Verification of Key Target Molecules for Intramuscular Fat Deposition and Screening of SNP Sites in Sheep from Small-Tail Han Sheep Breed and Its Cross with Suffolk
Abstract
:1. Introduction
2. Results
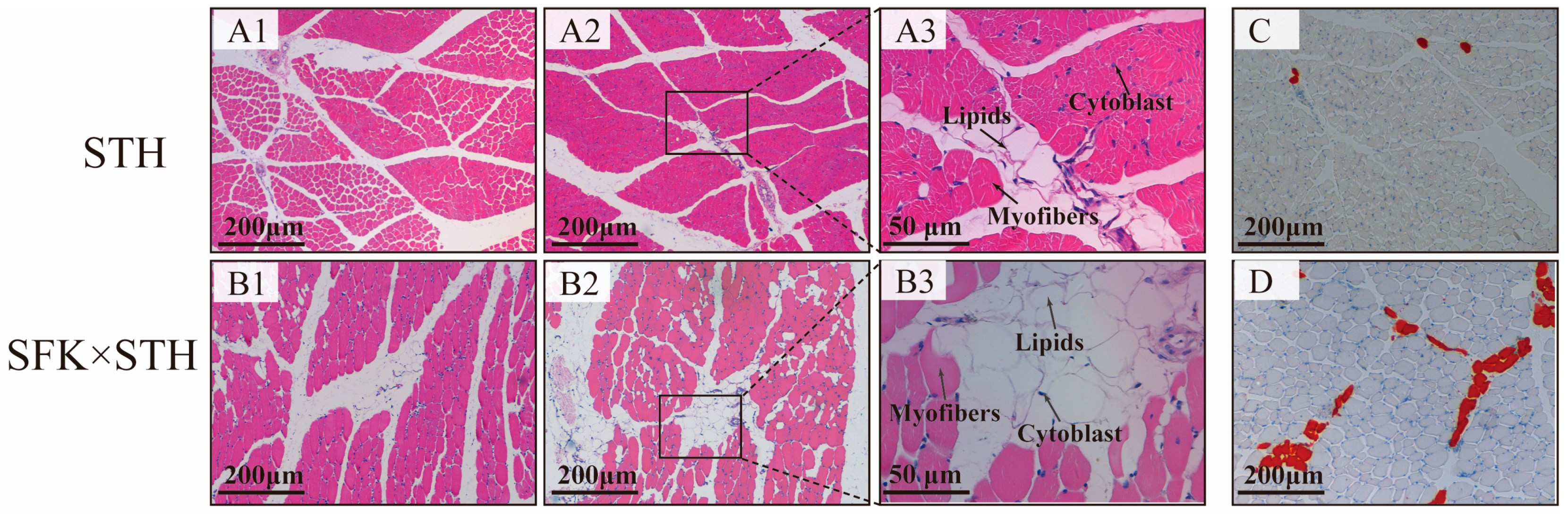
2.1. Comparison of IMF Content in the Longest Muscle Tissue of Different Groups of Sheep
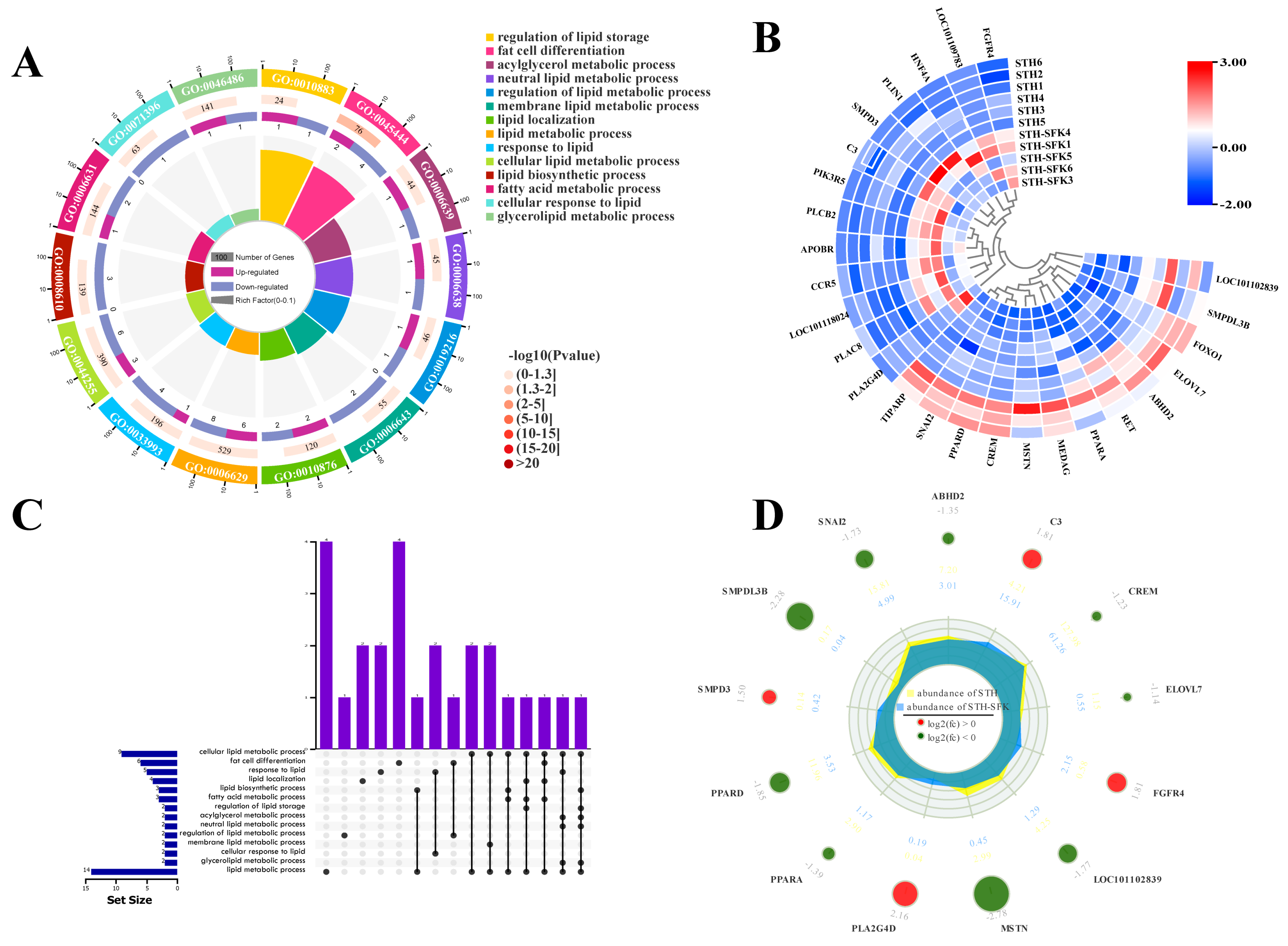
2.2. Differential Gene Identification and Screening Based on GO Analysis
2.3. Gene Co-Expression
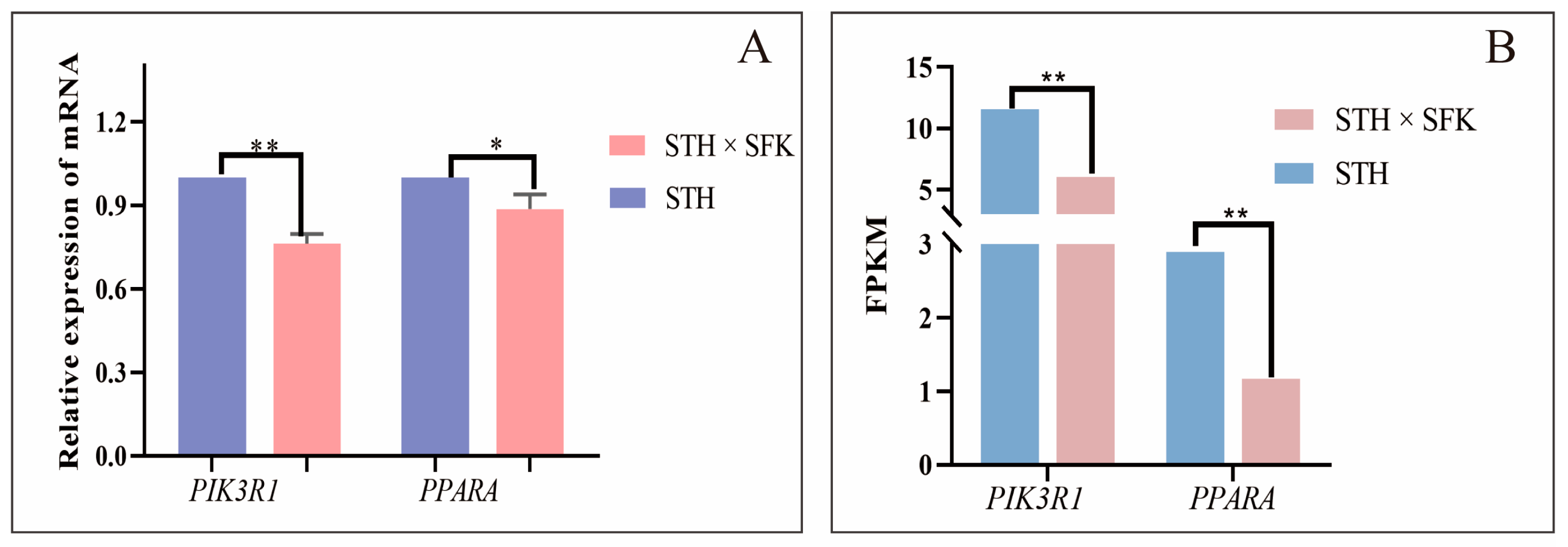
2.4. Functional Analysis of Candidate Genes Related to IMF
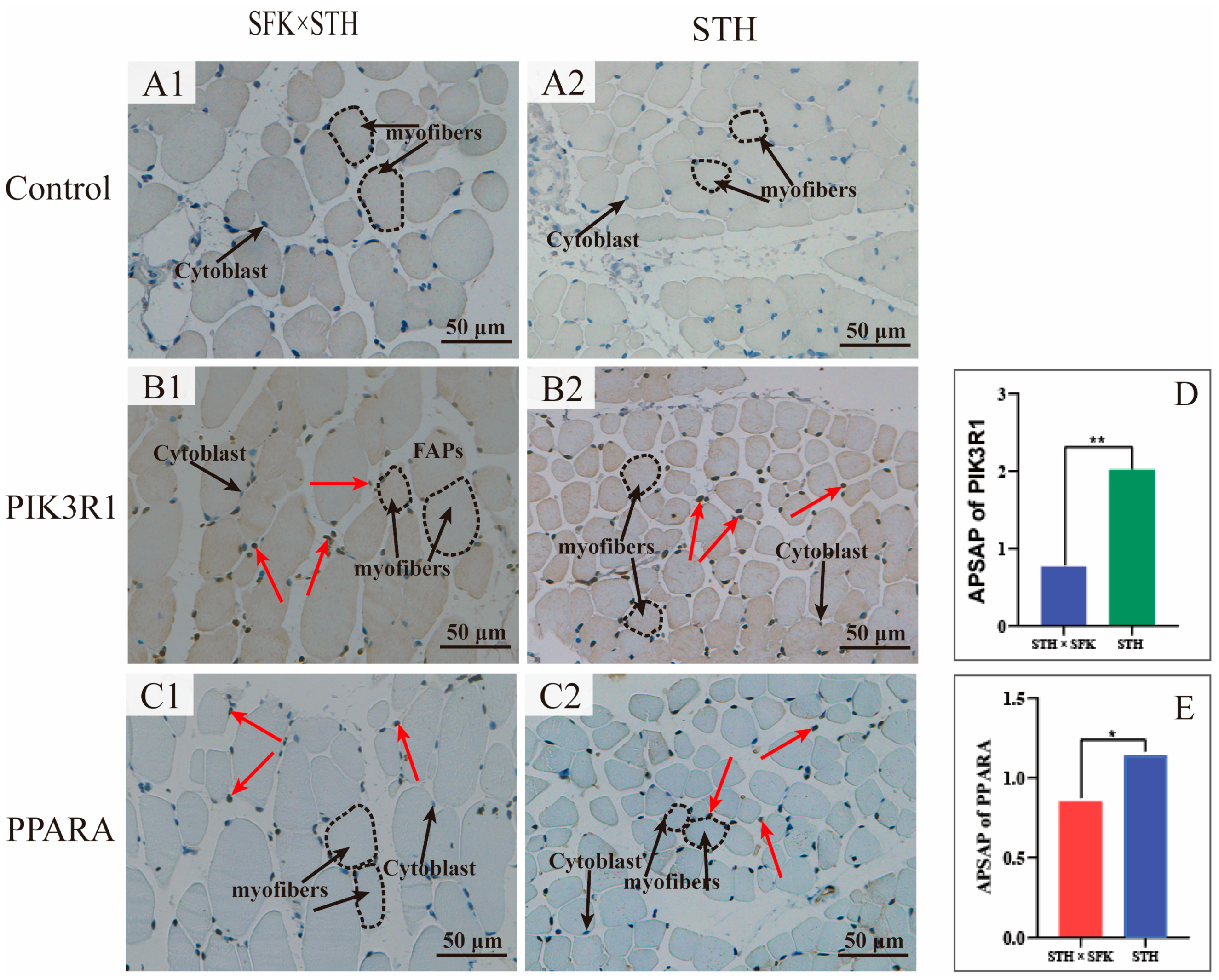
2.5. Immunohistochemistry
2.6. Co-Positioning
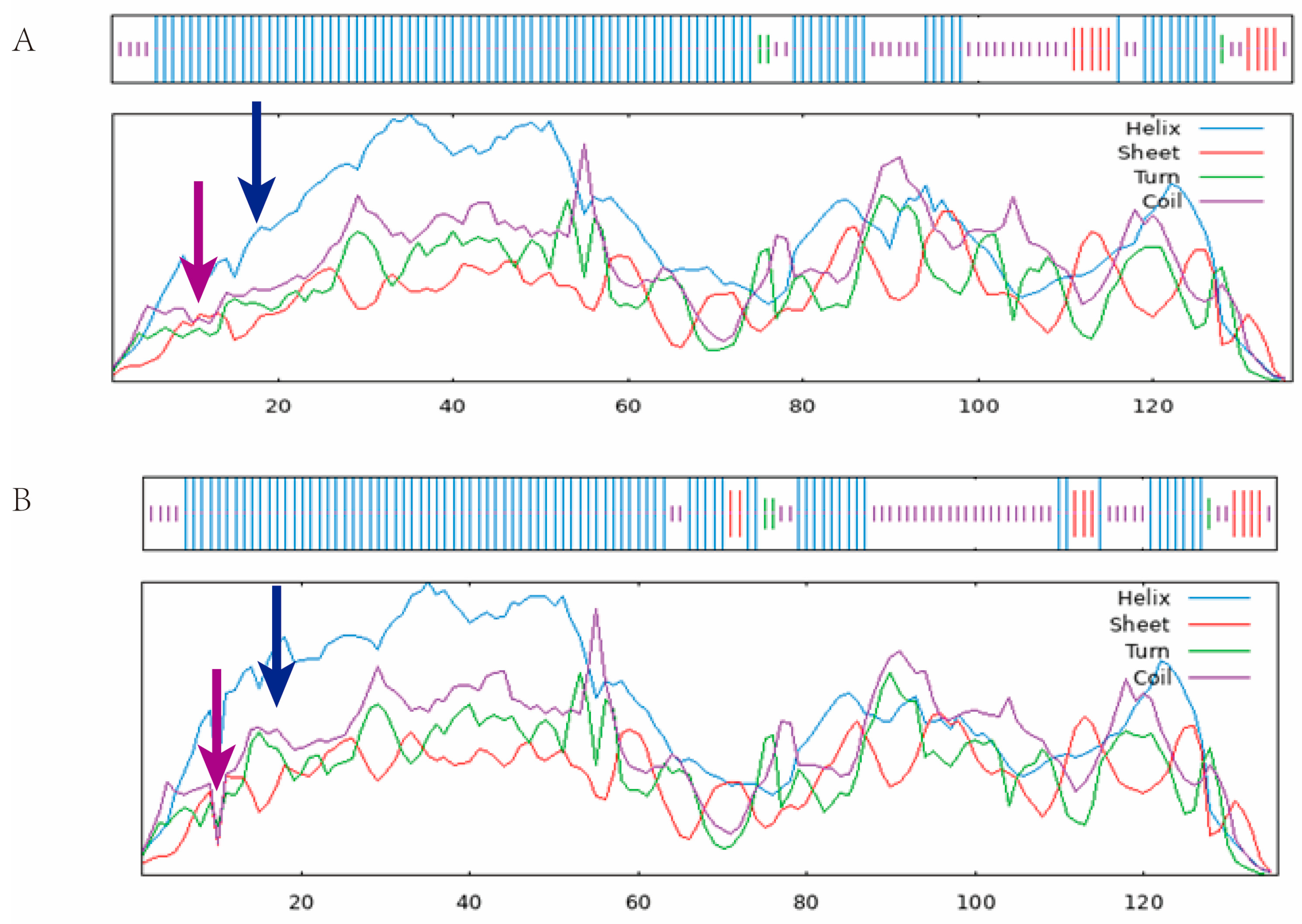
2.7. PIK3R1 Single Nucleotide Polymorphism Site
2.8. PIK3R1 Protein Physical and Chemical Parameters
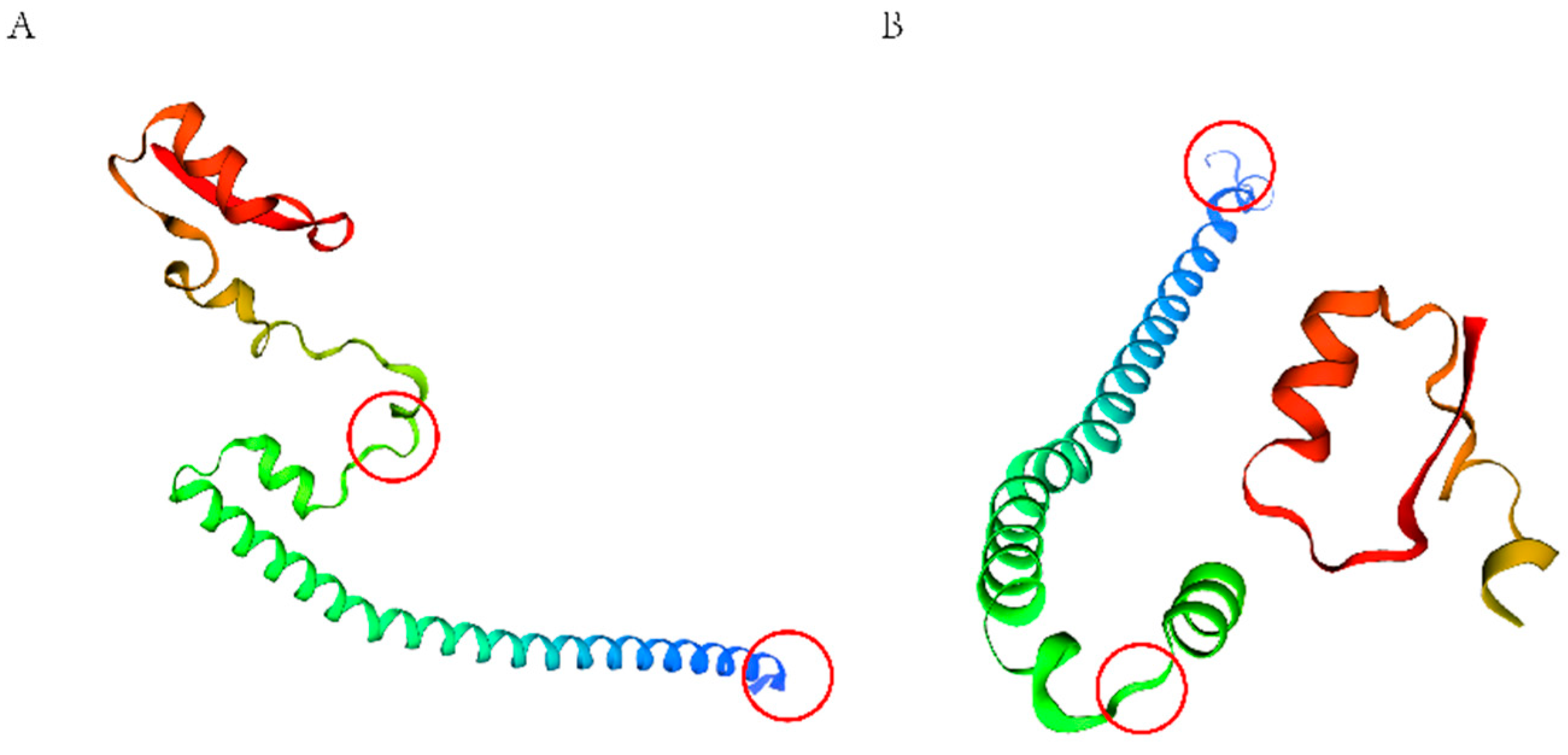
2.9. PIK3R1 Protein Structure Prediction
3. Discussion
4. Materials and Methods
4.1. Animal Welfare
4.2. Sample Collection and Preparation
4.3. Hematoxylin-Eosin (H&E) Staining Technique
4.4. Oil Red O Staining
4.5. Total RNA Preparation, cDNA Library, and Sequencing
4.6. Transcriptome Analysis and Differential Expression Gene Identification
4.7. Real-Time Fluorescence Quantitative PCR (qRT-PCR)
4.8. Immunohistochemistry Staining (IHC)
4.9. Immunofluorescence Staining of Tissues (IF)
4.10. Single Nucleotide Polymorphism Analysis and Bioinformatics Analysis of PIK3R1
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, H. Problems and Suggestions in the Development of Meat Sheep Industry in Jingyuan County. Gansu Livest. Vet. Med. 2020, 50, 76–78. [Google Scholar]
- Lai, J.; Liu, L.; Liu, D. Current Situation and Suggestions for the Development of Meat Sheep Industry in Gansu Province. Gansu Livest. Vet. Med. 2018, 48, 22–24. [Google Scholar]
- Yang, D.; Wu, J.; Liu, J.; Dou, X.; Meng, X.; Yang, G.; Ma, J. Geographical Indication Product Dongxiang Gongyang Mutton. Gansu Livest. Vet. Med. 2022, 52, 42–46. [Google Scholar]
- Qu, K.; Xu, X.; Fan, J.; Huo, C.; Hao, Q.; Shang, A. Analysis of the problems and suggestions for the development of small tailed Han sheep breeds. Shandong Anim. Hus. Vet. Med. 2022, 43, 28–31. [Google Scholar]
- Ilisiu, E.; Daraban, S.; Neascu, G.; Gabi-Neacsu, M.; Ilisiu, V.C.; Rahmann, G. Improvement of lamb production in Romania by crossbreeding of local Tsigai breed with high performance breeds. Landbauforsch. VTI Agric. For. Res. 2010, 60, 259–266. [Google Scholar]
- Chen, G.; Pan, X.; Yuan, Y. Study on Production Performance of the First Generation from Hu Sheep×Dorper Sheep Crossbreeding in Hexi Corridor. J. Anim. Hus. Vet. Med. 2017, 36, 32–36. [Google Scholar]
- Pan, L.; Li, J. Review of the Development of China’s Meat Sheep Industry during the 13th Five Year Plan and Prospects for the 14th Five Year Plan. Chin. J. Anim. Hus. 2022, 58, 262–267. [Google Scholar]
- Zhao, N.; An, R.X.; Li, Y.J.; Liang, J. Research progress on factors affecting the quality of lamb meat. Feed Ind. 2023, 44, 71–76. [Google Scholar]
- Eddinger, T.J.; Moss, R.L.; Cassens, R.G. Myosin-ATPase fibre typing of chemically skinned muscle fibres. Histochem. J. 1985, 17, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.F.; Khan, R.; Raza, S.H.A.; Nurgulsim, K.; Suhail, S.M.; Rahman, A.; Ahmed, I.; Ijaz, A.; Ahmad, I.; Zan, L.S. Transcriptional regulation of adipogenic marker genes for the improvement of intramuscular fat in Qinchuan beef cattle. Anim. Biotechnol. 2022, 33, 776–795. [Google Scholar]
- Qiao, Y.; Huang, J.C.; Chen, Y.L.; Chen, H.C.; Zhao, L.; Huang, M.; Zhou, G.H. Meat quality, fatty acid composition and sensory evaluation of Cherry Valley, Spent Layer and Crossbred ducks. Anim. Sci. J. 2017, 88, 156–165. [Google Scholar] [CrossRef]
- Hamill, R.M.; McBryan, J.; McGee, C.; Mullen, A.M.; Sweeney, T.; Talbot, A.; Cairns, M.T.; Davey, G.C. Functional analysis of muscle gene expression profiles associated with tenderness and intramuscular fat content in pork. Meat Sci. 2012, 92, 440–450. [Google Scholar] [CrossRef]
- Miao, Z.; Wei, P.; Khan, M.A.; Zhang, J.; Guo, L.; Liu, D.; Zhang, X.; Bai, Y.; Wang, S. Transcriptome analysis reveals differential gene expression in intramuscular adipose tissues of Jinhua and Landrace pigs. J. Vet. Med. Sci. 2018, 80, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Kurth, I.; Pamminger, T.; Hennings, J.C.; Soehendra, D.; Huebner, A.K.; Rotthier, A.; Baets, J.; Senderek, J.; Topaloglu, H.; Farrell, S.A.; et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat. Genet. 2009, 41, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Graugnard, D.E.; Piantoni, P.; Bionaz, M.; Berger, L.L.; Faulkner, D.B.; Loor, J.J. Adipogenic and energy metabolism gene networks in longissimus lumborum during rapid post-weaning growth in Angus and Angus x Simmental cattle fed high-starch or low-starch diets. BMC Genom. 2009, 10, 142. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, K.J.; Ehlers, L.B.; Aguirre, L.E.; Moore, R.L.; Jew, K.N.; Ortmeyer, H.K.; Hansen, B.C.; Reusch, J.E.; Draznin, B. Exercise training and calorie restriction increase SREBP-1 expression and intramuscular triglyceride in skeletal muscle. Am. J. Physiol. 2006, 291, E90–E98. [Google Scholar] [CrossRef] [PubMed]
- Kazala, E.C.; Petrak, J.L.; Lozeman, F.J.; Mir, P.S.; André, L.; Deng, J.; Weselake, R.J. Hormone-sensitive lipase activity in relation to fat content of muscle in wagyu hybrid cattle. Livest. Prod. Sci. 2003, 79, 87–96. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.; Kong, Y.; Li, F.; Yue, X. Effects of intramuscular fat on meat quality and its regulation mechanism in Tan sheep. Front. Nutr. 2022, 9, 908355. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wu, J.; Zhou, J.; Zhang, Y.; Qiao, M.; Sun, H.; Li, Z.; Li, L.; Chen, N.; Oyelami, F.O.; et al. Integration of ATAC-seq and RNA-seq analysis identifies key genes affecting intramuscular fat content in pigs. Front. Nutr. 2022, 9, 1016956. [Google Scholar] [CrossRef]
- Duckett, S.K.; Greene, M.A. Identification of microRNA Transcriptome Involved in Bovine Intramuscular Fat Deposition. Front. Vet. Sci. 2022, 9, 883295. [Google Scholar] [CrossRef]
- Luo, N.; Shu, J.; Yuan, X.; Jin, Y.; Cui, H.; Zhao, G.; Wen, J. Differential regulation of intramuscular fat and abdominal fat deposition in chickens. BMC Genom. 2022, 23, 308. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, K.; Wang, W.L.; Yin, T.T.; Dong, W.Q.; Xu, C.J. A high-throughput SNP discovery strategy for RNA-seq data. BMC Genom. 2019, 20, 160. [Google Scholar] [CrossRef]
- Li, M. Development and Application of High-Throughput Genotype Identification Technology. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2021. [Google Scholar]
- Li, P.; Fan, Y.; Zhang, N.; Gao, T. esearch progress of factors affecting intramuscular fat deposition in cattle and sheep. Heilongjiang Anim. Hus. Vet. Med. 2021, 617, 48–52. [Google Scholar]
- Gao, S.Z.; Zhao, S.M. Physiology, affecting factors and strategies for control of pig meat intramuscular fat. Recent Pat. Food Nutr. Agric. 2009, 1, 59–74. [Google Scholar]
- Xu, Y. Japan and Cattle × Qinchuan × Carcass Quality and Meat Quality Analysis of Angus Three Way Hybrid Snow Cattle. Master’s Thesis, Gansu Agricultural University, Lanzhou, China, 2014. [Google Scholar]
- da Silva, A.F.; McManus, C.; do Prado Paim, T.; Dallago, B.S.; Esteves, G.I.; Louvandini, H.; Neto, J.B.; Lucci, C.M. Production traits in F1 and F2 crosses with naturalized hair breed Santa Inês ewes. Springerplus 2014, 3, 66. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Zhirong, G.; Ming, W. The interaction between PPARD-87T>C and PPARA and PPARG single nucleotide polymorphisms on abdominal obesity. Chin. Med. J. 2012, 92, 5. [Google Scholar]
- Muñoz, M.; García-Casco, J.M.; Caraballo, C.; Fernández-Barroso, M.Á.; Sánchez-Esquiliche, F.; Gómez, F.; Rodríguez, M.D.C.; Silió, L. Identification of candidate genes and regulatory factors underlying intramuscular fat content through longissimus dorsi transcriptome analyses in heavy iberian Pigs. Front. Genet. 2018, 9, 608. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, M.; Kong, L.; Hao, M.; Gao, S.; Li, Y.; Zhao, S. Study on the Regulation of Lipid Metabolism in Pig Subcutaneous Pre adipocytes by Leptin Mediated AMPK Signal Pathway. Biotechnol. Bull. 2016, 32, 211–218. [Google Scholar]
- Zhao, X.; Hu, H.; Lin, H.; Wang, C.; Wang, Y.; Wang, J. Muscle transcriptome analysis reveals potential candidate genes and pathways affecting intramuscular fat content in Pigs. Front. Genet. 2020, 11, 877. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Y.; Wu, J.; Qiao, M.; Xu, Z.; Peng, X.; Mei, S. Proteomic and lipidomic analyses reveal saturated fatty acids, phosphatidylinositol, phosphatidylserine, and associated proteins contributing to intramuscular fat deposition. J. Proteom. 2021, 241, 104235. [Google Scholar] [CrossRef]
- Kuo, T.; Chen, T.C.; Lee, R.A.; Nguyen, N.H.T.; Broughton, A.E.; Zhang, D.; Wang, J.C. Pik3r1 is required for glucocorticoid-induced Perilipin 1 phosphorylation in lipid droplet for adipocyte lipolysis. Diabetes 2017, 66, 1601–1610. [Google Scholar] [CrossRef]
- Sun, G.; Jiao, D.; Xie, Z.; Zhou, P.; Yang, G. Research progress on the PI3K/AKT pathway in animal glucose metabolism. Chin. Bull. Life Sci. 2021, 5, 653–666. [Google Scholar]
- Hines, H.C.; Zikakis, J.P.; Haenlein GF, W.; Kiddy, C.A.; Trowbridge, C.L. Linkage relationships among loci of polymorphisms in blood and milk of cattle. J. Dairy Sci. 1981, 64, 71–76. [Google Scholar] [CrossRef]
- Wen, L.; Chen, Z.Y.; Chen, G.X.; Yuan, J.L. Preparation of paraffin sections and their immunohistochemical staining techniques. Anim. Husb. Vet. Sci. Technol. Inf. 2018, 5, 36. [Google Scholar]
- Ghosh, S.; Chan, C.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Dixit, D.; Ahmad, F.; Ghildiyal, R.; Joshi, S.D.; Sen, E. CK2 inhibition induced PDK4-AMPK axis regulates metabolic adaptation and survival responses in glioma. Exp. Cell Res. 2016, 344, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]




| SNPs | Groups | Number | Genotype | Gene | Ho | He | Ne | PIC | X2 (P) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1164T | CC | CT | TT | C | T | |||||||
| STH | 25 | 0.24 (6) | 0.48 (12) | 0.28 (7) | 0.48 | 0.52 | 0.50 | 0.50 | 1.99 | 0.63 | 0.04 (0.98) | |
| SFK × STH | 30 | 0.40 (12) | 0.47 (14) | 0.13 (4) | 0.64 | 0.36 | 0.47 | 0.53 | 1.85 | 0.65 | 0.01 (0.99) | |
| All | 55 | 0.33 (18) | 0.47 (26) | 0.20 (11) | 0.57 | 0.43 | 0.49 | 0.51 | 1.96 | 0.63 | 0.10 (0.95) | |
| Physical and Chemical Parameters | Before Mutation | After Mutation |
|---|---|---|
| Molecular weight | 16,443.35 | 16,464.27 |
| Theoretical pI | 6.02 | 6.39 |
| Total number of negatively charged residues (Asp + Glu) | 29 | 28 |
| Instability index: | 50.42 | 50.70 |
| Grand average of hydropathicity (GRAVY) | −1.379 | −1.354 |
| Secondary Protein Structure | Before Mutation | After Mutation |
|---|---|---|
| Alpha helix (Hh) | 69.12% | 61.76% |
| Random coil (Cc) | 22.06% | 29.41% |
| Sample | Live Weight (kg) | Carcass Weight (kg) | Dressing Percentages (%) | Ribeye Area (cm2) | Marbleization Grades |
|---|---|---|---|---|---|
| STH1 | 29.25 | 13.25 | 45.30 | 12.14 | 1.5 |
| STH2 | 26.68 | 12.05 | 45.16 | 10.62 | 1.5 |
| STH3 | 31.75 | 13.75 | 43.31 | 11.78 | 1.0 |
| STH4 | 27.60 | 12.85 | 46.56 | 12.02 | 2.0 |
| STH5 | 28.00 | 13.15 | 46.96 | 11.72 | 1.5 |
| STH6 | 27.20 | 12.45 | 45.77 | 12.28 | 2.0 |
| SFK × STH1 | 38.65 | 19.35 | 50.06 | 13.45 | 2.5 |
| SFK × STH2 | 39.25 | 19.45 | 49.55 | 14.74 | 2.0 |
| SFK × STH3 | 44.60 | 23.25 | 52.13 | 14.44 | 3.0 |
| SFK × STH4 | 40.05 | 21.75 | 54.31 | 15.68 | 3.5 |
| SFK × STH5 | 42.65 | 21.25 | 49.82 | 13.06 | 3.0 |
| SFK × STH6 | 39.45 | 20.85 | 52.85 | 14.75 | 2.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, L.; Shi, J.; Meng, Q.; Tang, Z.; Liu, T.; Zhang, Q.; Cheng, S. Verification of Key Target Molecules for Intramuscular Fat Deposition and Screening of SNP Sites in Sheep from Small-Tail Han Sheep Breed and Its Cross with Suffolk. Int. J. Mol. Sci. 2024, 25, 2951. https://doi.org/10.3390/ijms25052951
Fu L, Shi J, Meng Q, Tang Z, Liu T, Zhang Q, Cheng S. Verification of Key Target Molecules for Intramuscular Fat Deposition and Screening of SNP Sites in Sheep from Small-Tail Han Sheep Breed and Its Cross with Suffolk. International Journal of Molecular Sciences. 2024; 25(5):2951. https://doi.org/10.3390/ijms25052951
Chicago/Turabian StyleFu, Lingjuan, Jinping Shi, Quanlu Meng, Zhixiong Tang, Ting Liu, Quanwei Zhang, and Shuru Cheng. 2024. "Verification of Key Target Molecules for Intramuscular Fat Deposition and Screening of SNP Sites in Sheep from Small-Tail Han Sheep Breed and Its Cross with Suffolk" International Journal of Molecular Sciences 25, no. 5: 2951. https://doi.org/10.3390/ijms25052951





