Dysfunction in IGF2R Pathway and Associated Perturbations in Autophagy and WNT Processes in Beckwith–Wiedemann Syndrome Cell Lines
Abstract
:1. Introduction
2. Results
2.1. Imprinted and Imprinted-Related Gene Expression in BWS and Control Cell Lines
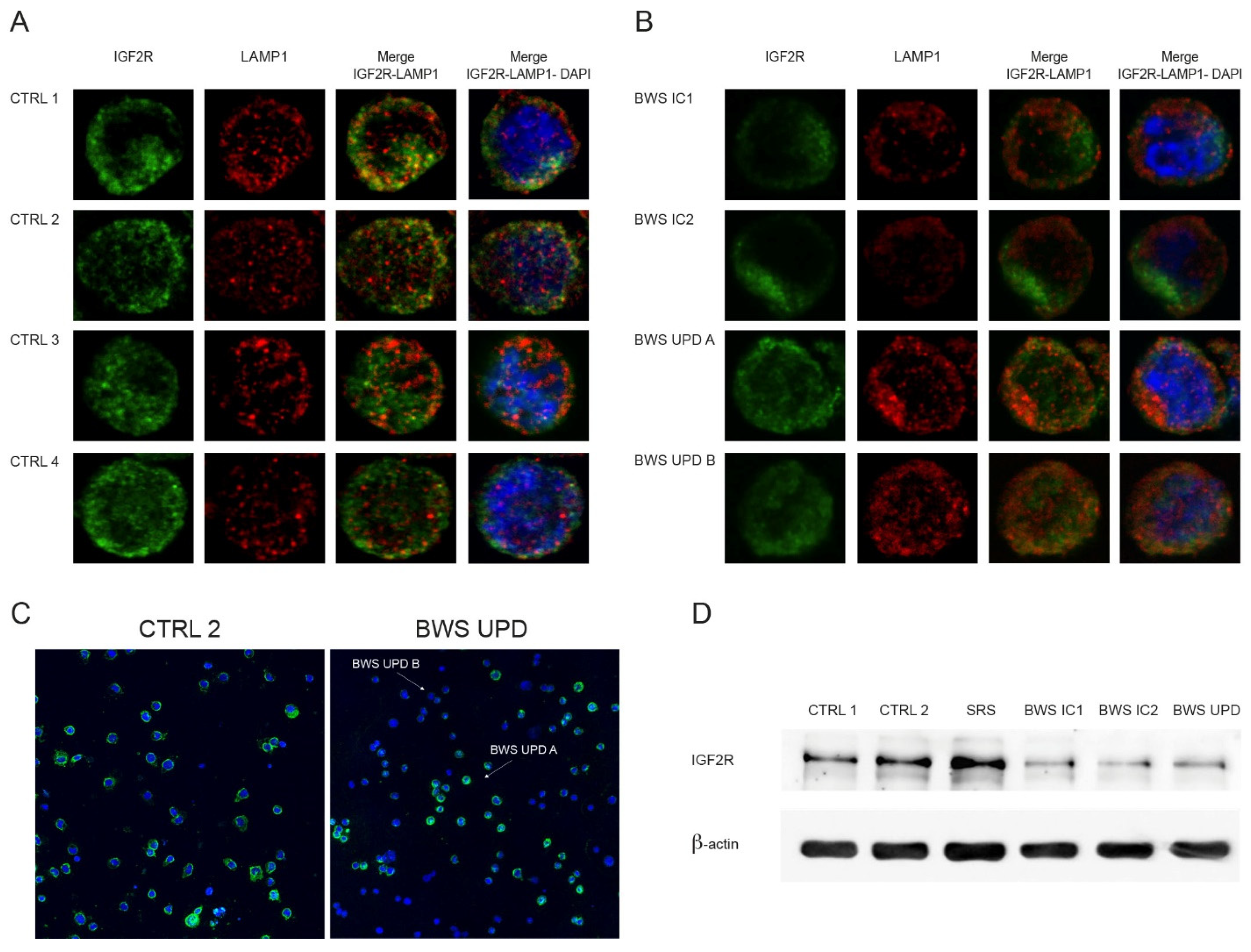
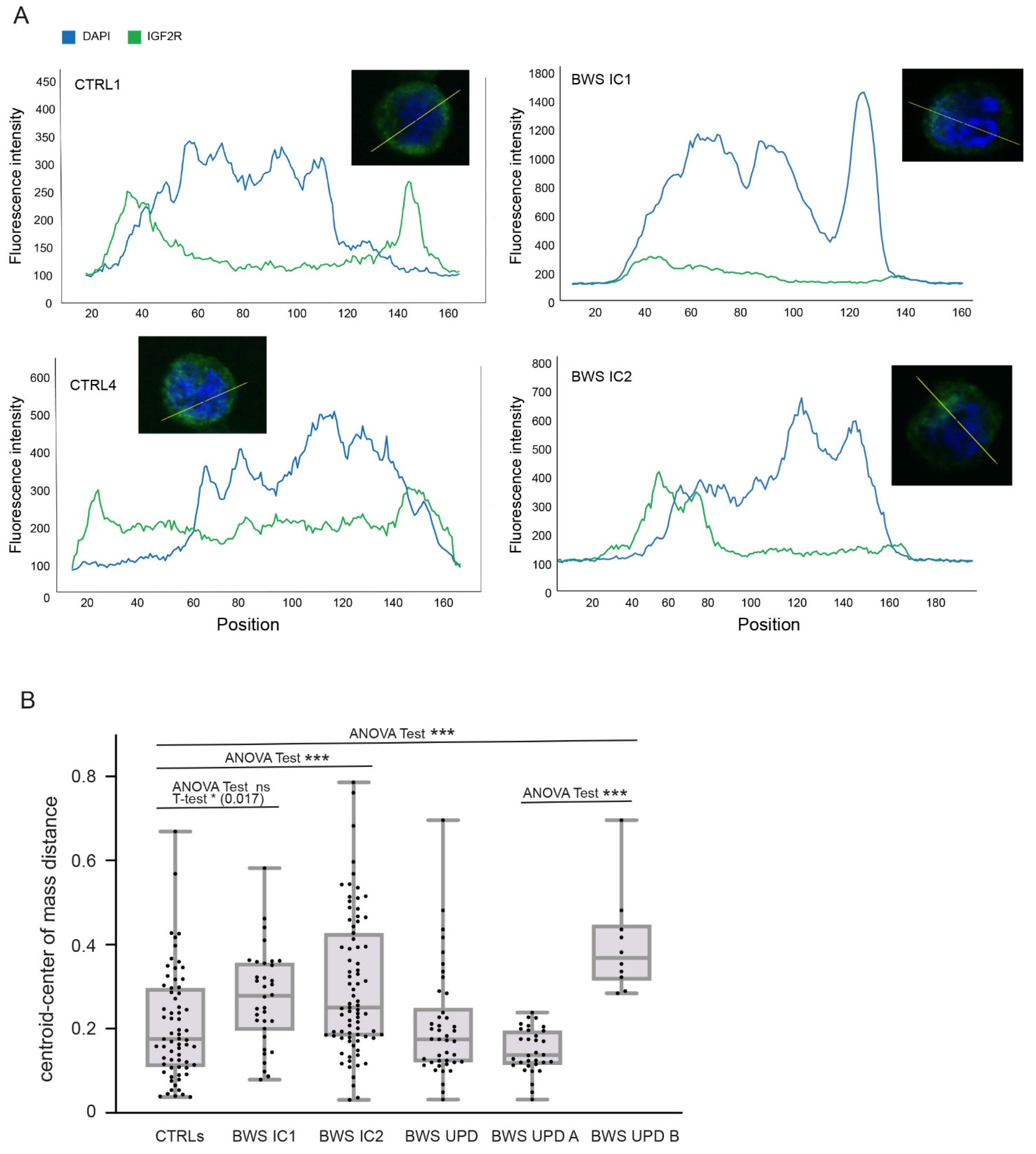
2.2. IGF2R Expression and Localization Analyses in BWS and Control Cell Lines
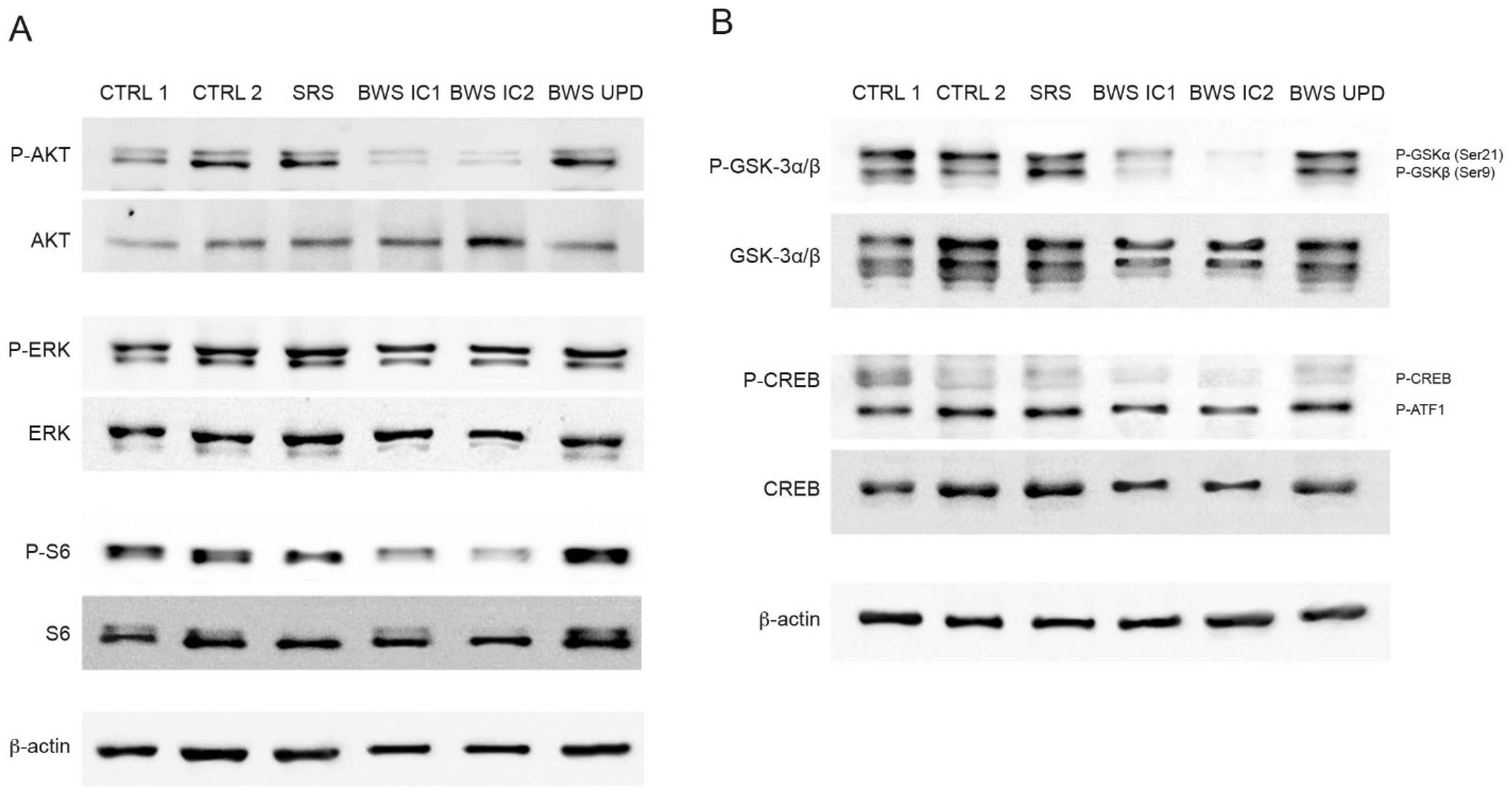
2.3. Analysis of IGF2R Targets in BWS and Control Cell Lines
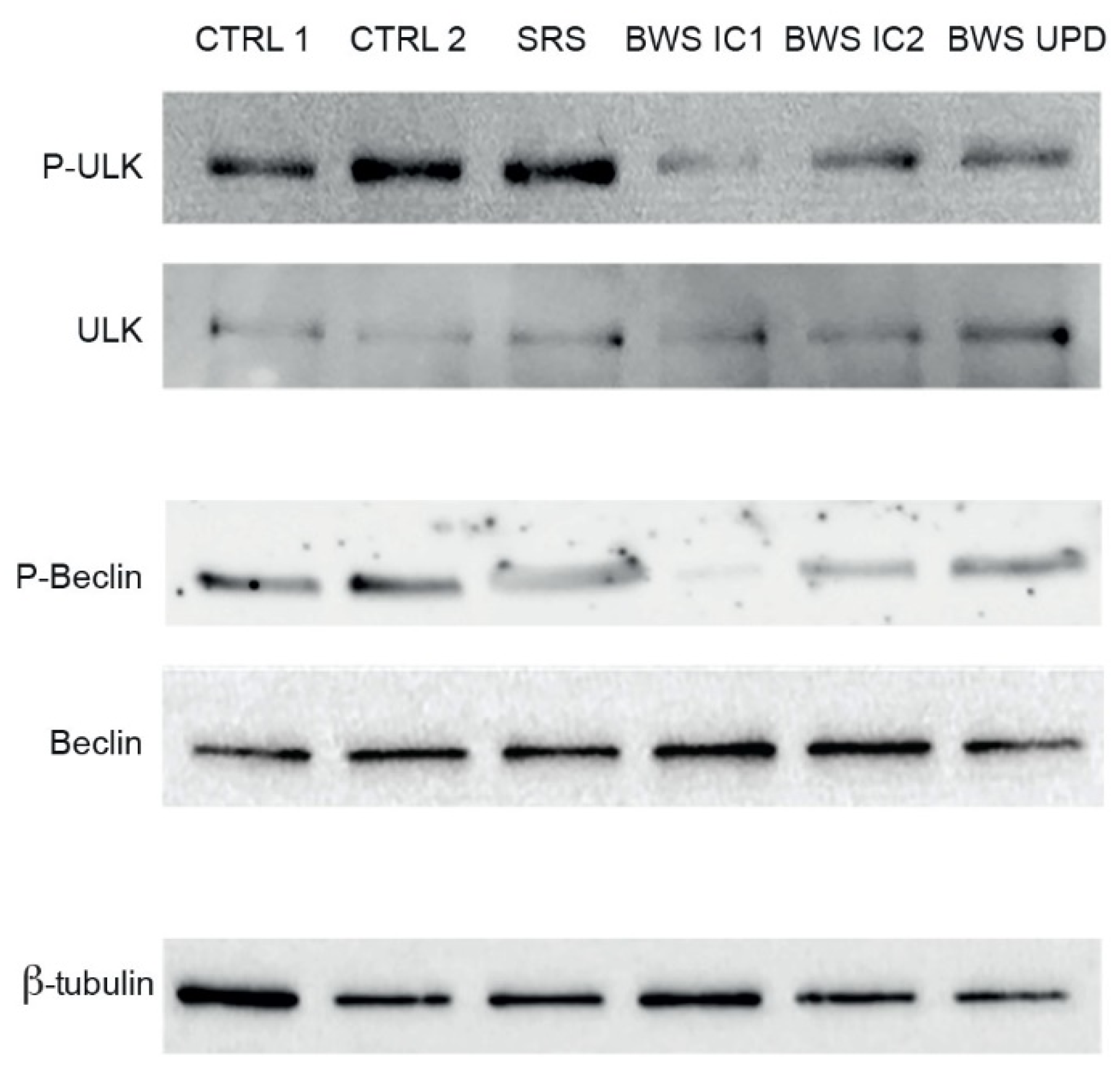
2.4. Analysis of Autophagy in BWS LCLs
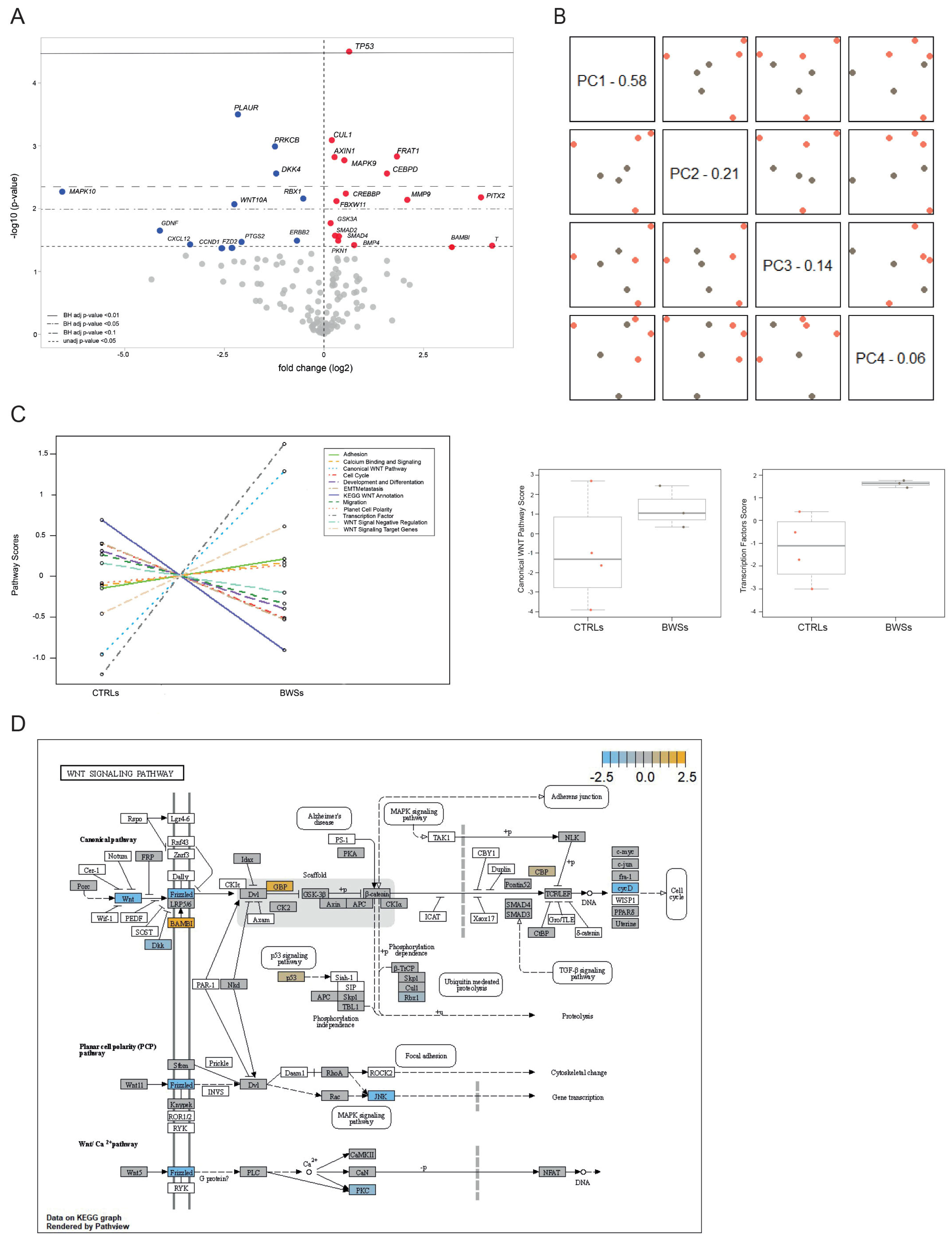
2.5. WNT Pathway Analysis in BWS and Control Cell Lines
3. Discussion
4. Materials and Methods
4.1. Lymphoblastoid Cell Lines
4.2. nCounter Analysis
4.3. Immunofluorescence Assay—Image Quantification and Statistics
4.4. Western Blot Analysis
4.5. Phospho-Array Profiler Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, K.H.; Kupa, J.; Duffy, K.A.; Kalish, J.M. Diagnosis and Management of Beckwith-Wiedemann Syndrome. Front. Pediatr. 2020, 7, 562. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Tabano, S.; Maitz, S.; Colapietro, P.; Garzia, E.; Gerli, A.G.; Sirchia, S.M.; Miozzo, M. Clinical and Molecular Diagnosis of Beckwith-Wiedemann Syndrome with Single- or Multi-Locus Imprinting Disturbance. Int. J. Mol. Sci. 2021, 22, 3445. [Google Scholar] [CrossRef]
- Borjas Mendoza, P.A.; Daley, J.O.; Mendez, M.D. Beckwith-Wiedemann Syndrome. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Mussa, A.; Russo, S.; De Crescenzo, A.; Freschi, A.; Calzari, L.; Maitz, S.; Macchiaiolo, M.; Molinatto, C.; Baldassarre, G.; Mariani, M.; et al. (Epi)genotype-phenotype correlations in Beckwith-Wiedemann syndrome. Eur. J. Hum. Genet. 2016, 24, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.W.; Ryan, E.; Kalish, J.M.; Ganguly, A. Prenatal molecular testing and diagnosis of Beckwith-Wiedemann syndrome. Prenat. Diagn. 2021, 41, 817–822. [Google Scholar] [CrossRef]
- Tüysüz, B.; Güneş, N.; Geyik, F.; Yeşil, G.; Celkan, T.; Vural, M. Investigation of (epi)genotype causes and follow-up manifestations in the patients with classical and atypical phenotype of Beckwith-Wiedemann spectrum. Am. J. Med. Genet. A 2021, 185, 1721–1731. [Google Scholar] [CrossRef]
- Brioude, F.; Netchine, I.; Praz, F.; Le Jule, M.; Calmel, C.; Lacombe, D.; Edery, P.; Catala, M.; Odent, S.; Isidor, B.; et al. Mutations of the Imprinted CDKN1C Gene as a Cause of the Overgrowth Beckwith-Wiedemann Syndrome: Clinical Spectrum and Functional Characterization. Hum. Mutat. 2015, 36, 894–902. [Google Scholar] [CrossRef]
- Jurkiewicz, D.; Skórka, A.; Ciara, E.; Kugaudo, M.; Pelc, M.; Chrzanowska, K.; Krajewska-Walasek, M. Rare clinical findings in three sporadic cases of Beckwith-Wiedemann syndrome due to novel mutations in the CDKN1C gene. Clin. Dysmorphol. 2020, 29, 28–34. [Google Scholar] [CrossRef]
- Russo, S.; Finelli, P.; Recalcati, M.P.; Ferraiuolo, S.; Cogliati, F.; Dalla Bernardina, B.; Tibiletti, M.G.; Agosti, M.; Sala, M.; Bonati, M.T.; et al. Molecular and genomic characterisation of cryptic chromosomal alterations leading to paternal duplication of the 11p15.5 Beckwith-Wiedemann region. J. Med. Genet. 2006, 43, e39. [Google Scholar] [CrossRef]
- Nativio, R.; Sparago, A.; Ito, Y.; Weksberg, R.; Riccio, A.; Murrell, A. Disruption of genomic neighborhoods at the imprinted IGF2-H19 locus in Beckwith-Wiedemann syndrome and Silver-Russell syndrome. Hum. Mol. Genet. 2011, 20, 1363–1374. [Google Scholar] [CrossRef]
- Wesseler, K.; Kraft, F.; Eggermann, T. Molecular and Clinical Opposite Findings in 11p15.5 Associated Imprinting Disorders: Characterization of Basic Mechanisms to Improve Clinical Management. Int. J. Mol. Sci. 2019, 20, 4219. [Google Scholar] [CrossRef]
- St-Pierre, J.; Hivert, M.F.; Perron, P.; Poirier, P.; Guay, S.P.; Brisson, D.; Bouchard, L. IGF2 DNA methylation is a modulator of newborn’s fetal growth and development. Epigenetics 2012, 7, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Forbes, B.E.; Blyth, A.J.; Wit, J.M. Disorders of IGFs and IGF-1R signaling pathways. Mol. Cell Endocrinol. 2020, 1, 518. [Google Scholar] [CrossRef] [PubMed]
- Massoner, P.; Ladurner-Rennau, M.; Eder, I.E.; Klocker, H. Insulin-like growth factors and insulin control a multifunctional signalling network of significant importance in cancer. Br. J. Cancer 2010, 103, 1479. [Google Scholar] [CrossRef]
- Maxia, C.; Isola, M.; Grecu, E.; Cuccu, A.; Scano, A.; Orrù, G.; Di Girolamo, N.; Diana, A.; Murtas, D. Synergic Action of Insulin-like Growth Factor-2 and miRNA-483 in Pterygium Pathogenesis. Int. J. Mol. Sci. 2023, 24, 4329. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Jones, E.Y.; Forbes, B.E. Interactions of IGF-II with the IGF2R/cation-independent mannose-6-phosphate receptor mechanism and biological outcomes. Vitam. Horm. 2009, 80, 699–719. [Google Scholar] [CrossRef]
- Rezgui, D.; Williams, C.; Savage, S.A.; Prince, S.N.; Zaccheo, O.J.; Jones, E.Y.; Crump, M.P.; Hassan, A.B. Structure and function of the human Gly1619Arg polymorphism of M6P/IGF2R domain 11 implicated in IGF2 dependent growth. J. Mol. Endocrinol. 2009, 42, 341–356. [Google Scholar] [CrossRef]
- LeRoith, D.; Holly, J.M.P.; Forbes, B.E. Insulin-like growth factors: Ligands, binding proteins, and receptors. Mol. Metab. 2021, 52, 101245. [Google Scholar] [CrossRef]
- Hughes, J.; Frago, S.; Bühnemann, C.; Carter, E.J.; Hassan, A.B. Maternal transmission of a humanised Igf2r allele results in an Igf2 dependent hypomorphic and non-viable growth phenotype. PLoS ONE 2013, 8, e57270. [Google Scholar] [CrossRef]
- Ghosh, P.; Dahms, N.M.; Kornfeld, S. Mannose 6-phosphate receptors: New twists in the tale. Nat. Rev. Mol. Cell Biol. 2003, 4, 202. [Google Scholar] [CrossRef]
- Sepúlveda, D.; Grunenwald, F.; Vidal, A.; Troncoso-Escudero, P.; Cisternas-Olmedo, M.; Villagra, R.; Vergara, P.; Aguilera, C.; Nassif, M.; Vidal, R.L. Insulin-like growth factor 2 and autophagy gene expression alteration arise as potential biomarkers in Parkinson’s disease. Sci. Rep. 2022, 12, 2038. [Google Scholar] [CrossRef]
- Liu, S.B.; Zhou, L.B.; Wang, H.F.; Li, G.; Xie, Q.P.; Hu, B. Loss of IGF2R indicates a poor prognosis and promotes cell proliferation and tumorigenesis in bladder cancer via AKT signaling pathway. Neoplasma 2020, 67, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, A.B.; Stern, S.A.; Kohtz, A.S.; Descalzi, G.; Alberini, C.M. Insulin-Like Growth Factor II Targets the mTOR Pathway to Reverse Autism-Like Phenotypes in Mice. J. Neurosci. 2018, 38, 1015–1029. [Google Scholar] [CrossRef]
- Wang, L.; Ye, X.; Zhao, T. The physiological roles of autophagy in the mammalian life cycle. Biol. Rev. Camb. Philos. Soc. 2019, 94, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef]
- Lorzadeh, S.; Kohan, L.; Ghavami, S.; Azarpira, N. Autophagy and the WNT signaling pathway: A focus on WNT/β-catenin signaling. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118926. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. WNT/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Vellai, T.; Bicsák, B.; Tóth, M.L.; Takács-Vellai, K.; Kovács, A.L. Regulation of cell growth by autophagy. Autophagy 2008, 4, 507–509. [Google Scholar] [CrossRef]
- Rousseau, A.; Bertolotti, A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 697–712. [Google Scholar] [CrossRef]
- Rovina, D.; La Vecchia, M.; Cortesi, A.; Fontana, L.; Pesant, M.; Maitz, S.; Tabano, S.; Bodega, B.; Miozzo, M.; Sirchia, S.M. Profound alterations of the chromatin architecture at chromosome 11p15.5 in cells from Beckwith-Wiedemann and Silver-Russell syndromes patients. Sci. Rep. 2020, 10, 8275. [Google Scholar] [CrossRef]
- Calvello, M.; Tabano, S.; Colapietro, P.; Maitz, S.; Pansa, A.; Augello, C.; Lalatta, F.; Gentilin, B.; Spreafico, F.; Calzari, L.; et al. Quantitative DNA methylation analysis improves epigenotype-phenotype correlations in Beckwith-Wiedemann syndrome. Epigenetics 2013, 8, 1053–1060. [Google Scholar] [CrossRef]
- Takeda, T.; Komatsu, M.; Chiwaki, F.; Komatsuzaki, R.; Nakamura, K.; Tsuji, K.; Kobayashi, Y.; Tominaga, E.; Ono, M.; Banno, K.; et al. Upregulation of IGF2R evades lysosomal dysfunction-induced apoptosis of cervical cancer cells via transport of cathepsins. Cell Death Dis. 2019, 10, 876. [Google Scholar] [CrossRef]
- Cheng, X.T.; Xie, Y.X.; Zhou, B.; Huang, N.; Farfel-Becker, T.; Sheng, Z.H. Revisiting LAMP1 as a marker for degradative autophagy-lysosomal organelles in the nervous system. Autophagy 2018, 14, 1472–1474. [Google Scholar] [CrossRef]
- Levra, S.; Rosani, U.; Gnemmi, I.; Brun, P.; Leonardi, A.; Carriero, V.; Bertolini, F.; Balbi, B.; Profita, M.; Ricciardolo, F.L.M.; et al. Impaired autophagy in the lower airways and lung parenchyma in sTable COPD. ERJ Open Res. 2023, 9, 00423–02023. [Google Scholar] [CrossRef]
- Russo, S.; Calzari, L.; Mussa, A.; Mainini, E.; Cassina, M.; Di Candia, S.; Clementi, M.; Guzzetti, S.; Tabano, S.; Miozzo, M.; et al. A multi-method approach to the molecular diagnosis of overt and borderline 11p15.5 defects underlying Silver-Russell and Beckwith-Wiedemann syndromes. Clin. Epigenetics 2016, 8, 23, Erratum in Clin. Epigenetics 2016, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Quadri, R.; Galli, M.; Galati, E.; Rotondo, G.; Gallo, G.R.; Panigada, D.; Plevani, P.; Muzi-Falconi, M. Haspin regulates Ras localization to promote Cdc24-driven mitotic depolarization. Cell Discov. 2020, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Thaunat, O.; Granja, A.G.; Barral, P.; Filby, A.; Montaner, B.; Collinson, L.; Martinez-Martin, N.; Harwood, N.E.; Bruckbauer, A.; Batista, F.D. Asymmetric segregation of polarized antigen on B cell division shapes presentation capacity. Science 2012, 335, 475–479. [Google Scholar] [CrossRef]
- Bohannon, K.P.; Jun, Y.; Gross, S.P.; Smith, G.A. Differential protein partitioning within the herpesvirus tegument and envelope underlies a complex and variable virion architecture. Proc. Natl. Acad. Sci. USA 2013, 110, E1613–E1620. [Google Scholar] [CrossRef] [PubMed]
- Sferruzzi-Perri, A.N.; Sandovici, I.; Constancia, M.; Fowden, A.L. Placental phenotype and the insulin-like growth factors: Resource allocation to fetal growth. J. Physiol. 2017, 595, 5057–5093. [Google Scholar] [CrossRef]
- Bernardelli, C.; Chiaramonte, E.; Ancona, S.; Sirchia, S.M.; Cerri, A.; Lesma, E. Primary TSC2−/meth Cells Induce Follicular Neogenesis in an Innovative TSC Mouse Model. Int. J. Mol. Sci. 2022, 23, 9713. [Google Scholar] [CrossRef]
- Bhat, S.A.; Henry, R.J.; Blanchard, A.C.; Stoica, B.A.; Loane, D.J.; Faden, A.I. Enhanced AKT/GSK-3β/CREB signaling mediates the anti-inflammatory actions of mGluR5 positive allosteric modulators in microglia and following traumatic brain injury in male mice. J. Neurochem. 2021, 156, 225–248. [Google Scholar] [CrossRef]
- Groenewald, W.; Lund, A.H.; Gay, D.M. The Role of WNT Pathway Mutations in Cancer Development and an Overview of Therapeutic Options. Cells 2023, 12, 990. [Google Scholar] [CrossRef] [PubMed]
- Pileggi, S.; La Vecchia, M.; Colombo, E.A.; Fontana, L.; Colapietro, P.; Rovina, D.; Morotti, A.; Tabano, S.; Porta, G.; Alcalay, M.; et al. Cohesin Mutations Induce Chromatin Conformation Perturbation of the H19/IGF2 Imprinted Region and Gene Expression Dysregulation in Cornelia de Lange Syndrome Cell Lines. Biomolecules 2021, 11, 1622. [Google Scholar] [CrossRef]
- Sobel Naveh, N.S.; Traxler, E.M.; Duffy, K.A.; Kalish, J.M. Molecular networks of hepatoblastoma predisposition and oncogenesis in Beckwith-Wiedemann syndrome. Hepatol. Commun. 2022, 6, 2132–2146. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.L.; Hassan, A.B. Cell survival and proliferation are modified by insulin-like growth factor 2 between days 9 and 10 of mouse gestation. Development 2001, 128, 3819–3830. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Chen, B.; Zhu, Z.; Du, C.; Gao, S.; Zhao, G.; Zhao, P.; Wang, Y.; Wang, A.; Schwartz, Z.; et al. Long noncoding RNA (lncRNA) H19: An essential developmental regulator with expanding roles in cancer, stem cell differentiation, and metabolic diseases. Genes Dis. 2023, 10, 1351–1366. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Song, S.; Gusscott, S.; Fu, Z.; VanderKolk, I.; Busscher, B.M.; Lau, K.H.; Brind’Amour, J.; Szabó, P.E. Establishment of paternal methylation imprint at the H19/Igf2 imprinting control region. Sci. Adv. 2023, 9, eadi2050. [Google Scholar] [CrossRef] [PubMed]
- Masunaga, Y.; Inoue, T.; Yamoto, K.; Fujisawa, Y.; Sato, Y.; Kawashima-Sonoyama, Y.; Morisada, N.; Iijima, K.; Ohata, Y.; Namba, N.; et al. IGF2 Mutations. J. Clin. Endocrinol. Metab. 2020, 105, dgz034. [Google Scholar] [CrossRef] [PubMed]
- Sandovici, I.; Georgopoulou, A.; Pérez-García, V.; Hufnagel, A.; López-Tello, J.; Lam, B.Y.H.; Schiefer, S.N.; Gaudreau, C.; Santos, F.; Hoelle, K.; et al. The imprinted Igf2-Igf2r axis is critical for matching placental microvasculature expansion to fetal growth. Dev. Cell 2022, 57, 63–79.e8. [Google Scholar] [CrossRef] [PubMed]
- Galal, M.A.; Alouch, S.S.; Alsultan, B.S.; Dahman, H.; Alyabis, N.A.; Alammar, S.A.; Aljada, A. Insulin Receptor Isoforms and Insulin Growth Factor-like Receptors: Implications in Cell Signaling, Carcinogenesis, and Chemoresistance. Int. J. Mol. Sci. 2023, 24, 15006. [Google Scholar] [CrossRef] [PubMed]
- Cruz, E.; Descalzi, G.; Steinmetz, A.; Scharfman, H.E.; Katzman, A.; Alberini, C.M. CIM6P/IGF-2 Receptor Ligands Reverse Deficits in Angelman Syndrome Model Mice. Autism Res. 2021, 14, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.W.; Pandey, K.; Katzman, A.C.; Alberini, C.M. A role for CIM6P/IGF2 receptor in memory consolidation and enhancement. eLife 2020, 9, e54781. [Google Scholar] [CrossRef]
- Steinmetz, A.B.; Johnson, S.A.; Iannitelli, D.E.; Pollonini, G.; Alberini, C.M. Insulin-like growth factor 2 rescues aging-related memory loss in rats. Neurobiol. Aging 2016, 44, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Lucas, M.; Viana da Silva, S.; Di Scala, M.; Garcia-Barroso, C.; González-Aseguinolaza, G.; Mulle, C.; Alberini, C.M.; Cuadrado-Tejedor, M.; Garcia-Osta, A. Insulin-like growth factor 2 reverses memory and synaptic deficits in APP transgenic mice. EMBO Mol. Med. 2014, 6, 1246–1262. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.; Yu, X.W.; Steinmetz, A.; Alberini, C.M. Autophagy coupled to translation is required for long-term memory. Autophagy 2021, 17, 1614–1635. [Google Scholar] [CrossRef]
- Sélénou, C.; Brioude, F.; Giabicani, E.; Sobrier, M.L.; Netchine, I. IGF2: Development, Genetic and Epigenetic Abnormalities. Cells 2022, 11, 1886. [Google Scholar] [CrossRef]
- Scalia, P.; Williams, S.J.; Fujita-Yamaguchi, Y. Human IGF2 Gene Epigenetic and Transcriptional Regulation: At the Core of Developmental Growth and Tumorigenic Behavior. Biomedicines 2023, 11, 1655. [Google Scholar] [CrossRef]
- Azzi, S.; Abi Habib, W.; Netchine, I. Beckwith-Wiedemann and Russell-Silver Syndromes: From new molecular insights to the comprehension of imprinting regulation. Curr. Opin. Endocrinol. Diabetes Obes. 2014, 21, 30–38. [Google Scholar] [CrossRef]
- Pandey, K.; Bessières, B.; Sheng, S.L.; Taranda, J.; Osten, P.; Sandovici, I.; Constancia, M.; Alberini, C.M. Neuronal activity drives IGF2 expression from pericytes to form long-term memory. Neuron 2023, 111, 3819–3836.e8. [Google Scholar] [CrossRef] [PubMed]
- Alberini, C.M. IGF2 in memory, neurodevelopmental disorders, and neurodegenerative diseases. Trends Neurosci. 2023, 46, 488–502. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, J. Insulin-like growth factor-II and bioactive proteins containing a part of the E-domain of pro-insulin-like growth factor-II. Biofactors 2020, 46, 563–578. [Google Scholar] [CrossRef]
- Atkins, R.J.; Dimou, J.; Paradiso, L.; Morokoff, A.P.; Kaye, A.H.; Drummond, K.J.; Hovens, C.M. Regulation of glycogen synthase kinase-3 beta (GSK-3beta) by the AKT pathway in gliomas. J. Clin. Neurosci. 2012, 19, 1558–1563. [Google Scholar] [CrossRef]
- Duda, P.; Akula, S.M.; Abrams, S.L.; Steelman, L.S.; Martelli, A.M.; Cocco, L.; Ratti, S.; Candido, S.; Libra, M.; Montalto, G.; et al. Targeting GSK3 and Associated Signaling Pathways Involved in Cancer. Cells 2020, 9, 1110. [Google Scholar] [CrossRef]
- Wen, A.Y.; Sakamoto, K.M.; Miller, L.S. The role of the transcription factor CREB in immune function. J. Immunol. 2010, 185, 6413–6419. [Google Scholar] [CrossRef]
- Romanelli, R.J.; LeBeau, A.P.; Fulmer, C.G.; Lazzarino, D.A.; Hochberg, A.; Wood, T.L. Insulin-like growth factor type-I receptor internalization and recycling mediate the sustained phosphorylation of Akt. J. Biol. Chem. 2007, 282, 22513–22524. [Google Scholar] [CrossRef]
- Park, K.-S.; Mitra, A.; Rahat, B.; Kim, K.; Pfeifer, K. Loss of imprinting mutations define both distinct and overlapping roles for misexpression of IGF2 and of H19 lncRNA. Nucleic Acids Res. 2017, 45, 12766–12779. [Google Scholar] [CrossRef]
- Marchand, B.; Arsenault, D.; Raymond-Fleury, A.; FM Boisvert, M.J. Boucher, Glycogen synthase kinase-3 (GSK3) inhibition induces prosurvival autophagic signals in human pancreatic cancer cells. J. Biol. Chem. 2015, 290, 5592–5605. [Google Scholar] [CrossRef] [PubMed]
- Veldman-Jones, M.H.; Brant, R.; Rooney, C.; Geh, C.; Emery, H.; Harbron, C.G.; Wappett, M.; Sharpe, A.; Dymond, M.; Barrett, J.C.; et al. Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res. 2015, 75, 2587–2593. [Google Scholar] [CrossRef] [PubMed]
- Gentien, D.; Piqueret-Stephan, L.; Henry, E.; Albaud, B.; Rapinat, A.; Koscielny, S.; Scoazec, J.Y.; Vielh, P. Digital Multiplexed Gene Expression Analysis of mRNA and miRNA from Routinely Processed and Stained Cytological Smears: A Proof-of-Principle Study. Acta Cytol. 2021, 65, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Bernardelli, C.; Ancona, S.; Lazzari, M.; Lettieri, A.; Selvaggio, P.; Massa, V.; Gervasini, C.; Di Marco, F.; Chiaramonte, R.; Lesma, E. LAM Cells as Potential Drivers of Senescence in Lymphangioleiomyomatosis Microenvironment. Int. J. Mol. Sci. 2022, 23, 7040. [Google Scholar] [CrossRef] [PubMed]
| LCLs Characterization | Study Design | ||||||
|---|---|---|---|---|---|---|---|
| Cell Line | LCLs Methylation Level | LCLs Methylation Status | SNP Array | nCounter Analysis | Immunofluorescence | Western Blot | |
| IC1 | IC2 | ||||||
| CTRL1 | 42% | 43% | NM | NA | + | + | + |
| CTRL2 | 46% | 44% | NM | NA | + | + | + |
| CTRL3 | 40% | 40% | NM | NA | + | + | − |
| CTRL4 | 41% | 42% | NM | NA | + | + | − |
| BWS-IC1 | 78% | 42% | IC1 GOM | NA | + | + | + |
| BWS-IC2 | 44% | 16% | IC2 LOM | NA | + | + | + |
| BWS-UPD | 60% | 27% | IC1 LOM/ IC2 GOM | 30% UPD cells | + | + | + |
| SRS | 28% | 50% | IC1 GOM | NA | − | − | + |
| Gene | Accession | Log2 Fold Change | p-Value | BH p-Value |
|---|---|---|---|---|
| IGF2R * | NM_000876.1:2605 | 0.487 | 0.0454 | 0.233 |
| PEG10 | NM_001040152.1:5000 | −1.18 | 0.115 | 0.321 |
| MEST | NM_177525.1:645 | −2.58 | 0.128 | 0.339 |
| IGF1R | NM_000875.4:4580 | 1.28 | 0.179 | 0.364 |
| GNAS-AS1 | NR_002785.2:1026 | −0.482 | 0.227 | 0.415 |
| GNAS | NM_080425.1:1910 | 0.127 | 0.294 | 0.502 |
| PLAGL1 | NM_006718.3:1872 | 0.226 | 0.435 | 0.652 |
| INSR | NM_000208.2:525 | −0.447 | 0.489 | 0.7 |
| KCNQ1OT1 | NR_002728.2:31875 | −0.146 | 0.659 | 0.845 |
| IGF1 | NM_000618.3:491 | −0.279 | 0.816 | 0.903 |
| FAM50B | NM_012135.1:1272 | 0.0365 | 0.836 | 0.915 |
| Gene | Accession | Log2 Fold Change | p-Value | BH p-Value | Pathway Annotation |
|---|---|---|---|---|---|
| TP53 | NM_000546.2:1330 | 0.632 | 2.81 × 10−5 | 0.00446 | KEGG WNT Annotation |
| PLAUR | NM_001005376.1:440 | −2.16 | 0.000288 | 0.0229 | Proteolysis |
| CUL1 | NM_003592.2:1487 | 0.196 | 0.000798 | 0.0377 | KEGG WNT Annotation |
| PRKCB | NM_212535.1:1750 | −1.23 | 0.00102 | 0.0377 | KEGG WNT Annotation |
| FRAT1 | NM_005479.3:1100 | 1.83 | 0.00146 | 0.0377 | Canonical WNT Pathway, KEGG WNT Annotation |
| AXIN1 | NM_181050.1:135 | 0.265 | 0.00149 | 0.0377 | Canonical WNT Pathway, KEGG WNT Annotation, WNT Signaling Negative Regulation |
| MAPK9 | NM_139068.2:365 | 0.51 | 0.00166 | 0.0377 | KEGG WNT Annotation |
| CEBPD | NM_005195.3:939 | 1.58 | 0.0027 | 0.0481 | Transcription Factors |
| DKK4 | NM_014420.2:640 | −1.2 | 0.00272 | 0.0481 | KEGG WNT Annotation |
| MAPK10 | NM_002753.2:2080 | −6.57 | 0.00526 | 0.079 | KEGG WNT Annotation |
| CREBBP | NM_001079846.1:4818 | 0.547 | 0.00573 | 0.079 | KEGG WNT Annotation |
| PITX2 | NM_000325.5:1381 | 3.94 | 0.00649 | 0.079 | Transcription Factors, WNT Signaling Target Genes |
| RBX1 | NM_014248.2:162 | −0.521 | 0.00682 | 0.079 | KEGG WNT Annotation |
| MMP9 | NM_004994.2:1530 | 2.09 | 0.00722 | 0.079 | Calcium Binding and Signaling, Development and Differentiation, Migration, Proteolysis |
| FBXW11 | NM_033645.2:3545 | 0.313 | 0.00745 | 0.079 | KEGG WNT Annotation, WNT Signaling Negative Regulation |
| WNT10A | NM_025216.2:2255 | −2.25 | 0.00837 | 0.0832 | Calcium Binding and Signaling, Canonical WNT Pathway, KEGG WNT Annotation |
| GSK3A | NM_019884.2:480 | 0.167 | 0.0168 | 0.157 | Canonical WNT Pathway |
| GDNF | NM_000514.2:580 | −4.12 | 0.0222 | 0.185 | Development and Differentiation, Migration |
| SMAD2 | NM_005901.5:1678 | 0.278 | 0.0268 | 0.206 | EMTMetastasis, KEGG WNT Annotation |
| SMAD4 | NM_005359.3:1370 | 0.374 | 0.0272 | 0.206 | KEGG WNT Annotation |
| ERBB2 | NM_001005862.1:1255 | −0.679 | 0.0319 | 0.222 | EMTMetastasis |
| PKN1 | NM_213560.1:2153 | 0.357 | 0.0322 | 0.222 | EMTMetastasis |
| PTGS2 | NM_000963.1:495 | −2.07 | 0.0335 | 0.222 | Calcium Binding and Signaling, Cell Cycle |
| CXCL12 | NM_000609.5:210 | −3.36 | 0.0367 | 0.226 | EMTMetastasis |
| BMP4 | NM_001202.3:395 | 0.757 | 0.0372 | 0.226 | Development and Differentiation |
| T | NM_003181.2:1836 | 4.22 | 0.0383 | 0.226 | Development and Differentiation, Transcription Factors |
| BAMBI | NM_012342.2:1010 | 3.2 | 0.0408 | 0.226 | Canonical WNT Pathway |
| CCND1 | NM_053056.2:690 | −2.39 | 0.0425 | 0.226 | Calcium Binding and Signaling, Cell Cycle, Development and Differentiation, KEGG WNT Annotation, WNT Signaling Negative Regulation, WNT Signaling Target Genes |
| FZD2 | NM_001466.2:845 | −2.36 | 0.0426 | 0.226 | Calcium Binding and Signaling, Canonical WNT Pathway, KEGG WNT Annotation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pileggi, S.; Colombo, E.A.; Ancona, S.; Quadri, R.; Bernardelli, C.; Colapietro, P.; Taiana, M.; Fontana, L.; Miozzo, M.; Lesma, E.; et al. Dysfunction in IGF2R Pathway and Associated Perturbations in Autophagy and WNT Processes in Beckwith–Wiedemann Syndrome Cell Lines. Int. J. Mol. Sci. 2024, 25, 3586. https://doi.org/10.3390/ijms25073586
Pileggi S, Colombo EA, Ancona S, Quadri R, Bernardelli C, Colapietro P, Taiana M, Fontana L, Miozzo M, Lesma E, et al. Dysfunction in IGF2R Pathway and Associated Perturbations in Autophagy and WNT Processes in Beckwith–Wiedemann Syndrome Cell Lines. International Journal of Molecular Sciences. 2024; 25(7):3586. https://doi.org/10.3390/ijms25073586
Chicago/Turabian StylePileggi, Silvana, Elisa A. Colombo, Silvia Ancona, Roberto Quadri, Clara Bernardelli, Patrizia Colapietro, Michela Taiana, Laura Fontana, Monica Miozzo, Elena Lesma, and et al. 2024. "Dysfunction in IGF2R Pathway and Associated Perturbations in Autophagy and WNT Processes in Beckwith–Wiedemann Syndrome Cell Lines" International Journal of Molecular Sciences 25, no. 7: 3586. https://doi.org/10.3390/ijms25073586






