mayer, F.; Haag, G.M.; Luley, K.; et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): A randomised, phase 2/3 trial. Lancet 2019, 393, 1948–1957. [Google Scholar]
- Ychou, M.; Boige, V.; Pignon, J.P.; Conroy, T.; Bouché, O.; Lebreton, G.; Ducourtieux, M.; Bedenne, L.; Fabre, J.M.; Saint-Aubert, B.; et al. Perioperative chemotherapy compared with surgery alone for resectable gastroesophageal adenocarcinoma: An FNCLCC and FFCD multicenter phase III trial. J. Clin. Oncol. 2011, 29, 1715–1721. [Google Scholar] [CrossRef]
- Ajani, J.A.; Bentrem, D.J.; Besh, S.; D’Amico, T.A.; Das, P.; Denlinger, C.; Fakih, M.G.; Fuchs, C.S.; Gerdes, H.; Glasgow, R.E.; et al. Gastric cancer, version 2.2013: Featured updates to the NCCN Guidelines. J. Natl. Compr. Canc. Netw. 2013, 11, 531–546. [Google Scholar] [CrossRef]
- Eom, S.S.; Choi, W.; Eom, B.W.; Park, S.H.; Kim, S.J.; Kim, Y.I.; Yoon, H.M.; Lee, J.Y.; Kim, C.G.; Kim, H.K.; et al. A Comprehensive and Comparative Review of Global Gastric Cancer Treatment Guidelines. J. Gastric Cancer 2022, 22, 3–23. [Google Scholar] [CrossRef]
- Yu, J. Il Role of Adjuvant Radiotherapy in Gastric Cancer. J. Gastric Cancer 2023, 23, 194–206. [Google Scholar] [CrossRef]
- van Boxel, G.I.; Ruurda, J.P.; van Hillegersberg, R. Robotic-assisted gastrectomy for gastric cancer: A European perspective. Gastric Cancer 2019, 22, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Hakkenbrak, N.A.G.; Jansma, E.P.; van der Wielen, N.; van der Peet, D.L.; Straatman, J. Laparoscopic versus open distal gastrectomy for gastric cancer: A systematic review and meta-analysis. Surgery 2022, 171, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Lou, S.; Yin, X.; Wang, Y.; Zhang, Y.; Xue, Y. Laparoscopic versus open gastrectomy for gastric cancer: A systematic review and meta-analysis of randomized controlled trials. Int. J. Surg. 2022, 102, 106678. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, G.P.; Esposito, G.; Magistri, P.; Serra, V.; Guidetti, C.; Olivieri, T.; Catellani, B.; Assirati, G.; Ballarin, R.; Di Sandro, S.; et al. Robotic versus laparoscopic gastrectomy for gastric cancer: The largest meta-analysis. Int. J. Surg. 2020, 82, 210–228. [Google Scholar] [CrossRef] [PubMed]
- Pennell, N.A.; Arcila, M.E.; Gandara, D.R.; West, H. Biomarker Testing for Patients with Advanced Non-Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am. Soc. Clin. Oncol. Educ. book. Am. Soc. Clin. Oncol. Annu. Meet. 2019, 39, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Berek, J.S.; Matias-Guiu, X.; Creutzberg, C.; Fotopoulou, C.; Gaffney, D.; Kehoe, S.; Lindemann, K.; Mutch, D.; Concin, N.; Berek, J.S.; et al. FIGO staging of endometrial cancer: 2023. Int. J. Gynaecol. Obstet. 2023, 162, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Arai, H.; Iwasa, S.; Boku, N.; Kawahira, M.; Yasui, H.; Masuishi, T.; Muro, K.; Minashi, K.; Hironaka, S.; Fukuda, N.; et al. Fluoropyrimidine with or without platinum as first-line chemotherapy in patients with advanced gastric cancer and severe peritoneal metastasis: A multicenter retrospective study. BMC Cancer 2019, 19, 652. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.J.G.; Al-Abdulla, R.; Lozano, E.; Briz, O.; Bujanda, L.; Banales, J.M.; Macias, R.I.R. Mechanisms of Resistance to Chemotherapy in Gastric Cancer. Anticancer. Agents Med. Chem. 2016, 16, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Yakir, R.; Luna, K.; Marc, W.; Tamar, S.; Avraham, R.; Ayala, H. The toxicity and outcomes of continuous 5-fluorouracil/cisplatin-based chemotherapy followed by chemoradiation in patients with resected high-risk gastric cancer: Results of a single institute. Ann. Acad. Med. Singapore 2008, 37, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Rivera, F.; Vega-Villegas, M.E.; López-Brea, M.F. Chemotherapy of advanced gastric cancer. Cancer Treat. Rev. 2007, 33, 315–324. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, M. Targeted therapies for cancer. BMC Med. 2022, 20, 90. [Google Scholar] [CrossRef]
- Liu, S.; Kurzrock, R. Toxicity of targeted therapy: Implications for response and impact of genetic polymorphisms. Cancer Treat. Rev. 2014, 40, 883–891. [Google Scholar] [CrossRef]
- Ajani, J.A.; D’Amico, T.A.; Bentrem, D.J.; Chao, J.; Cooke, D.; Corvera, C.; Das, P.; Enzinger, P.C.; Enzler, T.; Fanta, P.; et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 167–192. [Google Scholar] [CrossRef]
- Shinozaki-Ushiku, A.; Ishikawa, S.; Komura, D.; Seto, Y.; Aburatani, H.; Ushiku, T. The first case of gastric carcinoma with NTRK rearrangement: Identification of a novel ATP1B-NTRK1 fusion. Gastric Cancer 2020, 23, 944–947. [Google Scholar] [CrossRef]
- Tan, Y.K.; Fielding, J.W.L. Early diagnosis of early gastric cancer. Eur. J. Gastroenterol. Hepatol. 2006, 18, 821–829. [Google Scholar] [CrossRef]
- Onodera, H.; Tokunaga, A.; Yoshiyuki, T.; Kiyama, T.; Kato, S.; Matsukura, N.; Masuda, G.; Tajiri, T. Surgical outcome of 483 patients with early gastric cancer: Prognosis, postoperative morbidity and mortality, and gastric remnant cancer. Hepatogastroenterology 2004, 51, 82–85. [Google Scholar] [PubMed]
- Chiarello, M.M.; Fico, V.; Pepe, G.; Tropeano, G.; Adams, N.J.; Altieri, G.; Brisinda, G. Early gastric cancer: A challenge in Western countries. World J. Gastroenterol. 2022, 28, 693–703. [Google Scholar] [CrossRef]
- Liu, W.; Xu, J.; Zhang, C. Clinical usefulness of gastric adenocarcinoma predictive long intergenic noncoding RNA in human malignancies: A meta-analysis. Pathol. Res. Pract. 2019, 215, 152387. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.M.; Meng, Q.B.; Yu, J.C.; Ma, Z.Q.; Li, Z.T. Factors associated with early recurrence after curative surgery for gastric cancer. World J. Gastroenterol. 2015, 21, 5934. [Google Scholar] [CrossRef]
- Aurello, P.; Petrucciani, N.; Antolino, L.; Giulitti, D.; D’Angelo, F.; Ramacciato, G. Follow-up after curative resection for gastric cancer: Is it time to tailor it? World J. Gastroenterol. 2017, 23, 3379–3387. [Google Scholar] [CrossRef]
- Mokadem, I.; Dijksterhuis, W.P.M.; van Putten, M.; Heuthorst, L.; de Vos-Geelen, J.M.; Haj Mohammad, N.; Nieuwenhuijzen, G.A.P.; van Laarhoven, H.W.M.; Verhoeven, R.H.A. Recurrence after preoperative chemotherapy and surgery for gastric adenocarcinoma: A multicenter study. Gastric Cancer 2019, 22, 1263. [Google Scholar] [CrossRef] [PubMed]
- Nakauchi, M.; Vos, E.; Tang, L.H.; Gonen, M.; Janjigian, Y.Y.; Ku, G.Y.; Ilson, D.H.; Maron, S.B.; Yoon, S.S.; Brennan, M.F.; et al. Outcomes of Neoadjuvant Chemotherapy for Clinical Stages 2 and 3 Gastric Cancer Patients: Analysis of Timing and Site of Recurrence. Ann. Surg. Oncol. 2021, 28, 4829–4838. [Google Scholar] [CrossRef]
- Lavacchi, D.; Fancelli, S.; Buttitta, E.; Vannini, G.; Guidolin, A.; Winchler, C.; Caliman, E.; Vannini, A.; Giommoni, E.; Brugia, M.; et al. Perioperative Tailored Treatments for Gastric Cancer: Times Are Changing. Int. J. Mol. Sci. 2023, 24, 4877. [Google Scholar] [CrossRef]
- Sato, S.; Oshima, Y.; Matsumoto, Y.; Seto, Y.; Yamashita, H.; Hayano, K.; Kano, M.; Ono, H.A.; Mitsumori, N.; Fujisaki, M.; et al. The new prognostic score for unresectable or recurrent gastric cancer treated with nivolumab: A multi-institutional cohort study. Ann. Gastroenterol. Surg. 2021, 5, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Kuhara, Y.; Ninomiya, M.; Hirahara, S.; Doi, H.; Kenji, S.; Toyota, K.; Yano, R.; Kobayashi, H.; Hashimoto, Y.; Yokoyama, Y.; et al. A long-term survival case of unresectable gastric cancer with multidisciplinary therapy including immunotherapy and abscopal effect. Int. Cancer Conf. J. 2020, 9, 193–198. [Google Scholar] [CrossRef]
- Hu, H.M.; Tsai, H.J.; Ku, H.Y.; Lo, S.S.; Shan, Y.S.; Chang, H.C.; Chao, Y.; Chen, J.S.; Chen, S.C.; Chiang, C.J.; et al. Survival outcomes of management in metastatic gastric adenocarcinoma patients. Sci. Rep. 2021, 11, 23142. [Google Scholar] [CrossRef]
- Schütte, K.; Schulz, C.; Middelberg-Bisping, K. Impact of gastric cancer treatment on quality of life of patients. Best Pract. Res. Clin. Gastroenterol. 2021, 50–51. [Google Scholar] [CrossRef]
- Ha, G.-Y.; Yang, S.-H.; Kang, H.-J.; Lee, H.-L.; Kim, J.; Kim, Y.-J.; Yu, H.-J.; Lee, J.-I.; Jin, S.-H. Comparison of survival outcomes according of patients with metastatic gastric cancer receiving trastuzumab with systemic chemotherapy. Korean J. Clin. Oncol. 2020, 16, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Z.; Chen, Z. Comparison of Treatment Efficacy and Survival Outcomes Between Asian and Western Patients with Unresectable Gastric or Gastro-Esophageal Adenocarcinoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12, 831207. [Google Scholar] [CrossRef] [PubMed]
- Apicella, M.; Corso, S.; Giordano, S. Targeted therapies for gastric cancer: Failures and hopes from clinical trials. Oncotarget 2017, 8, 57654–57669. [Google Scholar] [CrossRef]
- Skórzewska, M.; Gęca, K.; Polkowski, W.P. A Clinical Viewpoint on the Use of Targeted Therapy in Advanced Gastric Cancer. Cancers 2023, 15, 5490. [Google Scholar] [CrossRef]
- Attia, H.; Smyth, E. Evolving therapies in advanced oesophago-gastric cancers and the increasing role of immunotherapy. Expert Rev. Anticancer Ther. 2021, 21, 535–546. [Google Scholar] [CrossRef]
- Sato, Y.; Okamoto, K.; Kawano, Y.; Kasai, A.; Kawaguchi, T.; Sagawa, T.; Sogabe, M.; Miyamoto, H.; Takayama, T. Novel Biomarkers of Gastric Cancer: Current Research and Future Perspectives. J. Clin. Med. 2023, 12, 4646. [Google Scholar] [CrossRef]
- Gao, J.P.; Xu, W.; Liu, W.T.; Yan, M.; Zhu, Z.G. Tumor heterogeneity of gastric cancer: From the perspective of tumor-initiating cell. World J. Gastroenterol. 2018, 24, 2567–2581. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, Y.; Qing, T.; Xing, K.; Li, J.; Zhen, T.; Zhu, S.; Zhan, X. Comprehensive analysis of metastatic gastric cancer tumour cells using single-cell RNA-seq. Sci. Rep. 2021, 11, 1141. [Google Scholar] [CrossRef]
- Chen, J.; Liu, K.; Luo, Y.; Kang, M.; Wang, J.; Chen, G.; Qi, J.; Wu, W.; Wang, B.; Han, Y.; et al. Single-Cell Profiling of Tumor Immune Microenvironment Reveals Immune Irresponsiveness in Gastric Signet-Ring Cell Carcinoma. Gastroenterology 2023, 165, 88–103. [Google Scholar] [CrossRef]
- Lee, H.H.; Kim, S.Y.; Jung, E.S.; Yoo, J.; Kim, T.M. Mutation heterogeneity between primary gastric cancers and their matched lymph node metastases. Gastric Cancer 2019, 22, 323–334. [Google Scholar] [CrossRef]
- Jiang, H.; Yu, D.; Yang, P.; Guo, R.; Kong, M.; Gao, Y.; Yu, X.; Lu, X.; Fan, X. Revealing the transcriptional heterogeneity of organ-specific metastasis in human gastric cancer using single-cell RNA Sequencing. Clin. Transl. Med. 2022, 12, e730. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Kim, K.T.; Shin, S.J.; Cheong, J.H.; Choi, Y.Y. Genomic and evolutionary characteristics of metastatic gastric cancer by routes. Br. J. Cancer 2023, 129, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Shitara, K. Trastuzumab deruxtecan and other HER2-targeting agents for the treatment of HER2-positive gastric cancer. Expert Rev. Anticancer Ther. 2021, 21, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.L.; He, Y.; Xu, R.H. Gastric cancer treatment: Recent progress and future perspectives. J. Hematol. Oncol. 2023, 16, 57. [Google Scholar] [CrossRef]
- Kim, M.; Seo, A.N. Molecular Pathology of Gastric Cancer. J. Gastric Cancer 2022, 22, 273–305. [Google Scholar] [CrossRef] [PubMed]
- Serra, O.; Galán, M.; Ginesta, M.M.; Calvo, M.; Sala, N.; Salazar, R. Comparison and applicability of molecular classifications for gastric cancer. Cancer Treat. Rev. 2019, 77, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Sohn, B.H.; Hwang, J.E.; Jang, H.J.; Lee, H.S.; Oh, S.C.; Shim, J.J.; Lee, K.W.; Kim, E.H.; Yim, S.Y.; Lee, S.H.; et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin. Cancer Res. 2017, 23, 4441–4449. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, Q.; Liu, Y.; Guo, H.; Ren, Y.; Li, J.; Zhao, Q. Clinical characteristics and prognostic significance of TCGA and ACRG classification in gastric cancer among the Chinese population. Mol. Med. Rep. 2020, 22, 828–840. [Google Scholar] [CrossRef]
- Nshizirungu, J.P.; Bennis, S.; Mellouki, I.; Sekal, M.; Benajah, D.A.; Lahmidani, N.; El Bouhaddouti, H.; Ibn Majdoub, K.; Ibrahimi, S.A.; Celeiro, S.P.; et al. Reproduction of the Cancer Genome Atlas (TCGA) and Asian Cancer Research Group (ACRG) Gastric Cancer Molecular Classifications and Their Association with Clinicopathological Characteristics and Overall Survival in Moroccan Patients. Dis. Markers 2021, 2021, 9980410. [Google Scholar] [CrossRef]
- Patel, T.H.; Cecchini, M. Targeted Therapies in Advanced Gastric Cancer. Curr. Treat. Options Oncol. 2020, 21, 70. [Google Scholar] [CrossRef]
- Tan, I.B.; Ivanova, T.; Lim, K.H.; Ong, C.W.; Deng, N.; Lee, J.; Tan, S.H.; Wu, J.; Lee, M.H.; Ooi, C.H.; et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology 2011, 141, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Tan, I.B.; Das, K.; Deng, N.; Zouridis, H.; Pattison, S.; Chua, C.; Feng, Z.; Guan, Y.K.; Ooi, C.H.; et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology 2013, 145, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ding, Y.; Chen, Y.; Jiang, J.; Chen, Y.; Lu, J.; Kong, M.; Mo, F.; Huang, Y.; Zhao, W.; et al. A novel genomic classification system of gastric cancer via integrating multidimensional genomic characteristics. Gastric Cancer 2021, 24, 1227–1241. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.A.; Khanin, R.; Tang, L.; Janjigian, Y.Y.; Klimstra, D.S.; Gerdes, H.; Kelsen, D.P. Molecular Classification of Gastric Cancer: A new paradigm. Clin. Cancer Res. 2011, 17, 2693. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Lucas, F.A.M.; Cristovam, S.N. HER2 testing in gastric cancer: An update. World J. Gastroenterol. 2016, 22, 4619. [Google Scholar]
- Furukawa, K.; Hatakeyama, K.; Terashima, M.; Nagashima, T.; Urakami, K.; Ohshima, K.; Notsu, A.; Sugino, T.; Yagi, T.; Fujiya, K.; et al. Molecular classification of gastric cancer predicts survival in patients undergoing radical gastrectomy based on project HOPE. Gastric Cancer 2022, 25, 138–148. [Google Scholar] [CrossRef]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.N.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.; Profitós-Pelejà, N.; Sánchez-Vinces, S.; Roué, G. RHOA Therapeutic Targeting in Hematological Cancers. Cells 2023, 12, 433. [Google Scholar] [CrossRef]
- Cheong, J.H.; Wang, S.C.; Park, S.; Porembka, M.R.; Christie, A.L.; Kim, H.; Kim, H.S.; Zhu, H.; Hyung, W.J.; Noh, S.H.; et al. Development and validation of a prognostic and predictive 32-gene signature for gastric cancer. Nat. Commun. 2022, 13, 774. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yuan, Y.; Lu, H.; Li, X.; Liu, Z.; Gan, J.; Yue, Z.; Wu, J.; Sheng, J.; Xin, L. Cancer functional states-based molecular subtypes of gastric cancer. J. Transl. Med. 2023, 21, 80. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yang, W.; Ruan, X.; Xu, L.; Cheng, W.; Zhao, M.; Wang, X.; Chen, X.; Cai, D.; Li, G.; et al. Metabolism-associated molecular classification of gastric adenocarcinoma. Front. Oncol. 2022, 12, 1024985. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, J. Molecular characterization of metabolic subtypes of gastric cancer based on metabolism-related lncRNA. Sci. Rep. 2021, 11, 21491. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Pan, X.; Zhao, L.; Yang, C.; Zhang, Z.; Wang, B.; Gao, Z.; Jiang, K.; Ye, Y.; Wang, S.; et al. Immune cell infiltration signatures identified molecular subtypes and underlying mechanisms in gastric cancer. npj Genom. Med. 2021, 6, 83. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Feng, M.; Shen, H.; Shen, X.; Hu, J.; Liu, J.; Yang, Y.; Li, Y.; Yang, M.; Wang, W.; et al. Prediction of Two Molecular Subtypes of Gastric Cancer Based on Immune Signature. Front. Genet. 2022, 12, 793494. [Google Scholar] [CrossRef]
- Li, X.; Wu, W.K.K.; Xing, R.; Hwong, S.; Liu, Y.; Fang, X.; Zhang, Y.; Wang, M.; Wang, J.; Li, L.; et al. Distinct Subtypes of Gastric Cancer Defined by Molecular Characterization Include Novel Mutational Signatures with Prognostic Capability. Cancer Res. 2016, 76, 1724–1732. [Google Scholar] [CrossRef]
- Wei, C.; Li, M.; Lin, S.; Xiao, J. Characterization of Tumor Mutation Burden-Based Gene Signature and Molecular Subtypes to Assist Precision Treatment in Gastric Cancer. Biomed Res. Int. 2022, 2022, 4006507. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.; Li, M.; Deng, J.; Xu, H.; Ren, Y.; Zhou, Z.; Wang, L.; Zhang, Y.; Xing, Z.; Li, L.; et al. Epigenetically regulated gene expression profiles decipher four molecular subtypes with prognostic and therapeutic implications in gastric cancer. Clin. Epigenetics 2023, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, C.; Wang, B.; Xu, F.; Ma, F.; Qu, Y.; Jiang, D.; Li, K.; Feng, J.; Tian, S.; et al. Proteomic characterization of gastric cancer response to chemotherapy and targeted therapy reveals potential therapeutic strategies. Nat. Commun. 2022, 13, 5723. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Xia, X.; Ding, C.; Zhen, B.; Zhou, Q.; Feng, J.; Yuan, J.; Chen, R.; Li, Y.; Ge, Z.; et al. A proteomic landscape of diffuse-type gastric cancer. Nat. Commun. 2018, 9, 1012. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Chiwaki, F.; Kojima, S.; Kawazu, M.; Komatsu, M.; Ueno, T.; Inoue, S.; Sekine, S.; Matsusaki, K.; Matsushita, H.; et al. Multi-omic profiling of peritoneal metastases in gastric cancer identifies molecular subtypes and therapeutic vulnerabilities. Nat. Cancer 2021, 2, 962–977. [Google Scholar] [CrossRef]
- Raab, S.S. The cost-effectiveness of immunohistochemistry. Arch. Pathol. Lab. Med. 2000, 124, 1185–1191. [Google Scholar] [CrossRef]
- Magaki, S.; Hojat, S.A.; Wei, B.; So, A.; Yong, W.H. An Introduction to the Performance of Immunohistochemistry. Methods Mol. Biol. 2019, 1897, 289–298. [Google Scholar]
- Duraiyan, J.; Govindarajan, R.; Kaliyappan, K.; Palanisamy, M. Applications of immunohistochemistry. J. Pharm. Bioallied Sci. 2012, 4, 307. [Google Scholar]
- Bonacho, T.; Rodrigues, F.; Liberal, J. Immunohistochemistry for diagnosis and prognosis of breast cancer: A review. Biotech. Histochem. 2020, 95, 71–91. [Google Scholar] [CrossRef]
- Jensen, E. Technical review: In situ hybridization. Anat. Rec. 2014, 297, 1349–1353. [Google Scholar] [CrossRef]
- Ahn, S.; Woo, J.W.; Lee, K.; Park, S.Y. HER2 status in breast cancer: Changes in guidelines and complicating factors for interpretation. J. Pathol. Transl. Med. 2020, 54, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.Y.; Sy, K.; Brezden-Masley, C.; Streutker, C.J. Histo- and immunohistochemistry-based estimation of the TCGA and ACRG molecular subtypes for gastric carcinoma and their prognostic significance: A single-institution study. PLoS ONE 2019, 14, e0224812. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.F.K.P.; Pereira, M.A.; de Mello, E.S.; Cirqueira, C.d.S.; Zilberstein, B.; Alves, V.A.F.; Ribeiro-Junior, U.; Cecconello, I. Gastric cancer molecular classification based on immunohistochemistry and in situ hybridization: Analysis in western patients after curative-intent surgery. World J. Clin. Oncol. 2021, 12, 688–701. [Google Scholar] [CrossRef]
- Zhao, C.; Feng, Z.; He, H.; Zang, D.; Du, H.; Huang, H.; Du, Y.; He, J.; Zhou, Y.; Nie, Y. Protein expression-based classification of gastric cancer by immunohistochemistry of tissue microarray. PLoS ONE 2020, 15, e0238836. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.P.; Córdova-Delgado, M.; Retamal, I.N.; Muñoz-Medel, M.; Bravo, M.L.; Durán, D.; Villanueva, F.; Sanchez, C.; Acevedo, F.; Mondaca, S.; et al. A Molecular Stratification of Chilean Gastric Cancer Patients with Potential Clinical Applicability. Cancers 2020, 12, 1863. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.S.; Messing, S.; Tu, X.; McMahon, L.A.; Whitney-Miller, C.L. Immunohistochemistry as a surrogate for molecular subtyping of gastric adenocarcinoma. Hum. Pathol. 2016, 56, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Birkman, E.M.; Mansuri, N.; Kurki, S.; Ålgars, A.; Lintunen, M.; Ristamäki, R.; Sundström, J.; Carpén, O. Gastric cancer: Immunohistochemical classification of molecular subtypes and their association with clinicopathological characteristics. Virchows Arch. 2018, 472, 369–382. [Google Scholar] [CrossRef]
- Díaz del Arco, C.; Estrada Muñoz, L.; Molina Roldán, E.; Cerón Nieto, M.Á.; Ortega Medina, L.; García Gómez de las Heras, S.; Fernández Aceñero, M.J. Immunohistochemical classification of gastric cancer based on new molecular biomarkers: A potential predictor of survival. Virchows Arch. 2018, 473, 687–695. [Google Scholar] [CrossRef]
- Shimozaki, K.; Nakayama, I.; Hirota, T.; Yamaguchi, K. Current Strategy to Treat Immunogenic Gastrointestinal Cancers: Perspectives for a New Era. Cells 2023, 12, 1049. [Google Scholar] [CrossRef]
- Wang, J.B.; Qiu, Q.Z.; Zheng, Q.L.; Zhao, Y.J.; Xu, Y.; Zhang, T.; Wang, S.H.; Wang, Q.; Jin, Q.W.; Ye, Y.H.; et al. Tumor Immunophenotyping-Derived Signature Identifies Prognosis and Neoadjuvant Immunotherapeutic Responsiveness in Gastric Cancer. Adv. Sci. 2023, 10, e2207417. [Google Scholar] [CrossRef]
- Xu, H.Y.; Xu, W.L.; Wang, L.Q.; Chen, M.B.; Shen, H.L. Relationship between p53 status and response to chemotherapy in patients with gastric cancer: A meta-analysis. PLoS ONE 2014, 9, e95371. [Google Scholar] [CrossRef] [PubMed]
- O’Day, V.; Adler, A.; Kuchinsky, A.; Bouch, A. When Worlds Collide: Molecular Biology as Interdisciplinary Collaboration. In Proceedings of the Seventh European Conference on Computer-Supported Cooperative Work, Bonn, Germany, 16–20 September 2001; pp. 399–418. [Google Scholar]




| Subtypes | Upregulated | Downregulated |
|---|---|---|
| Proximal non-diffuse (vs. diffuse) | TRIM32, PRF1, CXCL9, CXCL10, IF144L, PLA2G2A | PSCA, PGA3, XIST, SST, ABCA8 |
| Proximal non-diffuse (vs. distal non-diffuse) | PF4V1, HMBO1, CYP2J2, DSC3, S100A12 | MSLN, IGJ, ENPP4, PLA2G2A |
| Diffuse (vs. distal non-diffuse) | ABCA8, HMBOX1, COCH, S100A12, CYP2J2 | IFI44L, HOXA9, MSLN, ENPP4 |
| Ye et al. | Molecular Features | Immune and Clinicopathological Features |
|---|---|---|
| Immune—suppressed (C1) | Pathways: HIPPO, WNT, NOTCH, cell cycle | Neutrophil-induced immune suppression Intestinal/tubular tumors No MSI a |
| Metabolic (C2) | Pathways: MYC, NRF2 | Abundance of T cells (Th1, cytotoxic, Tcm), B cells and dendritic cells |
| Mesenchymal—immune exhausted (C3) | Pathways: TGF-β, angiogenesis Enhanced EMT b | Diffuse/poorly cohesive tumors Lower HER2 expression Worse prognosis |
| Hypermutated (C4) | Pathways: cell cycle, TP53, PI3K MSI CIMP c | Abundance of Th2 cells EBV d-related tumors Intestinal/tubular tumors Response to immunotherapy Better prognosis |
| Authors | Year | Molecular Categories |
|---|---|---|
| TCGA a [42] | 2014 | MSI b, genomically stable, EBV c positive, chromosomal instability |
| ACRG d [43] | 2015 | MSI, mesenchymal-like, TP53 active, TP53 inactive |
| Tan et al. [155] | 2011 | Intrinsic subtypes: G-INT and G-DIF |
| Lei et al. [156] | 2013 | Mesenchymal, proliferative, metabolic |
| Wang et al. [157] | 2021 | Subtypes 1–4 |
| Shah et al. [158] | 2011 | Proximal non-diffuse, diffuse, distal non-diffuse |
| Furukawa et al. [161] | 2022 | Hypermutators, T-cell inflamed, EMT e-high, EMT-low |
| Wang et al. [162] | 2014 | MSI, stable associated with EBV, stable non-EBV |
| Cheong et al. [164] | 2022 | Groups 1–4 |
| Zhou et al. [165] | 2023 | Clusters 1–3 (functional status-based) |
| Ye et al. [156] | 2022 | Immune-suppressed (C1), metabolic (C2), mesenchymal-immune exhausted (C3), hypermutated (C4) |
| Li et al. [167] | 2021 | C1 and C2 types (metabolism-based) |
| Lin et al. [168] | 2021 | High and low immune microenvironment score |
| Wu et al. [169] | 2022 | Non-activated (C1) and immune activated (C2) |
| Li et al. [170] | 2016 | Regular, hypermutated |
| Wei et al. [171] | 2022 | High TMB f, low TMB |
| Weng et al. [172] | 2023 | C1-C4 types (epigenetic-based) |
| Li et al. [174] | 2022 | PX1-3 types (proteomic-based) |
| Tanaka et al. [175] | 2021 | Two types of ascited-disseminated gastric cancer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz del Arco, C.; Fernández Aceñero, M.J.; Ortega Medina, L. Molecular Classifications in Gastric Cancer: A Call for Interdisciplinary Collaboration. Int. J. Mol. Sci. 2024, 25, 2649. https://doi.org/10.3390/ijms25052649
Díaz del Arco C, Fernández Aceñero MJ, Ortega Medina L. Molecular Classifications in Gastric Cancer: A Call for Interdisciplinary Collaboration. International Journal of Molecular Sciences. 2024; 25(5):2649. https://doi.org/10.3390/ijms25052649
Chicago/Turabian StyleDíaz del Arco, Cristina, María Jesús Fernández Aceñero, and Luis Ortega Medina. 2024. "Molecular Classifications in Gastric Cancer: A Call for Interdisciplinary Collaboration" International Journal of Molecular Sciences 25, no. 5: 2649. https://doi.org/10.3390/ijms25052649






