Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses
Abstract
:1. Introduction
2. Results
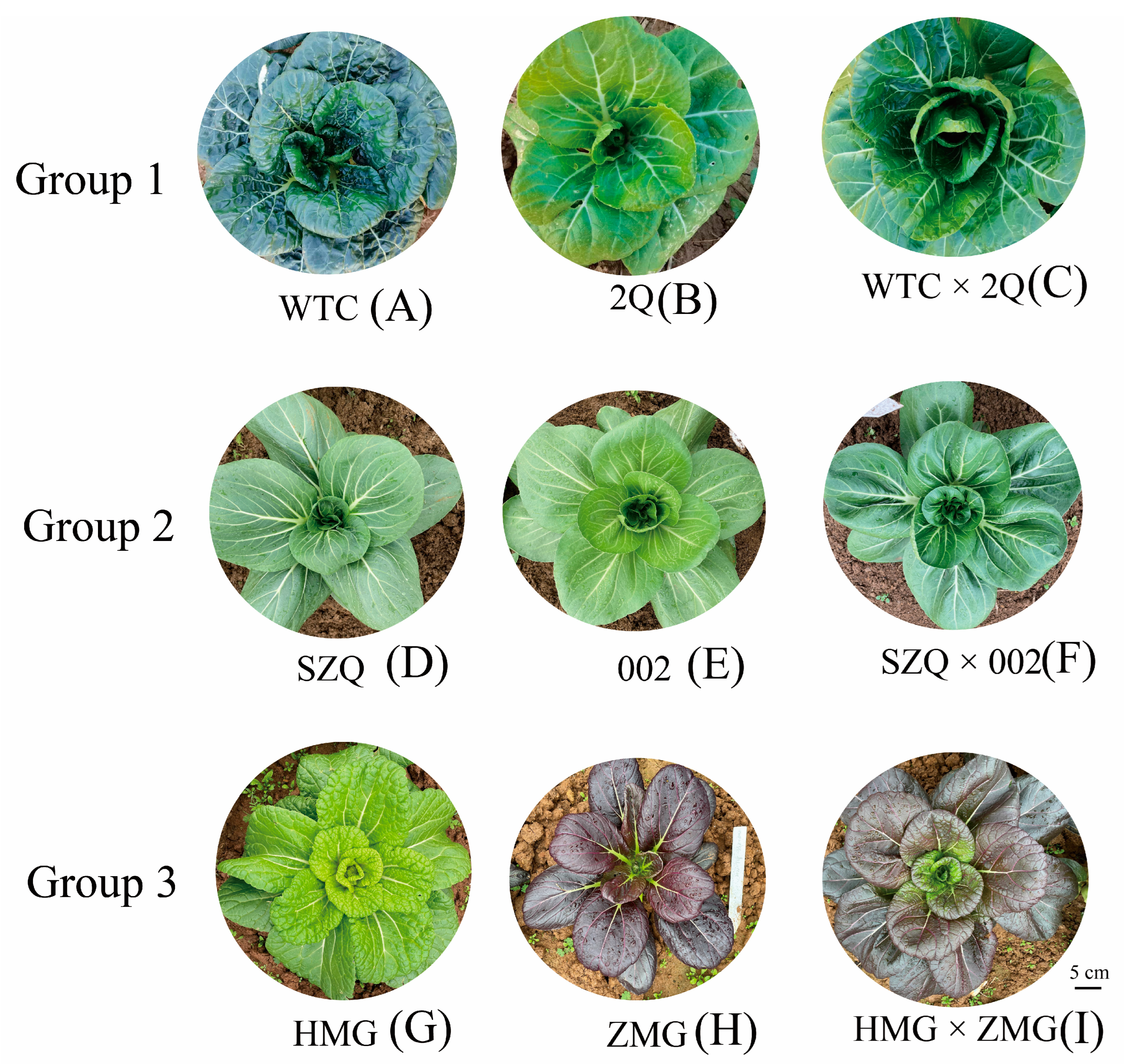
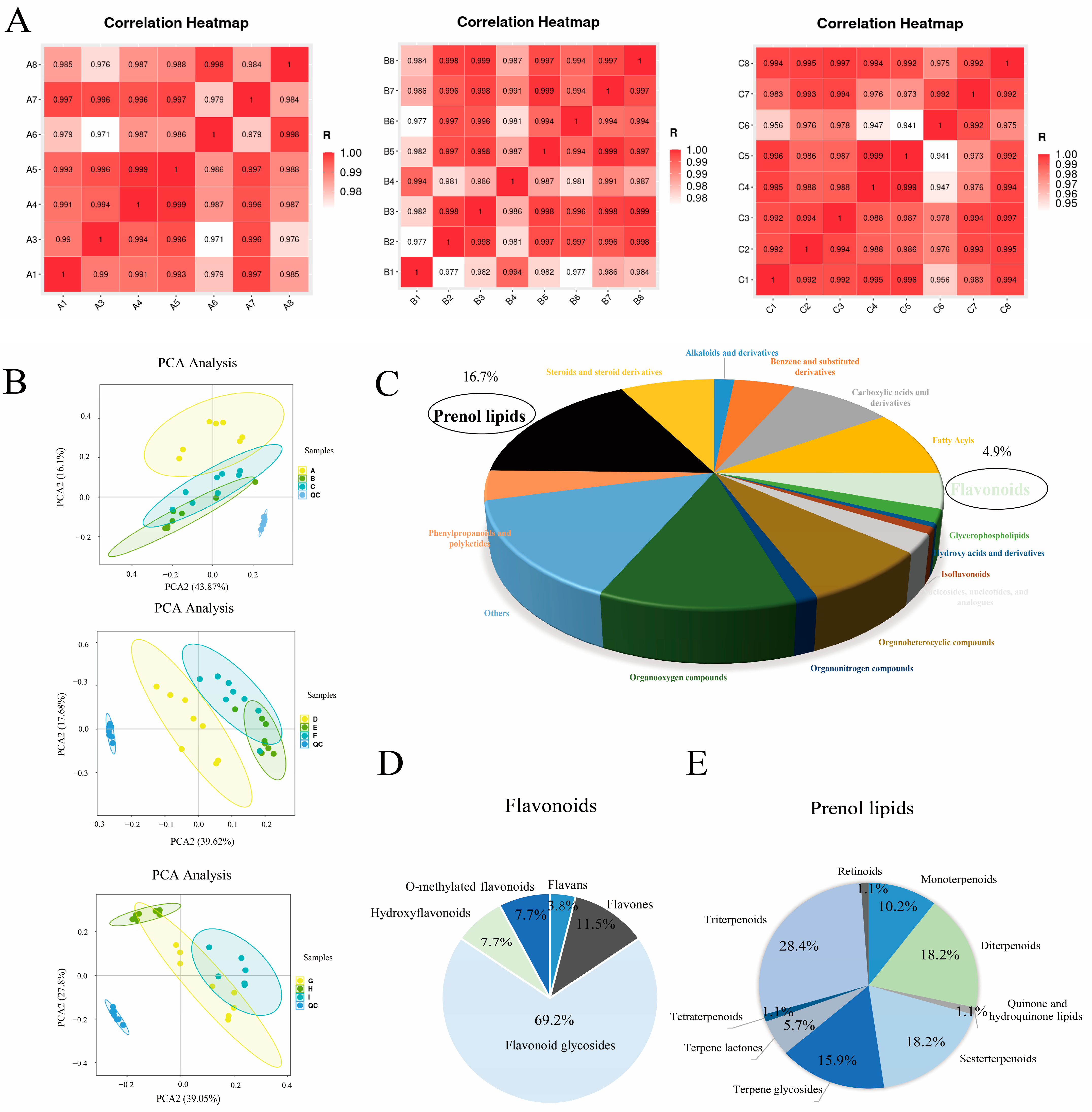
2.1. Analysis of Metabolites in Pak Choi Leaves
2.2. Transcriptome Analysis in Pak Choi Leaves
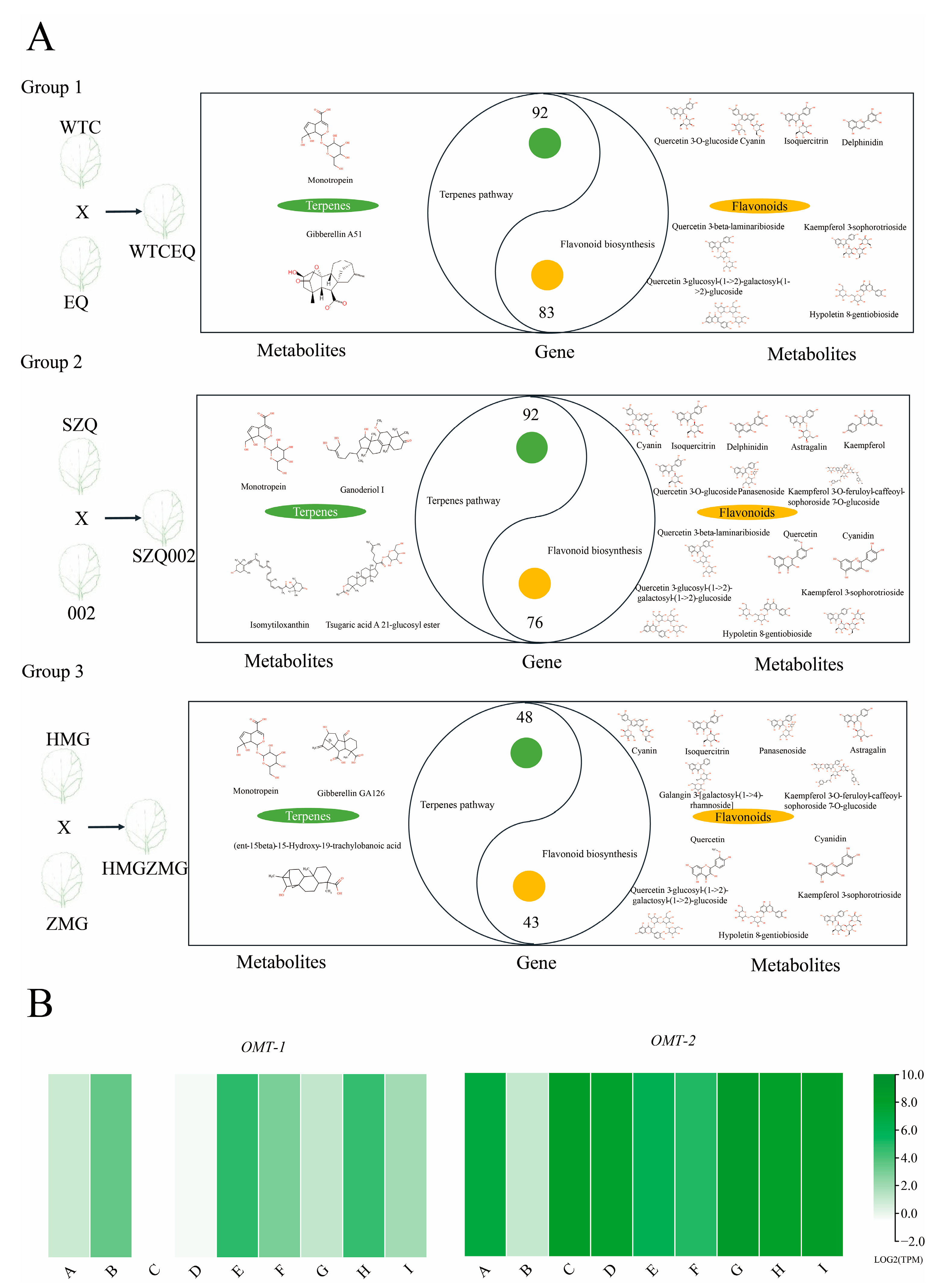
2.3. Association Analysis of Metabolic and Transcriptomic Data
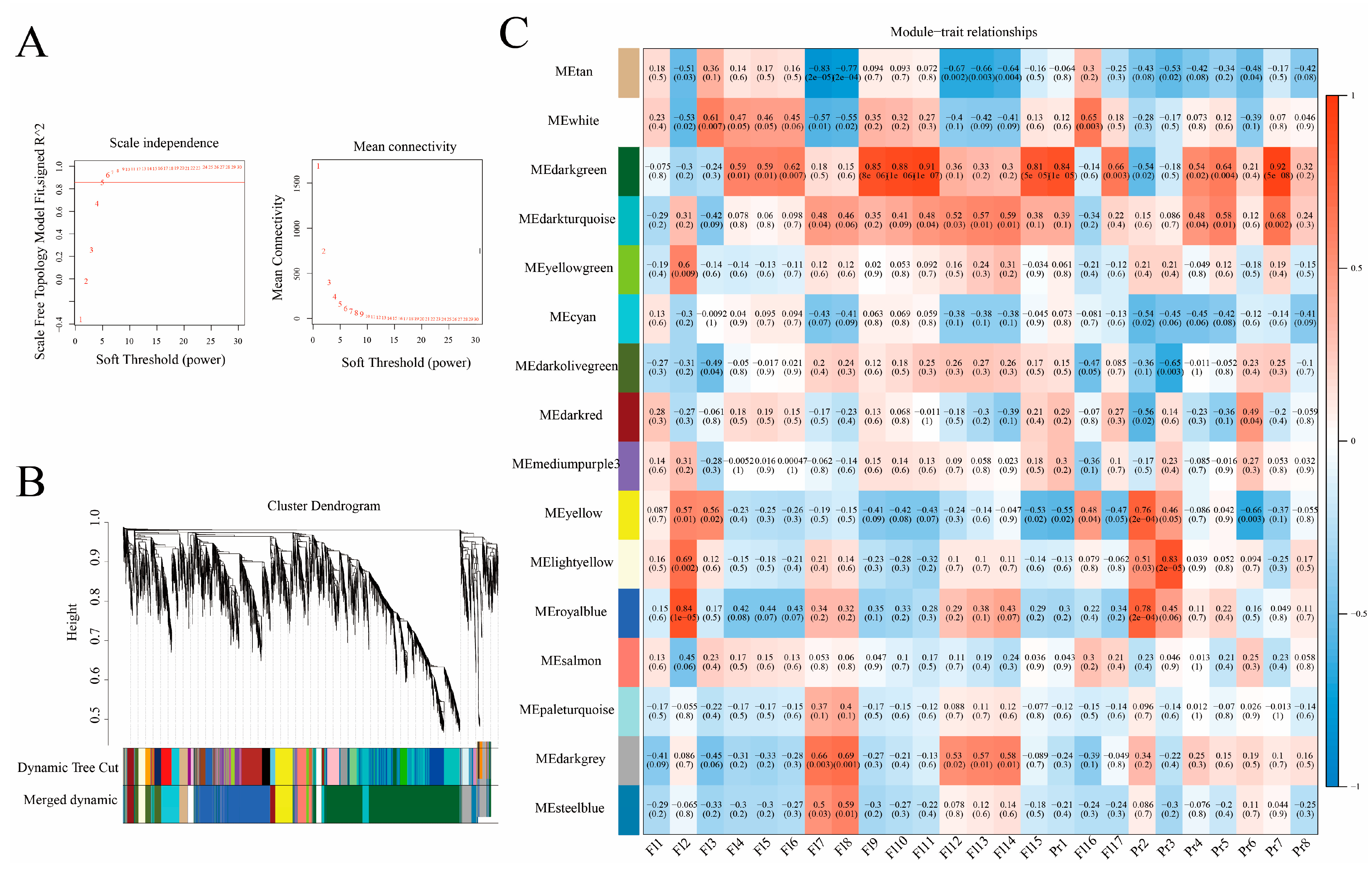
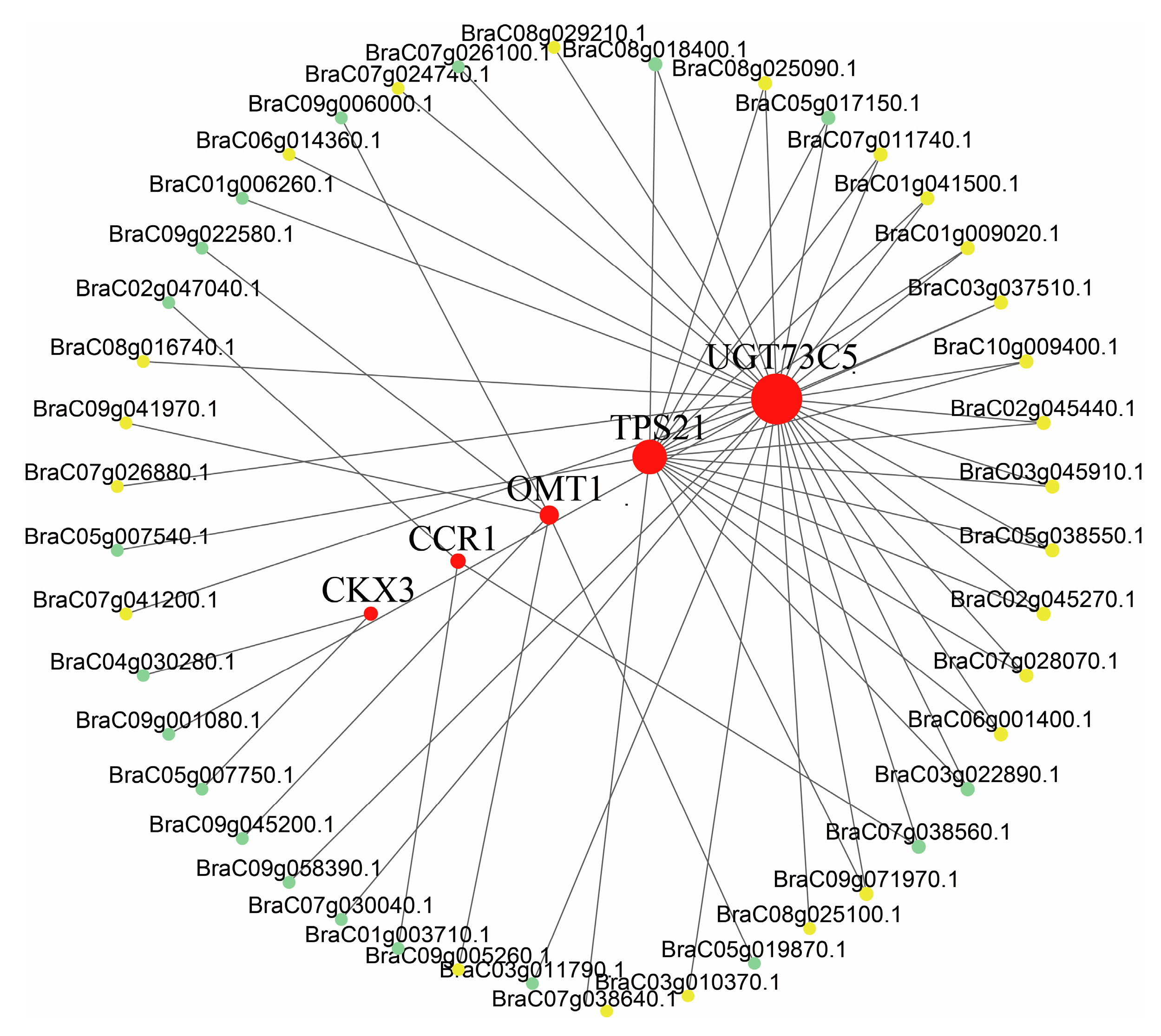
2.4. Co-Expression Analysis
2.5. Flavonoid and Terpenoid Regulation Networks in Pak Choi Leaves
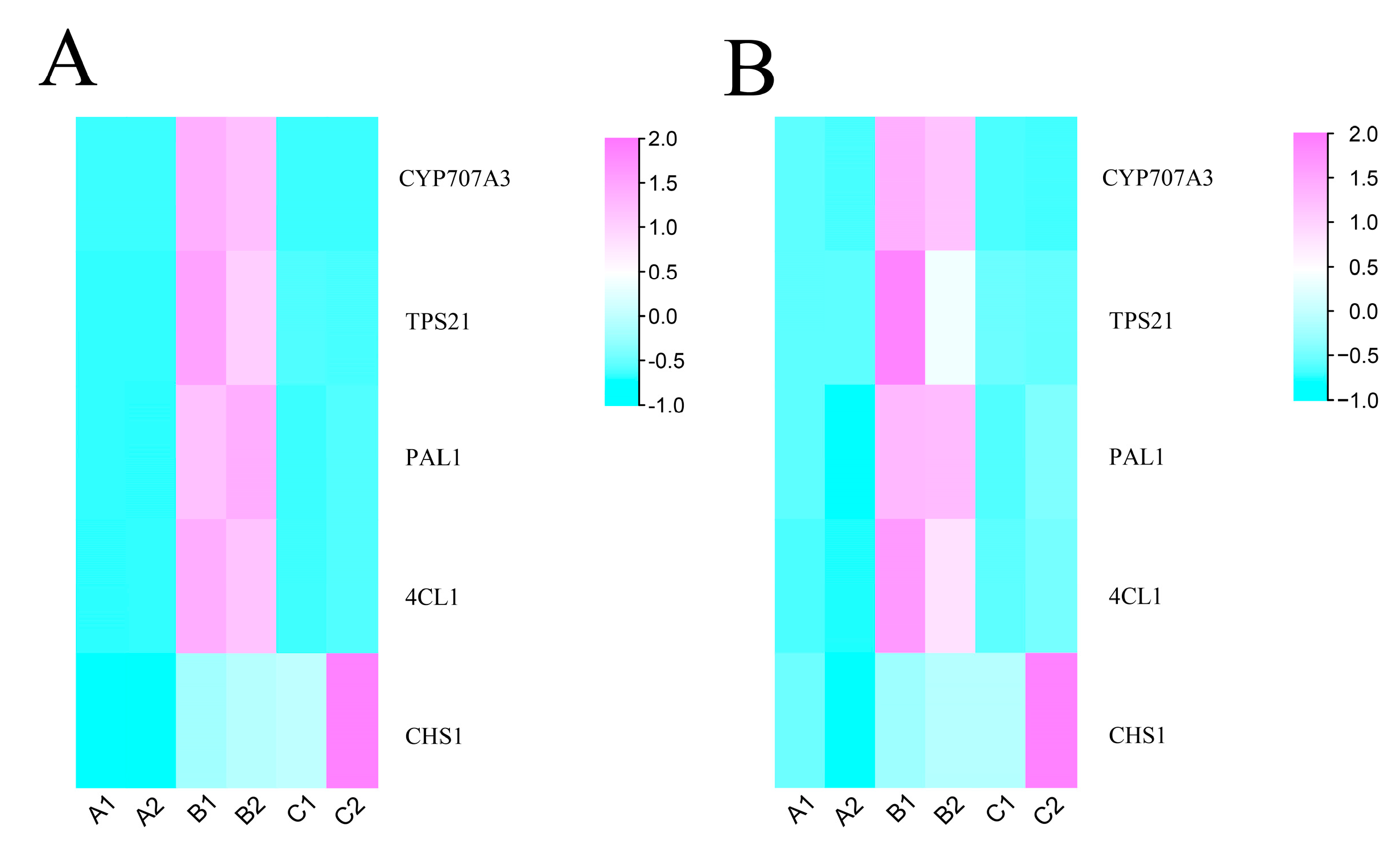
2.6. Validation of the Key DEGs Involved in Flavonoids and Terpenoids with qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Metabolite Extraction and Detection
4.3. Transcriptomic Analysis
4.4. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.5. Quantitative Real-Time PCR (qPCR) Expression Analyses
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.; Liao, X.; Yu, L.; Rao, S.; Chen, Q.; Zhu, Z.; Cong, X.; Zhang, W.; Ye, J.; Cheng, S.; et al. Combined metabolome and transcriptome analysis reveal the mechanism of selenate influence on the growth and quality of cabbage (Brassica oleracea var. capitata L.). Food Res. Int. 2022, 156, 111135. [Google Scholar] [CrossRef]
- Xie, J.; Wang, W.; Yang, T.; Zhang, Q.; Zhang, Z.; Zhang, X.; Li, N.; Zhi, L.; Ma, X.; Zhang, S.; et al. Large-scale genomic and transcriptomic profiles of rice hybrids reveal a core mechanism underlying heterosis. Genome Biol. 2022, 23, 264. [Google Scholar] [CrossRef]
- Thole, V.; Bassard, J.E.; Ramírez-González, R.; Trick, M.; Afshar, B.G.; Breitel, D.; Hill, L.; Foito, A.; Shepherd, L.; Freitag, S.; et al. RNA-seq, de novo transcriptome assembly and flavonoid gene analysis in 13 wild and cultivated berry fruit species with high content of phenolics. BMC Genom. 2019, 20, 995. [Google Scholar] [CrossRef]
- Feng, Z.; Sun, L.; Dong, M.; Fan, S.; Shi, K.; Qu, Y.; Zhu, L.; Shi, J.; Wang, W.; Liu, Y.; et al. Novel players in organogenesis and flavonoid biosynthesis in cucumber glandular trichomes. Plant Physiol. 2023, 192, 2723–2736. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, D.; Ma, F.N.; Yang, L.; Wu, B.; Xing, W.; Sun, P.; Chen, D.; Xu, B.; Song, S. Identification of key genes involved in flavonoid and terpenoid biosynthesis and the pathway of triterpenoid biosynthesis in Passiflora edulis. J. Integr. Agric. 2023, 22, 1412–1423. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, L.; Yang, J.; Hu, H.; Wei, G.; Cui, J.; Xu, J. Transcriptome and metabolome analyses reveal differences in terpenoid and flavonoid biosynthesis in Cryptomeria fortunei needles across different seasons. Front. Plant Sci. 2022, 13, 862746. [Google Scholar] [CrossRef] [PubMed]
- Apel, L.; Kammerer, D.; Stintzing, F.; Spring, O. Comparative metabolite profiling of triterpenoid saponins and flavonoids in flower color mutations of Primula veris L. Int. J. Mol. Sci. 2017, 18, 153. [Google Scholar] [CrossRef]
- Xue, T.; Zheng, X.; Chen, D.; Zhang, T.; Chen, Y.; Zhong, Q.; Chen, B.; Li, B. Metabolome and whole transcriptome analyses reveal the molecular mechanisms underlying terpenoids biosynthesis in Sapindus mukorossi fruits. Ind. Crops Prod. 2022, 181, 114810. [Google Scholar] [CrossRef]
- Khan, W.A.; Hu, H.; Cuin, T.A.; Hao, Y.; Ji, X.; Wang, J.; Hu, C. Untargeted metabolomics and comparative flavonoid analysis reveal the nutritional aspects of pak choi. Food Chem. 2022, 383, 132375. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, Y.; Guo, F.; Wang, G. Proteomic and metabolomic analyses reveal stage- and tissue-specific flavonoid accumulation in Ginkgo biloba. LWT 2022, 171, 114111. [Google Scholar] [CrossRef]
- Rao, S.; Gou, Y.; Yu, T.; Cong, X.; Gui, J.; Zhu, Z.; Zhang, W.; Liao, Y.; Ye, J.; Cheng, S.; et al. Effects of selenate on Se, flavonoid, and glucosinolate in broccoli florets by combined transcriptome and metabolome analyses. Food Res. Int. 2021, 146, 110463. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, H.; Lv, X.; Zheng, C.; Wu, Z.; Wang, N.; Wang, J.; Chen, H.; Wei, F. Profiling and spatial distribution of phenolic compounds in rapeseed by two-step extraction strategy and targeted metabolomics combined with chemometrics. Food Chem. 2023, 401, 134151. [Google Scholar] [CrossRef]
- Wang, P.; Gu, M.; Shao, S.; Chen, X.; Hou, B.; Ye, N.; Zhang, X. Changes in non-volatile and volatile metabolites associated with heterosis in tea plants (Camellia sinensis). J. Agric. Food Chem. 2022, 70, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Byrne, P.F.; McMullen, M.D.; Snook, M.E.; Musket, T.A.; Theuri, J.M.; Widstrom, N.W.; Wiseman, B.R.; Coe, E.H. Quantitative trait loci and metabolic pathways: Genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc. Natl. Acad. Sci. USA 1996, 93, 8820–8825. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Lee, Y.S.; Nguyen, V.B.; Giang, V.N.L.; Koo, H.J.; Park, H.-S.; Mohanan, P.; Song, Y.H.; Ryu, B.; Kang, K.B.; et al. Comparative transcriptome and metabolome analyses of four Panax species explore the dynamics of metabolite biosynthesis. J. Ginseng Res. 2022, 47, 44–53. [Google Scholar] [CrossRef]
- Qu, C.; Zhu, M.; Hu, R.; Niu, Y.; Chen, S.; Zhao, H.; Li, C.; Wang, Z.; Yin, N.; Sun, F.; et al. Comparative genomic analyses reveal the genetic basis of the yellow-seed trait in Brassica napus. Nat. Commun. 2023, 14, 5194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, C.; Chao, H.; Long, Y.; Wu, J.; Li, Z.; Ge, X.; Xia, H.; Yin, Y.; Batley, J.; et al. Integration of metabolome and transcriptome reveals flavonoid accumulation in the intergeneric hybrid between Brassica rapa and Raphanus sativus. Sci. Rep. 2019, 9, 18368. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yao, L.; Pecoraro, L.; Liu, C.; Wang, J.; Huang, L.; Gao, W. Cold stress regulates accumulation of flavonoids and terpenoids in plants by phytohormone, transcription process, functional enzyme, and epigenetics. Crit. Rev. Biotechnol. 2023, 43, 680–697. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Y.; Gao, F.; Jin, W.; Li, S.; Kimani, S.; Yang, S.; Bao, T.; Gao, X.; Wang, L. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 4140–4158. [Google Scholar] [CrossRef]
- Shan, X.; Li, Y.; Yang, S.; Yang, Z.; Qiu, M.; Gao, R.; Han, T.; Meng, X.; Xu, Z.; Wang, L.; et al. The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytol. 2020, 228, 1864–1879. [Google Scholar] [CrossRef]
- Li, P.; Xia, E.; Fu, J.; Xu, Y.; Zhao, X.; Tong, W.; Tang, Q.; Tadege, M.; Fernie, A.R.; Zhao, J. Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants (Camellia sinensis). Plant J. 2022, 110, 1144–1165. [Google Scholar] [CrossRef]
- Singh, V.V.; Balbeer; Sharma, H.K.; Priyamedha; Ram, B.; Meena, H.S.; Rai, P.K. Heterosis and gene action studies for agro-physiological traits in Indian mustard (Brassica juncea L.). Vegetos 2022, 35, 803–809. [Google Scholar] [CrossRef]
- Yi, G.; Shin, H.; Park, H.R.; Park, J.E.; Ahn, J.H.; Lim, S.; Lee, J.G.; Lee, E.J.; Huh, J.H. Revealing biomass heterosis in the allodiploid ×Brassicoraphanus, a hybrid between Brassica rapa and Raphanus sativus, through integrated transcriptome and metabolites analysis. BMC Plant Biol. 2020, 20, 252. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lu, X.; Zhu, Y.; Pan, J.; Zhou, S.; Zhang, X.; Zhu, G.; Shang, Y.; Huang, S.; Zhang, C. The multi-omics basis of potato heterosis. J. Integr. Plant Biol. 2022, 64, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Arceo, M.; Gomez-Lopez, I.; Carr-Ugarte, H.; Eseberri, I.; Gonzalez, M.; Cano, M.P.; Portillo, M.P.; Gomez-Zorita, S. Anti-obesity effects of isorhamnetin and isorhamnetin conjugates. Int. J. Mol. Sci. 2022, 24, 299. [Google Scholar] [CrossRef]
- Wang, B.; Hou, M.; Shi, J.; Ku, L.; Song, W.; Li, C.; Ning, Q.; Li, X.; Li, C.; Zhao, B.; et al. De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nat. Genet. 2023, 55, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yue, M.; Liu, Y.; Zhang, N.; Lin, Y.; Zhang, Y.; Wang, Y.; Li, M.; Luo, Y.; Zhang, Y.; et al. A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × ananassa). Plant Biotechnol. J. 2023, 21, 1140–1158. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, B.; Yao, T.; Shen, C.; Wen, T.; Zhang, R.; Li, Y.; Le, Y.; Li, Z.; Zhang, X.; et al. Re enhances anthocyanin and proanthocyanidin accumulation to produce red foliated cotton and brown fiber. Plant Physiol. 2022, 189, 1466–1481. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zhao, C.; Wang, S.; Gao, N.; Wang, X.; Na, X.; Wang, X.; Bi, Y. PIF4-PAP1 interaction affects MYB-bHLH-WD40 complex formation and anthocyanin accumulation in Arabidopsis. J Plant Physiol. 2022, 268, 153558. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, X.; Gong, Q.; Cao, J.; Shen, W.; Yin, X.; Grierson, D.; Zhang, B.; Xu, C.; Li, X.; et al. Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 2021, 19, 671–688. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Zhou, L.; You, S.; Deng, H.; Chen, Y.; Alseekh, S.; Yuan, Y.; Fu, R.; Zhang, Z.; et al. Microtom metabolic network: Rewiring tomato metabolic regulatory network throughout the growth cycle. Mol. Plant. 2020, 13, 1203–1218. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021, 105, 167–181. [Google Scholar] [CrossRef]
- Mo, R.; Han, G.; Zhu, Z.; Essemine, J.; Dong, Z.; Li, Y.; Deng, W.; Qu, M.; Zhang, C.; Yu, C. The ethylene response factor ERF5 regulates anthocyanin biosynthesis in ‘Zijin’ mulberry fruits by interacting with MYBA and F3H Genes. Int. J. Mol. Sci. 2022, 23, 7615. [Google Scholar] [CrossRef]
- Wei, C.; Li, M.; Cao, X.; Jin, Z.; Zhang, C.; Xu, M.; Chen, K.; Zhang, B. Linalool synthesis related PpTPS1 and PpTPS3 are activated by transcription factor PpERF61 whose expression is associated with DNA methylation during peach fruit ripening. Plant Sci. 2022, 317, 111200. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, M.; Zhang, M.; Yang, M.; Dai, S.; Meng, Q.; Lv, W.; Zhuang, K. ETHYLENE-INSENSITIVE 3-LIKE 2 regulates beta-carotene and ascorbic acid accumulation in tomatoes during ripening. Plant Physiol. 2023, 192, 2067–2080. [Google Scholar] [CrossRef]
- Zhang, Y.; Ji, A.; Xu, Z.; Luo, H.; Song, J. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol. Biol. 2019, 100, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gao, M.; Zhao, Y.; Chen, Y.; Wu, L.; Yin, H.; Xiong, S.; Wang, S.; Wang, J.; Yang, Y.; et al. LcERF19, an AP2/ERF transcription factor from Litsea cubeba, positively regulates geranial and neral biosynthesis. Hortic. Res. 2022, 9, uhac093. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Han, T.; Bai, A.; Xu, H.; Wang, J.; Hou, X.; Li, Y. Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses. Int. J. Mol. Sci. 2024, 25, 3587. https://doi.org/10.3390/ijms25073587
Wang H, Han T, Bai A, Xu H, Wang J, Hou X, Li Y. Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses. International Journal of Molecular Sciences. 2024; 25(7):3587. https://doi.org/10.3390/ijms25073587
Chicago/Turabian StyleWang, Haibin, Tiantian Han, Aimei Bai, Huanhuan Xu, Jianjun Wang, Xilin Hou, and Ying Li. 2024. "Potential Regulatory Networks and Heterosis for Flavonoid and Terpenoid Contents in Pak Choi: Metabolomic and Transcriptome Analyses" International Journal of Molecular Sciences 25, no. 7: 3587. https://doi.org/10.3390/ijms25073587




