Design, Synthesis, Anti-Inflammatory Activity, DFT Modeling and Docking Study of New Ibuprofen Derivatives
Abstract
:1. Introduction
2. Results and Discussion
2.1. IA Description
2.2. Complex Characterization
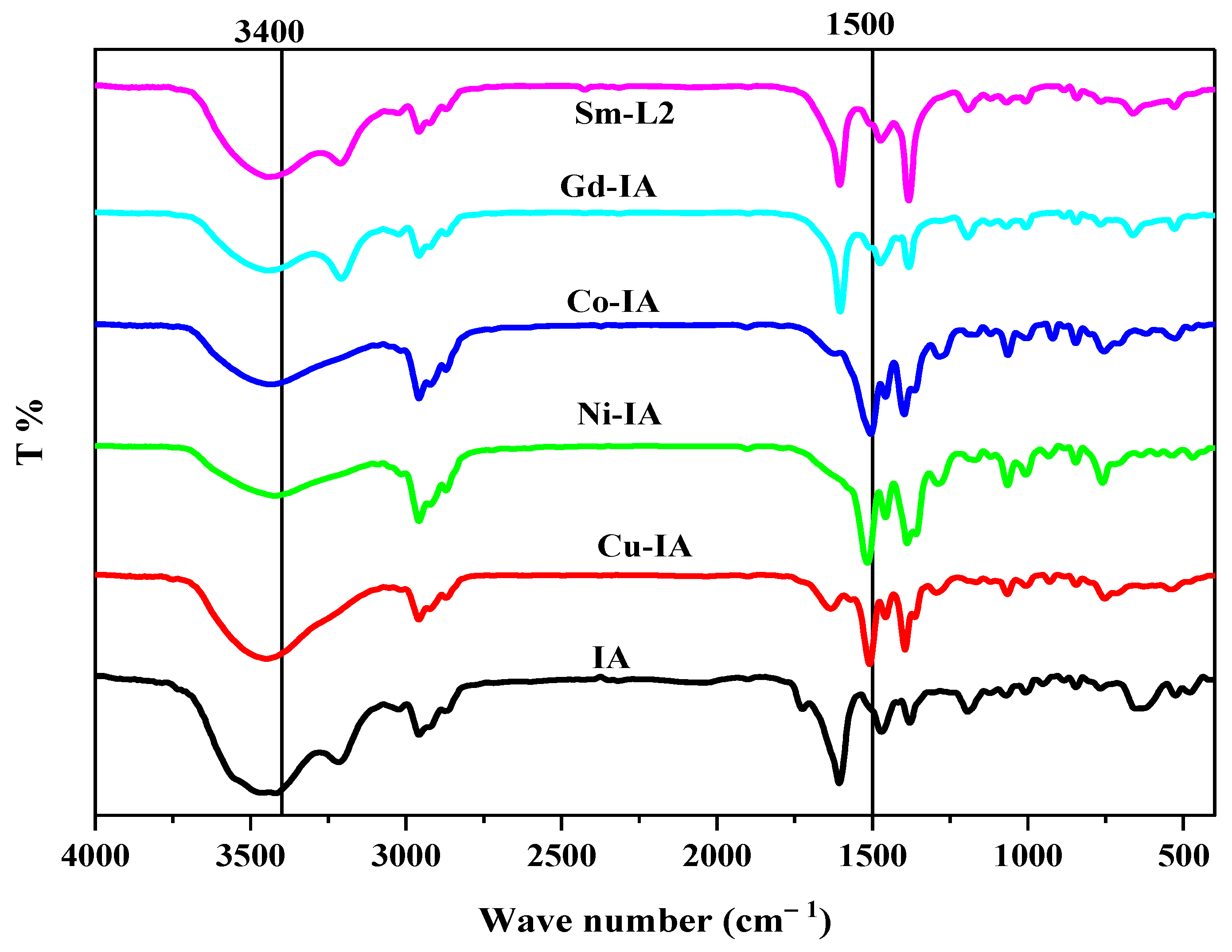
2.2.1. FTIR Spectra
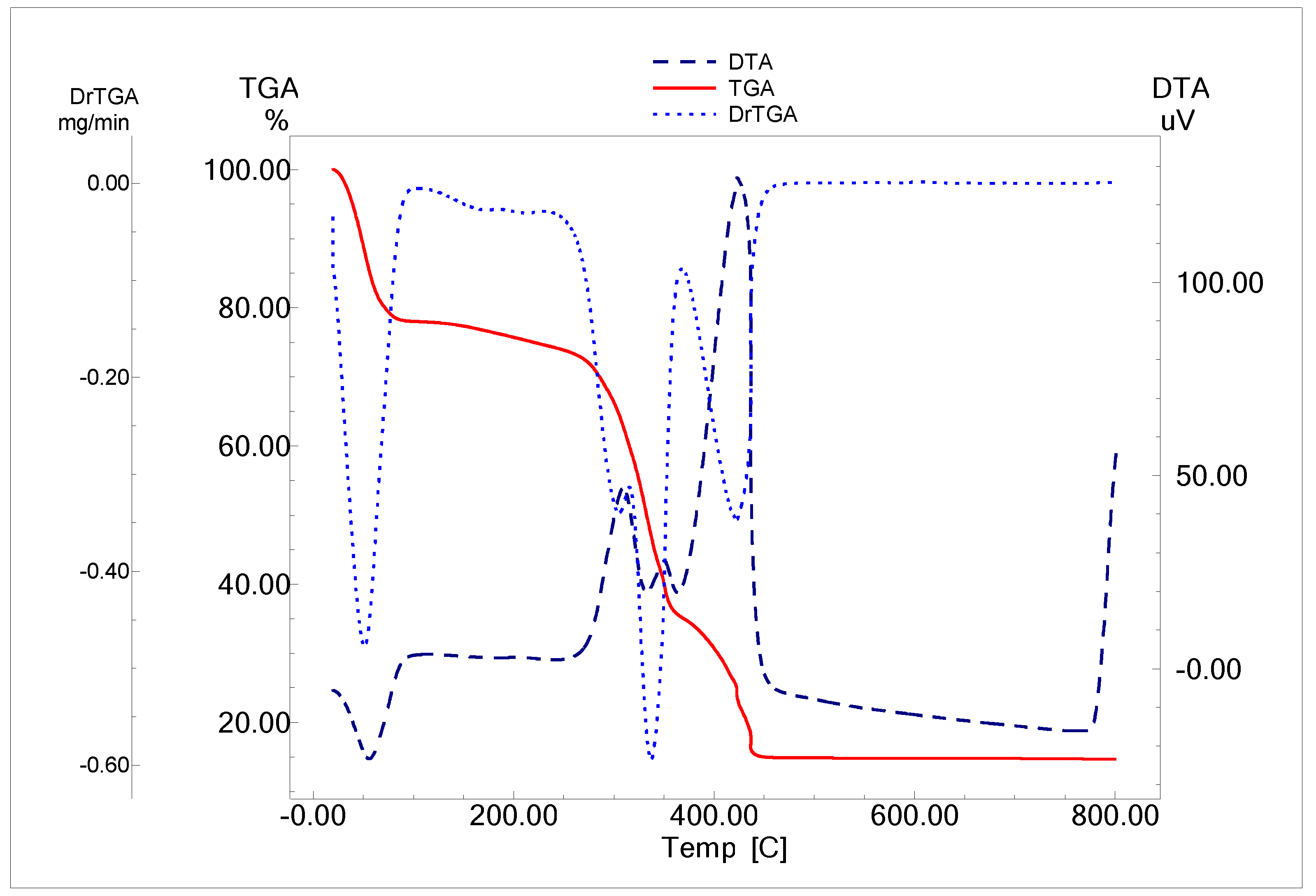
2.2.2. Thermal Analysis
2.2.3. Kinetics and Thermodynamic Parameters
2.2.4. Magnetic Moments and UV–Vis Spectroscopy
2.3. Computational Chemistry
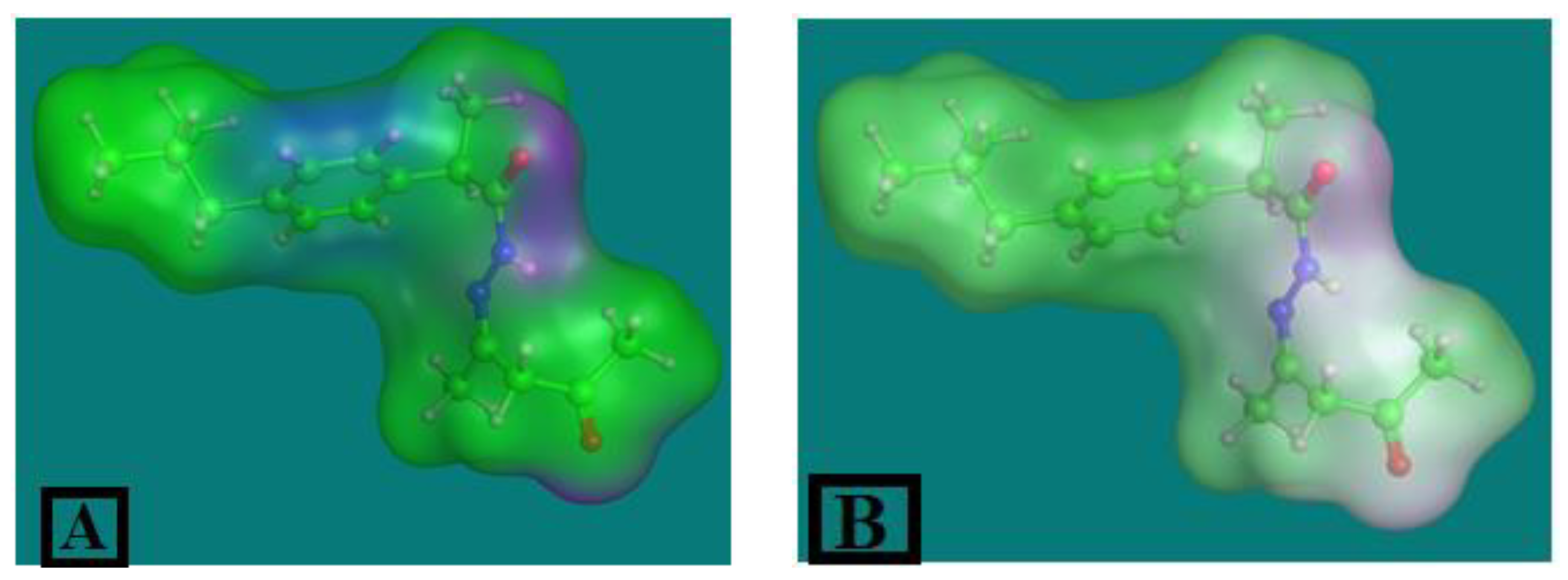
2.3.1. The IA Ligand Surface Properties
2.3.2. The DFT Calculation for Ibuprofen and IA
- The key equations describing the transition between ground states provide a robust framework for defining local, global, and non-local hardness and softness functions.
- Under chemical potential, it has been shown that interactions between two systems evolve towards a state of maximum hardness. Additionally, both soft–soft and hard–hard interactions are observed to be preferentially established.
- Lastly, it has been demonstrated that a system’s ground-state energy tends to decrease as its hardness increases, at least to a significant extent. These general principles could be instrumental in understanding the behavior of molecules in general and in discerning their reactions with various chemical types.
2.4. Molecular Docking Analyses
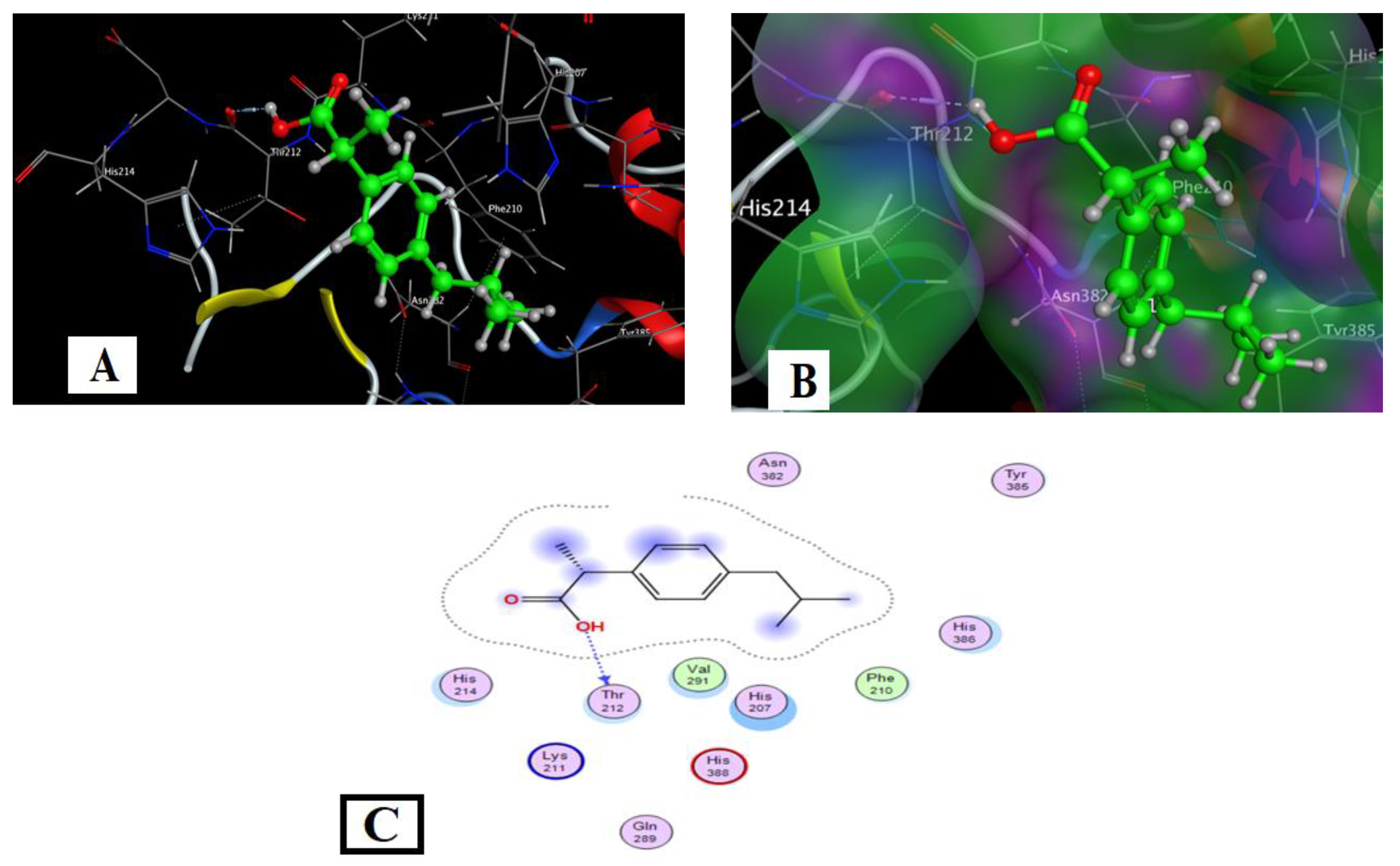
2.4.1. Ibuprofen Docking with COX2 (PDB Code: 5IKT)
2.4.2. IA Docking of with COX2
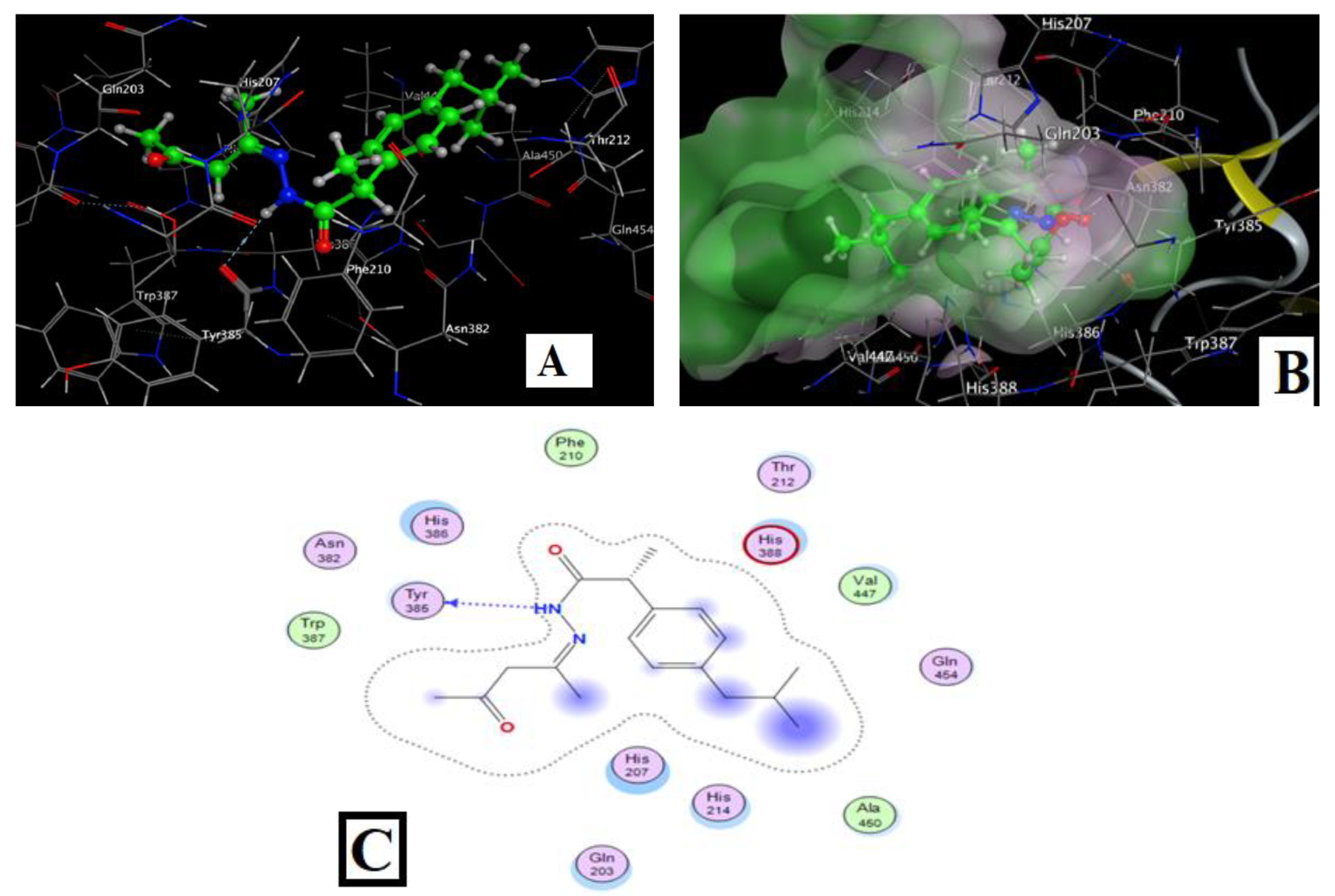
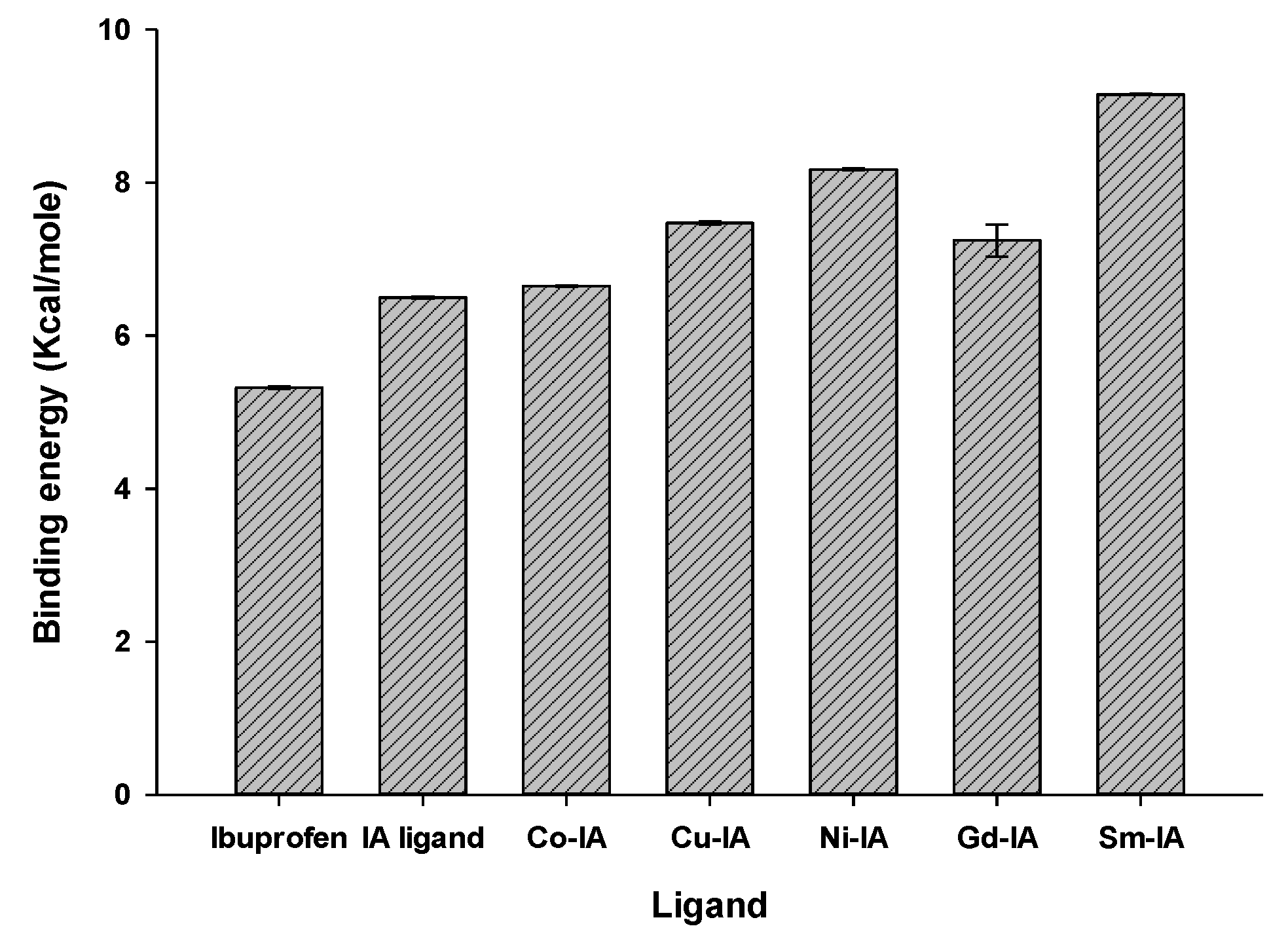
2.4.3. Molecular Docking of the Complexes
2.5. In Silico Studies
2.5.1. Prediction of Target–Ligand Interactions
2.5.2. Bioavailability Prediction
2.6. In Vitro: Anti-Inflammatory Action
2.6.1. IA and Its Complexes: Cyclooxygenase Inhibition
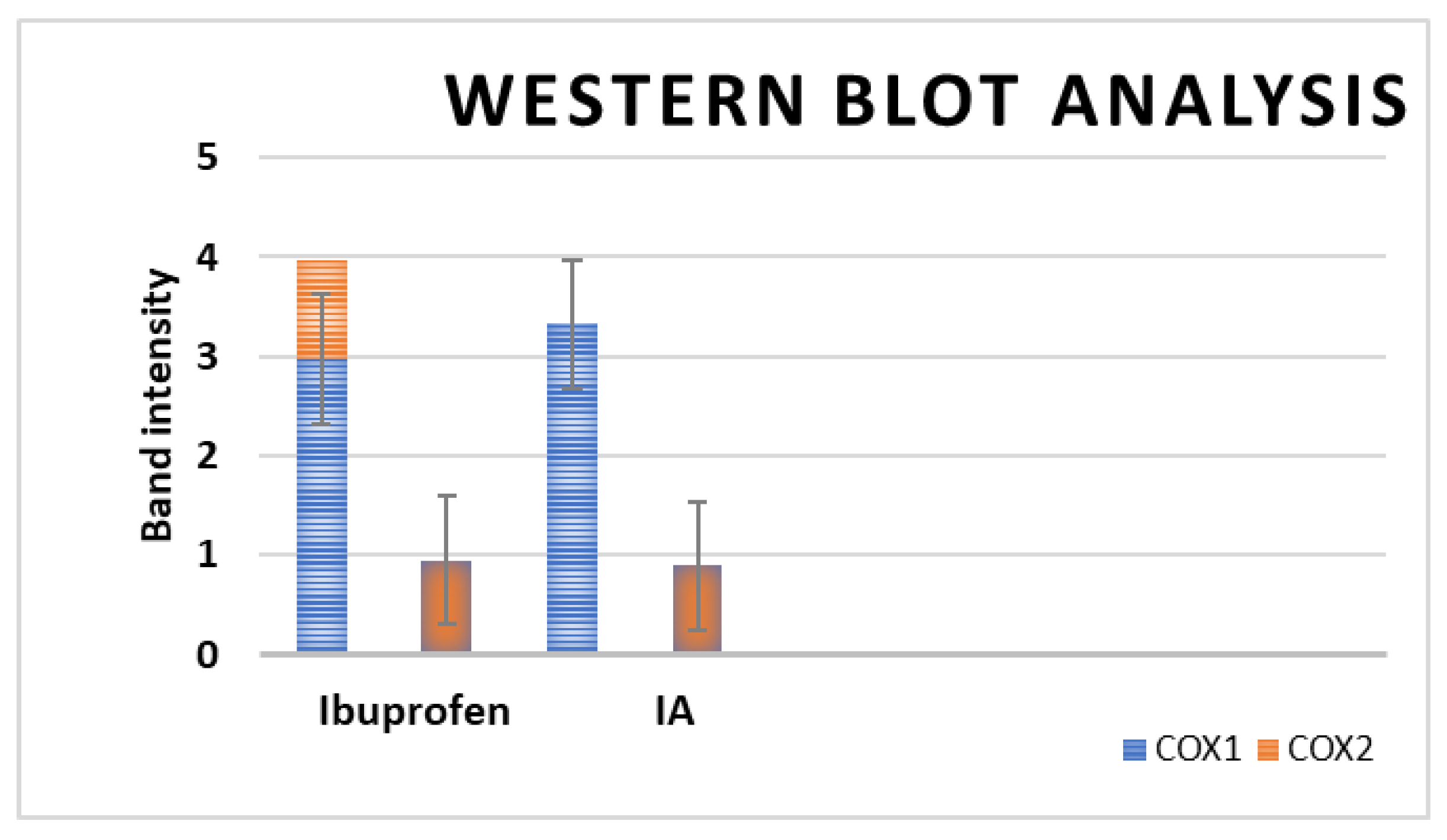
2.6.2. Western Blotting Assay
2.6.3. The MTT Test
3. Materials and Methods
3.1. Materials
3.2. Instrumentation
3.3. General Synthesis
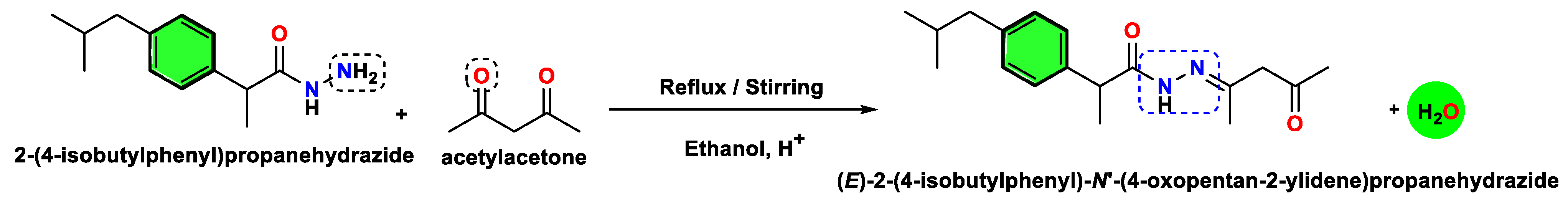
3.3.1. Synthesis of IA
3.3.2. Synthesis of Metal Complexes
3.4. Anti-Inflammatory Action
3.4.1. ELISA Test
3.4.2. Western Blot Analysis
3.4.3. The MTT Assay
4. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adams, S.; Cliffe, E.; Lessel, B.; Nicholson, J. Some biological properties of 2-(4-isobutylphenyl)-propionic acid. J. Pharm. Sci. 1967, 56, 1686. [Google Scholar] [CrossRef]
- Mousa, E.F.; Jassim, I.K. Preparation and characterization of oxadiazoles derived from ibuprofen. J. Pharm. Sci. Res. 2018, 10, 2297–2304. [Google Scholar]
- Adam, R.W. Synthesis and Characterization of some New synthesis of N-(pyrimidin-2-yl) benzenesulfonamide derivatives combined with oxaimidizolidine. J. Pharm. Sci. Res. 2018, 10, 3103. [Google Scholar]
- Belkheiri, N.; Bouguerne, B.; Bedos-Belval, F.; Duran, H.; Bernis, C.; Salvayre, R.; Nègre-Salvayre, A.; Baltas, M. Synthesis and antioxidant activity evaluation of a syringic hydrazones family. Eur. J. Med. Chem. 2010, 45, 3019–3026. [Google Scholar] [CrossRef] [PubMed]
- Vasincu, I.M.; Apotrosoaei, M.; Panzariu, A.-T.; Buron, F.; Routier, S.; Profire, L. Synthesis and biological evaluation of new 1, 3-thiazolidine-4-one derivatives of 2-(4-isobutylphenyl) propionic acid. Molecules 2014, 19, 15005–15025. [Google Scholar] [CrossRef] [PubMed]
- Fiori, A.T.M.; Lustri, W.R.; Magalhães, A.; Corbi, P.P. Chemical, spectroscopic characterization and antibacterial activities in vitro of a novel gold (I)–ibuprofen complex. Inorg. Chem. Commun. 2011, 14, 738–740. [Google Scholar] [CrossRef]
- Etcheverry, S.B.; Barrio, D.A.; Cortizo, A.M.; Williams, P.A.M. Three new vanadyl (IV) complexes with non-steroidal anti-inflammatory drugs (Ibuprofen, Naproxen and Tolmetin). Bioactivity on osteoblast-like cells in culture. J. Inorg. Biochem. 2002, 88, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Bonora, S.; Pisi, A.; Ottani, S.; Cesini, D.; Maris, A.; Di Foggia, M. Raman and SERS study on ibuprofen metal complexes with biomedical interest. Vib. Spectrosc. 2014, 73, 45–55. [Google Scholar] [CrossRef]
- e Silva, I.M.P.; de Moraes Profirio, D.; de Paiva, R.E.F.; Lancellotti, M.; Formiga, A.L.B.; Corbi, P.P. A silver complex with ibuprofen: Synthesis, solid state characterization, DFT calculations and antibacterial assays. J. Mol. Struct. 2013, 1049, 1–6. [Google Scholar] [CrossRef]
- Khodov, I.; Efimov, S.; Klochkov, V.; Alper, G.; De Carvalho, L.B. Determination of preferred conformations of ibuprofen in chloroform by 2D NOE spectroscopy. Eur. J. Pharm. Sci. 2014, 65, 65–73. [Google Scholar] [CrossRef]
- Khodov, I.; Dyshin, A.; Efimov, S.; Ivlev, D.; Kiselev, M. High-pressure NMR spectroscopy in studies of the conformational composition of small molecules in supercritical carbon dioxide. J. Mol. Liq. 2020, 309, 113113. [Google Scholar] [CrossRef]
- Abbas, A.M.; Fisal, S.R.; Orabi, A.S. Novel β-lactam antibiotic derivative and is complexes: DFT, frontier energy levels, DNA interaction, docking, physicochemical and antimicrobial properties. J. Mol. Struct. 2020, 1218, 128487. [Google Scholar] [CrossRef]
- Morgan, S.M.; Diab, M.; El-Sonbati, A. Supramolecular assembly of hydrogen bonding, ESR studies and theoretical calculations of Cu (II) complexes. Appl. Organomet. Chem. 2018, 32, e4504. [Google Scholar] [CrossRef]
- Varghese, H.T.; Panicker, C.Y.; Philip, D.; Mannekutla, J.R.; Inamdar, S. IR, Raman and SERS studies of methyl salicylate. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2007, 66, 959–963. [Google Scholar] [CrossRef]
- Wang, L.; Cao, C.; Cao, C. Substituent effects on the stretching vibration of C=N in multi-substituted benzylideneanilines. J. Phys. Org. Chem. 2019, 32, e3969. [Google Scholar] [CrossRef]
- Montazerozohori, M.; Musavi, S.; Masoudiasl, A.; Naghiha, A.; Dusek, M.; Kucerakova, M. Synthesis, spectral, crystal structure, thermal behavior, antimicrobial and DNA cleavage potential of two octahedral cadmium complexes: A supramolecular structure. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 137, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Kandil, S.S.; El-Hefnawy, G.B.; Baker, E.A. Thermal and spectral studies of 5-(phenylazo)-2-thiohydantoin and 5-(2-hydroxyphenylazo)-2-thiohydantoin complexes of cobalt (II), nickel (II) and copper (II). Thermochim. Acta 2004, 414, 105–113. [Google Scholar] [CrossRef]
- Chang, E.L.; Simmers, C.; Knight, D.A. Cobalt complexes as antiviral and antibacterial agents. Pharmaceuticals 2010, 3, 1711–1728. [Google Scholar] [CrossRef] [PubMed]
- Orabi, A.S. Complexes derived from some biologically active ligands. J. Coord. Chem. 2008, 61, 1294–1305. [Google Scholar] [CrossRef]
- Lal, R.A.; Singh, M.N.; Das, S. Complexes of Uranyl Nitrate, Uranyl Acetate, Uranyl Thiocyanate and Uranyl Chloride with Benzoyl, Salicyloyl and Isonicotinoyl Hydrazines. Synth. React. Inorg. Met.-Org. Chem. 1986, 16, 513–525. [Google Scholar] [CrossRef]
- Di Stefano, R.; Scopelliti, M.; Pellerito, C.; Fiore, T.; Vitturi, R.; Colomba, M.; Gianguzza, P.; Stocco, G.; Consiglio, M.; Pellerito, L. Organometallic complexes with biological molecules: XVII. Triorganotin (IV) complexes with amoxicillin and ampicillin. J. Inorg. Biochem. 2002, 89, 279–292. [Google Scholar] [CrossRef]
- Bravo, A.; Anacona, J. Synthesis and characterization of metal complexes with ampicillin. J. Coord. Chem. 1998, 44, 173–182. [Google Scholar] [CrossRef]
- El-Gamel, N.E. Metal chelates of ampicillin versus amoxicillin: Synthesis, structural investigation, and biological studies. J. Coord. Chem. 2010, 63, 534–543. [Google Scholar] [CrossRef]
- Wyllie, G.R.; Munro, O.Q.; Schulz, C.E.; Scheidt, W.R. Structural and physical characterization of (nitrato) iron (III) porphyrinates [Fe (por)(NO3)]–Variable coordination of nitrate. Polyhedron 2007, 26, 4664–4672. [Google Scholar] [CrossRef]
- Barszcz, B. Coordination properties of didentate N, O heterocyclic alcohols and aldehydes towards Cu (II), Co (II), Zn (II) and Cd (II) ions in the solid state and aqueous solution. Coord. Chem. Rev. 2005, 249, 2259–2276. [Google Scholar] [CrossRef]
- Coats, A.W.; Redfern, J. Kinetic parameters from thermogravimetric data. Nature 1964, 201, 68–69. [Google Scholar] [CrossRef]
- Sallam, S.; Orabi, A.; Abbas, A. DNA interaction with octahedral and square planar Ni (II) complexes of aspartic-acid Schiff-bases. J. Mol. Struct. 2011, 1006, 272–281. [Google Scholar] [CrossRef]
- Lever, A.B.P. Inorganic & Electronic Spectroscopy, 1st ed.; Elsevier: Amsterdam, The Netherlands, 1968; pp. 249–360. [Google Scholar]
- Dholakiya, P.P.; Patel, M. Preparation, magnetic, spectral, and biocidal studies of some transition metal complexes with 3, 5-dibromosalicylideneaniline and neutral bidentate ligands. Synth. React. Inorg. Met.-Org. Chem. 2002, 32, 819–829. [Google Scholar] [CrossRef]
- Bunker, P.R.; Jensen, P. Molecular Symmetry and Spectroscopy; NRC Research Press: Ottawa, ON, Canada, 2006; Volume 46853. [Google Scholar]
- Hamer, A.M.; Livingstone, S.E. The magnetic moments and electronic spectra of lanthanide chelates of 2-thenoyltrifluoroacetone. Transit. Met. Chem. 1983, 8, 298–304. [Google Scholar] [CrossRef]
- Abbas, A.M.; Fisal, S.R.; Orabi, A.S. Enhancement of the biochemical activity of some market antibiotics by chemical modification: Synthesis, characterization, and biochemical evaluation. J. Chin. Chem. Soc. 2021, 68, 131–149. [Google Scholar] [CrossRef]
- Tong, M.M.; Brewer, D. Nature of the Coordination Bond in Metal Complexes of Substituted Pyridine Derivatives. VI. Electronic Spectra of Some Complexes of Copper (II). Can. J. Chem. 1971, 49, 102–104. [Google Scholar] [CrossRef]
- Abbas, A.M.; Fisal, S.R.; Radwan, A.; Makhlouf, M.; Orabi, A.S. Novel action for ampicillin derivative and its complexes: Physicochemical, thermal analysis, DNA interaction, docking with FabH protein, in silico, and in vitro studies. J. Mol. Liq. 2022, 351, 118333. [Google Scholar] [CrossRef]
- Hisaindee, S.; Al-Kaabi, L.; Ajeb, S.; Torky, Y.; Iratni, R.; Saleh, N.i.; AbuQamar, S.F. Antipathogenic effects of structurally-related Schiff base derivatives: Structure–activity relationship. Arab. J. Chem. 2015, 8, 828–836. [Google Scholar] [CrossRef]
- Tyagi, P.; Tyagi, M.; Agrawal, S.; Chandra, S.; Ojha, H.; Pathak, M. Synthesis, characterization of 1, 2, 4-triazole Schiff base derived 3d-metal complexes: Induces cytotoxicity in HepG2, MCF-7 cell line, BSA binding fluorescence and DFT study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 171, 246–257. [Google Scholar] [CrossRef]
- Rauf, A.; Shah, A.; Munawar, K.S.; Khan, A.A.; Abbasi, R.; Yameen, M.A.; Khan, A.M.; Khan, A.R.; Qureshi, I.Z.; Kraatz, H.-B. Synthesis, spectroscopic characterization, DFT optimization and biological activities of Schiff bases and their metal (II) complexes. J. Mol. Struct. 2017, 1145, 132–140. [Google Scholar] [CrossRef]
- Gázquez, J.L. Hardness and softness in density functional theory. In Chemical Hardness; Springer: Berlin/Heidelberg, Germany, 1993; pp. 27–43. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Cheng, T.; Zhao, Y.; Li, X.; Lin, F.; Xu, Y.; Zhang, X.; Li, Y.; Wang, R.; Lai, L. Computation of octanol− water partition coefficients by guiding an additive model with knowledge. J. Chem. Inf. Model. 2007, 47, 2140–2148. [Google Scholar] [CrossRef]
- Kishk, S.M.; McLean, K.J.; Sood, S.; Helal, M.A.; Gomaa, M.S.; Salama, I.; Mostafa, S.M.; de Carvalho, L.P.S.; Munro, A.W.; Simons, C. Synthesis and biological evaluation of novel cYY analogues targeting Mycobacterium tuberculosis CYP121A1. Bioorg. Med. Chem. 2019, 27, 1546–1561. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Wildman, S.A.; Crippen, G.M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Brito, M.A.d. Pharmacokinetic study with computational tools in the medicinal chemistry course. Braz. J. Pharm. Sci. 2011, 47, 797–805. [Google Scholar] [CrossRef]
- Lequin, R.M. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef]
- Kurien, B.T.; Scofield, R.H. Western blotting: An introduction. In Western Blotting: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2015; pp. 17–30. [Google Scholar]
- Mahmood, T.; Yang, P.-C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429. [Google Scholar] [PubMed]
- Morgan, D.M. Tetrazolium (MTT) assay for cellular viability and activity. In Polyamine Protocols; Springer: Berlin/Heidelberg, Germany, 1998; pp. 179–184. [Google Scholar]
- Lee, R.H.; Griswold, E.; Kleinberg, J. Studies on the stepwise controlled decomposition of 2, 2′-bipyridine complexes of cobalt (II) and nickel (II) chlorides. Inorg. Chem. 1964, 3, 1278–1283. [Google Scholar] [CrossRef]
- Kansara, S.; Pandit, R.; Bhawe, V. Synthesis of some new Ibuprofen derivatives containing chief heterocyclic moiety like s-Triazine and evaluated for their analgesic activity. Rasayan J Chem 2009, 2, 699–705. [Google Scholar]
- Pânzariu, A.-T.; Apotrosoaei, M.; Vasincu, I.M.; Drăgan, M.; Constantin, S.; Buron, F.; Routier, S.; Profire, L.; Tuchilus, C. Synthesis and biological evaluation of new 1, 3-thiazolidine-4-one derivatives of nitro-l-arginine methyl ester. Chem. Cent. J. 2016, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.M.; Aboelmagd, A.; Kishk, S.M.; Nasrallah, H.H.; Boyd, W.C.; Kalil, H.; Orabi, A.S. A Novel Ibuprofen Derivative and Its Complexes: Physicochemical Characterization, DFT Modeling, Docking, In Vitro Anti-Inflammatory Studies, and DNA Interaction. Molecules 2022, 27, 7540. [Google Scholar] [CrossRef]
- Fernandes, C.; Palmeira, A.; Ramos, I.I.; Carneiro, C.; Afonso, C.; Tiritan, M.E.; Cidade, H.; Pinto, P.C.; Saraiva, M.L.M.; Reis, S. Chiral derivatives of xanthones: Investigation of the effect of enantioselectivity on inhibition of cyclooxygenases (COX-1 and COX-2) and binding interaction with human serum albumin. Pharmaceuticals 2017, 10, 50. [Google Scholar] [CrossRef]
- COX (Ovine/Human) Inhibitor Screening Assay Kit|Cayman Chemical. Available online: https://www.caymanchem.com/product/560131 (accessed on 25 April 2021).
- Yang, P.-C.; Liu, Z.-Q.; Mahmood, T. Western blot: Technique, theory and trouble shooting. North Am. J. Med. Sci. 2014, 6, 160. [Google Scholar] [CrossRef]
- Bahuguna, A.; Khan, I.; Bajpai, V.K.; Kang, S.C. MTT assay to evaluate the cytotoxic potential of a drug. Bangladesh J. Pharmacol. 2017, 12, 115–118. [Google Scholar] [CrossRef]
- ElKhazendar, M.; Chalak, J.; El-Huneidi, W.; Vinod, A.; Abdel-Rahman, W.M.; Abu-Gharbieh, E. Antiproliferative and proapoptotic activities of ferulic acid in breast and liver cancer cell lines. Trop. J. Pharm. Res. 2019, 18, 2571–2576. [Google Scholar]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the MTT assay. Cold Spring Harb. Protoc. 2018, 2018, pdb.prot095505. [Google Scholar] [CrossRef]
- Fernandes, T.J.; Costa, J.; Oliveira, M.B.P.; Mafra, I. Exploiting 16S rRNA gene for the detection and quantification of fish as a potential allergenic food: A comparison of two real-time PCR approaches. Food Chem. 2018, 245, 1034–1041. [Google Scholar] [CrossRef] [PubMed]








| Compound | Molecular Weight | Color | Melting Point (°C) | Ω * (µS) | CHNM% | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| C% | H% | N% | M% ** | |||||||
| Calc. | Calc. | Calc. | A | B | Calc. | |||||
| Found | Found | Found | ||||||||
| IA (C18H26N2O2) | 302.5 | Canary-yellow | 135 | --- | 71.42 71.33 | 8.59 8.55 | 10.58 10.61 | --- | --- | --- |
| [Cu(IA)·(H2O)·Cl2]6H2O | 562.97 | Dirty green | >280 | 13 | 47.53 47.44 | 6.20 6.22 | 6.16 6.20 | 12.16 | 11.59 | 11.29 |
| [Ni(IA)·(H2O)2·Cl]Cl·H2O | 486.06 | Violet | >280 | 100 | 49.98 49.88 | 6.99 6.88 | 6.48 6.51 | 12.28 | 12.08 | 12.08 |
| [Co(IA)·(H2O)·Cl2]2H2O | 468.29 | Brick brown | >280 | 15 | 44.46 44.55 | 6.63 6.71 | 5.76 5.66 | 12.25 | 12.19 | 12.12 |
| [Gd(IA)2·(NO3)2·(H2O)]NO3·2H2O | 1002.25 | Yellowish white | >280 | 10 | 47.23 47.22 | 5.89 5.92 | 9.44 9.50 | --- | 15.75 | 15.69 |
| [Sm(IA)2·(NO3)2]NO3·3H2O | 995.15 | Yellowish white | >280 | 100 | 48.59 48.61 | 5.83 5.91 | 9.71 9.80 | --- | 15.09 | 15.15 |
| Compound | υ (NH, H2O, OH) | υ(C=O) | υ(C=N) | υ(NO3−) | υ (M-O) | υ(M-N) |
|---|---|---|---|---|---|---|
| IA(C18H26N2O2) | 3720–3000 m, br | 1725 s 1660 w | 1611 s | --- | --- | --- |
| [Cu(IA)Cl2·(H2O)]6H2O | 3780–3000 m, br | 1641w | 1528 sh | --- | 636 w | 540 w |
| [Ni(IA)·(H2O)3]Cl2·H2O | 3760–3070 m, br | 1608 w | 1510 sh | --- | 622 w | 521 w |
| [Co(IA)·(H2O)·Cl]Cl·H2O | 3760–3070 m, br | 1612 w | 1508 sh | --- | 617 w | 526 w |
| [Gd(IA)2·(H2O)·(NO3)]2H2O | 3790–3070 m, br | 1612 w | 1528 sh | 1384 | 651 w | 531 w |
| [Sm(IA)2·(H2O)·NO3](NO3)2·2H2O | 3760–3080 m, br | 1617 w | 1518 sh | 1384 | 680 w | 516 w |
| Compound | Molecular Weight | Temp. Range °C | DTG Temp. °C | Mass Loss % | Process | Expected Products | Residue | M% | |
|---|---|---|---|---|---|---|---|---|---|
| Found | Calcd. | (Calcd.) Found | |||||||
| [Cu(IA)·(H2O)·Cl2]6H2O | 562.97 | 36–64 | 49 | 19.70 | 19.20 | Dehydration | 6H2O | CuO (14.15) 14.50 | 11.59 |
| 180–310 | 304 | 15.14 | 15.16 | Coordination sphere + ligand decomposition | 2HCl+ H2O+ 0.15 IA | ||||
| 330–348 | 338 | 30.84 | 30.81 | ligand decomposition | 0.43 IA | ||||
| 395–460 | 423 | 20.75 | 20.56 | Complete decomposition | 0.37 IA | ||||
| [Ni(IA)·(H2O)2·Cl]Cl·H2O | 486.06 | 38–58 | 49 | 3.65 | 3.71 | Dehydration | H2O | NiO (15.37) 12.41 | 12.08 |
| 289–347 | 341 | 50.46 | 50.01 | Coordination sphere + ligand decomposition | 2HCl + 2H2O + 0.51 IA | ||||
| 385–401 | 393 | 30.62 | 30.61 | Complete decomposition | 0.49 IA | ||||
| [Co(IA)·(H2O)·Cl2]2H2O | 486.29 | 28–199 | 53 | 7.35 | 7.41 | Dehydration | 2H2O | CoO (15.40) 18.99 | 12.12 |
| 263–321 | 271 | 46.62 | 46.56 | Coordination sphere + ligand decomposition | 2HCl+ H2O 0.49 IA | ||||
| 296–356 | 353 | 30.50 | 30.52 | Complete decomposition | 0.51 IA | ||||
| [Gd(IA)2·(NO3)2·(H2O)] NO3·2H2O | 1002.25 | 24–35 | 38 | 4.21 | 3.59 | Dehydration | 2H2O | Gd2O3 (36.17) 33.25 | 15.75 |
| 253–317 | 280 | 33.64 | 33.65 | Coordination sphere + Ligand decomposition | H2O+ 2HNO3 0.55 IA | ||||
| 398–434 | 420 | 15.82 | 15.92 | Coordination sphere + ligand decomposition | HNO3 + 0.30 IA | ||||
| 481–607 | 497 548 590 | 9.97 | 9.98 | Complete decomposition | 0.17 IA | ||||
| [Sm(IA)2·(NO3)2]NO3·3H2O | 995.25 | 69–134 | 98 | 5.45 | 5.34 | Dehydration | 2H2O | Sm2O3 (35.03) 30.01 | 15.09 |
| 260–299 | 279 | 53.06 | 53.11 | Coordination sphere + ligand decomposition | 2HNO3 0.80 IA | ||||
| 453–476 | 460 | 6.62 | 6.63 | NO3 liberation + ligand decomposition | HNO3+ 0.16 IA | ||||
| 651–670 | 660 | 0.39 | 0.38 | Complete decomposition | 0.092 IA | ||||
| Complex | Step | R2 | Order (n) | Ts (K) | Ea (J/mol) | Z (s−1) | ΔS‡ (J/K·mol) | ΔH‡ (kJ/mol) | ΔG‡ (kJ/mol) |
|---|---|---|---|---|---|---|---|---|---|
| [Cu(IA)(H2O)Cl2]6H2O | Complete ligand decomposition | 0.97 | 2.00 | 696 | 231.38 | 2.87 × 1017 | +82.26 | −5.555 | −62.81 |
| [Ni(IA)(H2O)2Cl]Cl·H2O | Coordination sphere+ ligand decomposition | 0.99 | 2.00 | 614 | 32.69 | 212.24 | −206.41 | −5.072 | 121.05 |
| [Co(IA)(H2O)Cl2]2H2O | Complete ligand decomposition | 0.99 | 2.00 | 626 | 45.83 | 1.37 × 104 | −172.10 | −5.0159 | 102.58 |
| [Gd(IA)2(NO3)2(H2O)]NO3·2H2O | NO3 liberation and ligand decomposition | 0.98 | 2.00 | 693 | 172.49 | 1.02 × 1013 | −2.88 | −5.589 | −3.59 |
| [Sm(IA)2(NO3)2]NO3·3H2O | NO3 liberation and ligand decomposition | 0.99 | 2.00 | 552 | 115.17 | 1.19 × 1010 | −57.16 | −4.474 | 27.08 |
| Compound | Ligand Sites | Receptor Sites | Type of Interaction | Bond Distance (Å) | Binding Energy (kcal/mol) | (S) (kcal/mol) |
|---|---|---|---|---|---|---|
| Ibuprofen | O(14) | THR 212 | H-donor | 2.85 | −2.9 | −5.33 |
| IA | N(14) | TYR 385 | H- donor | 3.05 | −0.7 | −6.48 |
| Co–IA | C(1) | HIS214 | H- acceptor | 3.19 | −5.2 | −6.65 |
| Ph | HIS207 | H-pi | 3.78 | −0.6 | ||
| Ph | HIS388 | H-pi | 3.70 | 0 | ||
| Cu–IA | Cl | SER 451 | H- acceptor | 3.47 | −0.9 | −7.45 |
| Ph | TYR385 | H-pi | 4.6 | −0.6 | ||
| Ni–IA | O(21) | PHE 210 | H- donor | 2.73 | −2.2 | −8.16 |
| O(22) | ASN382 | H- donor | 3.19 | −1.3 | ||
| Ph | HIS 386 | H-pi | 4.69 | −0.6 | ||
| Gd–IA | Ph | HIS 386 | H-pi | 4.42 | −0.9 | −7.41 |
| Sm–IA | Ph | HIS 386 | H-pi | 4.33 | −1 | −9.14 |
| Ph | HIS 207 | H-pi | 4.86 | −0.7 |
| Compound | Ibuprofen | IA | Co–IA |
|---|---|---|---|
| Heavy atoms | 15 | 22 | 26 |
| Rotatable bonds | 4 | 8 | 4 |
| H-bond donor | 1 | 1 | 1 |
| H-bond acceptor | 2 | 3 | 4 |
| Fraction Csp3 | 0.46 | 0.50 | 0.39 |
| LogS | −3.44 | −5.27 | −6.13 |
| XLogP3 | 3.50 | 3.35 | 6.82 |
| Molar refractivity | 62.18 | 91.33 | 111.07 |
| TPSA (Å2) | 37.30 | 58.53 | 43.29 |
| Log Kp (skin permeation) (cm/s) | −5.07 | −5.77 | −4.19 |
| GI absorption | High | High | High |
| P-gp substrate | No | No | Yes |
| Compound | IC50 (a) (µM) | ||
|---|---|---|---|
| COX-1 | COX-2 | COX-2 (b) S.I. | |
| Ibuprofen | 12.9 | 31.4 | 0.4 |
| Indomethacin | 0.4 | 0.1 | 5.0 |
| Diclofenac sodium | 3.8 | 0.8 | 4.5 |
| IA | 10.2 | 3.6 | 2.8 |
| Compound | IC50 (µM) | ||
|---|---|---|---|
| COX-1 | COX-2 | COX-2 S.I. | |
| Ibuprofen | 2.975 | 3.325 | 0.895 |
| IA | 0.946 | 0.894 | 1.058 |
| Compound | Fibroblast Cells |
|---|---|
| Ibuprofen | 31.4 |
| IA | 3.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, A.M.; Nasrallah, H.H.; Aboelmagd, A.; Kishk, S.M.; Boyd, W.C.; Kalil, H.; Orabi, A.S. Design, Synthesis, Anti-Inflammatory Activity, DFT Modeling and Docking Study of New Ibuprofen Derivatives. Int. J. Mol. Sci. 2024, 25, 3558. https://doi.org/10.3390/ijms25063558
Abbas AM, Nasrallah HH, Aboelmagd A, Kishk SM, Boyd WC, Kalil H, Orabi AS. Design, Synthesis, Anti-Inflammatory Activity, DFT Modeling and Docking Study of New Ibuprofen Derivatives. International Journal of Molecular Sciences. 2024; 25(6):3558. https://doi.org/10.3390/ijms25063558
Chicago/Turabian StyleAbbas, Abbas M., Hossam H. Nasrallah, Ahmed Aboelmagd, Safaa M. Kishk, W. Christopher Boyd, Haitham Kalil, and Adel S. Orabi. 2024. "Design, Synthesis, Anti-Inflammatory Activity, DFT Modeling and Docking Study of New Ibuprofen Derivatives" International Journal of Molecular Sciences 25, no. 6: 3558. https://doi.org/10.3390/ijms25063558








