Convergent Mutations and Single Nucleotide Variants in Mitochondrial Genomes of Modern Humans and Neanderthals
Abstract
1. Introduction
2. Results
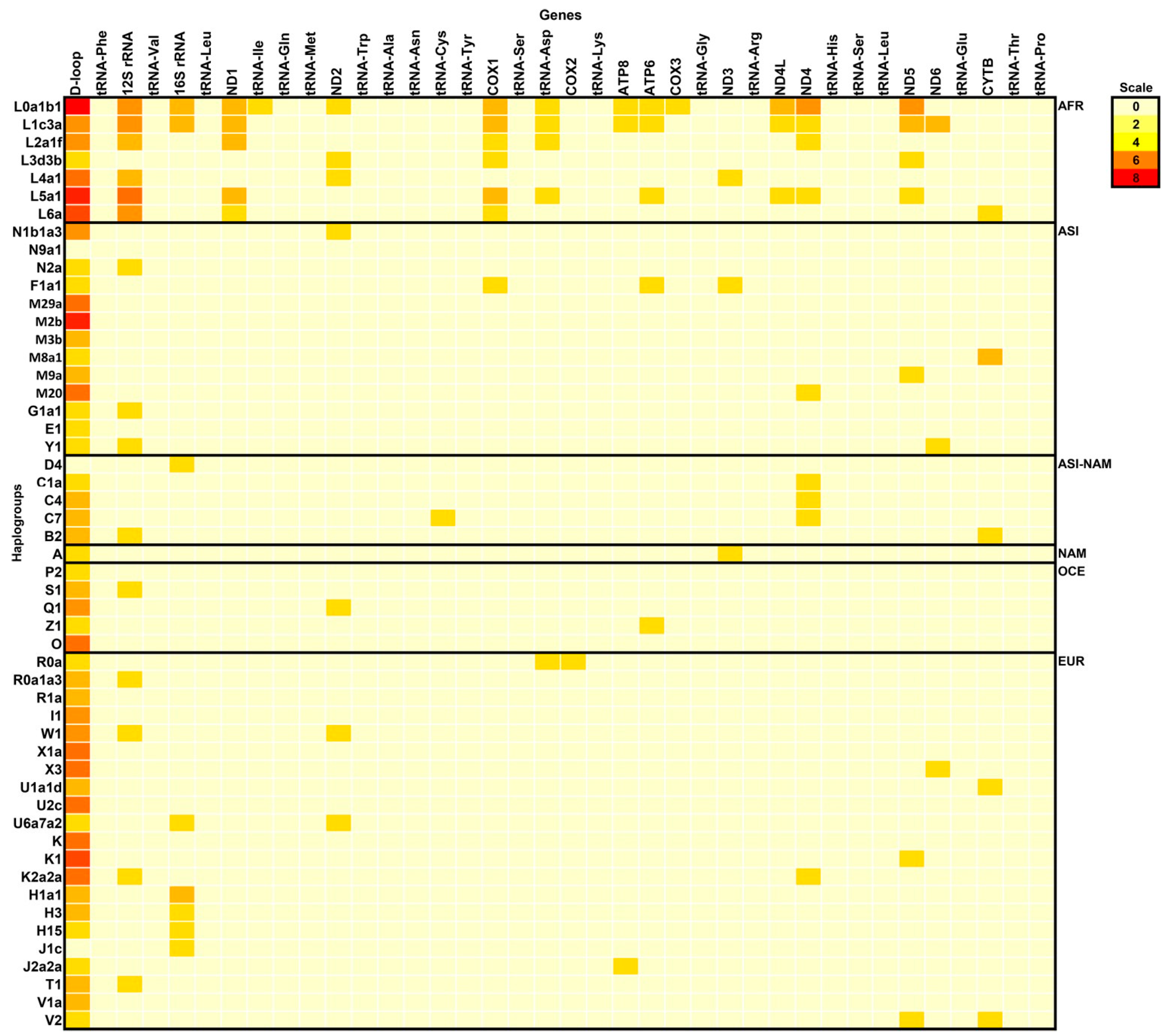
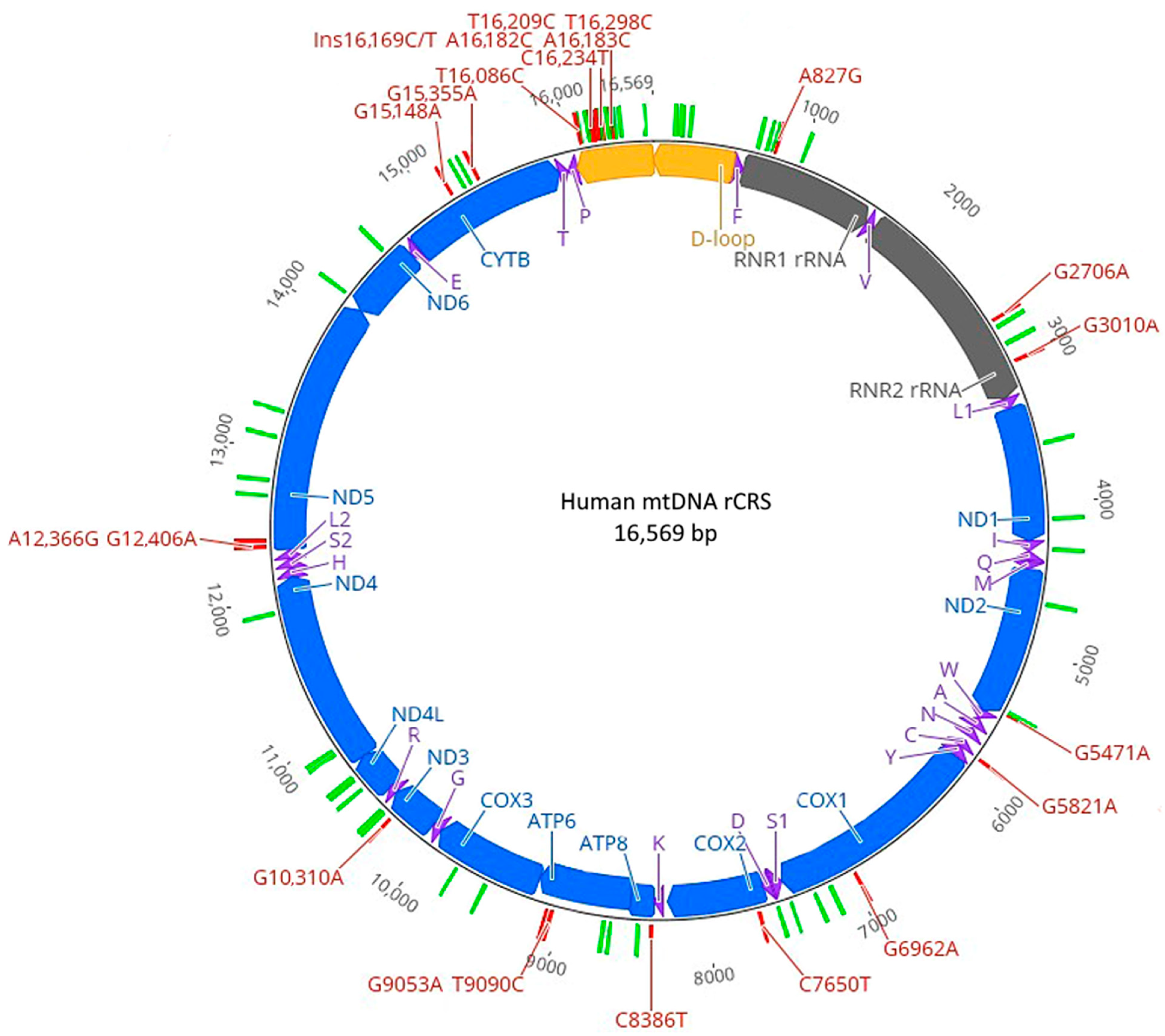
2.1. Distribution of Neanderthal SNVs
2.2. Disease-Associated N-SNVs
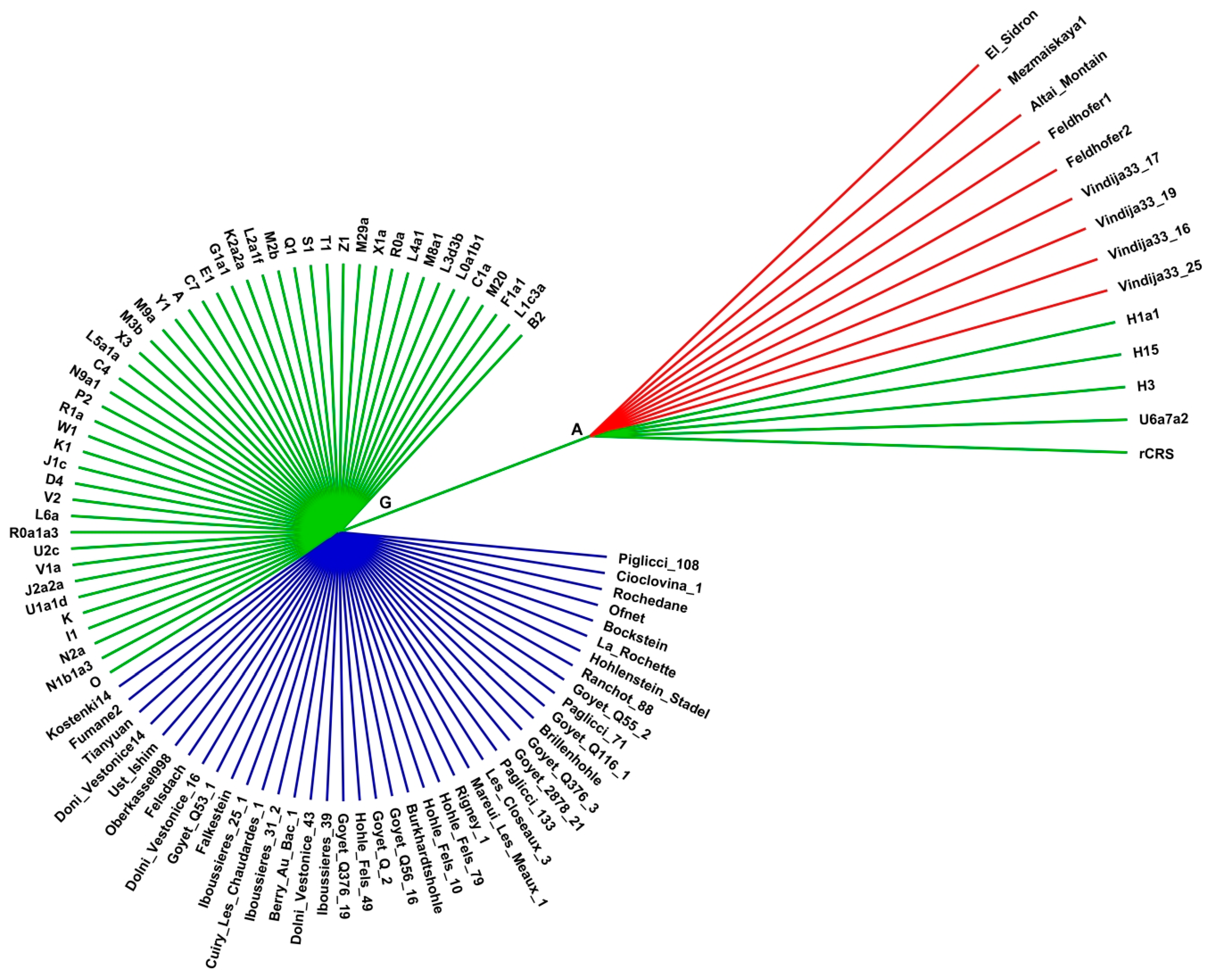
2.3. Haplogroups of Paleogenomes
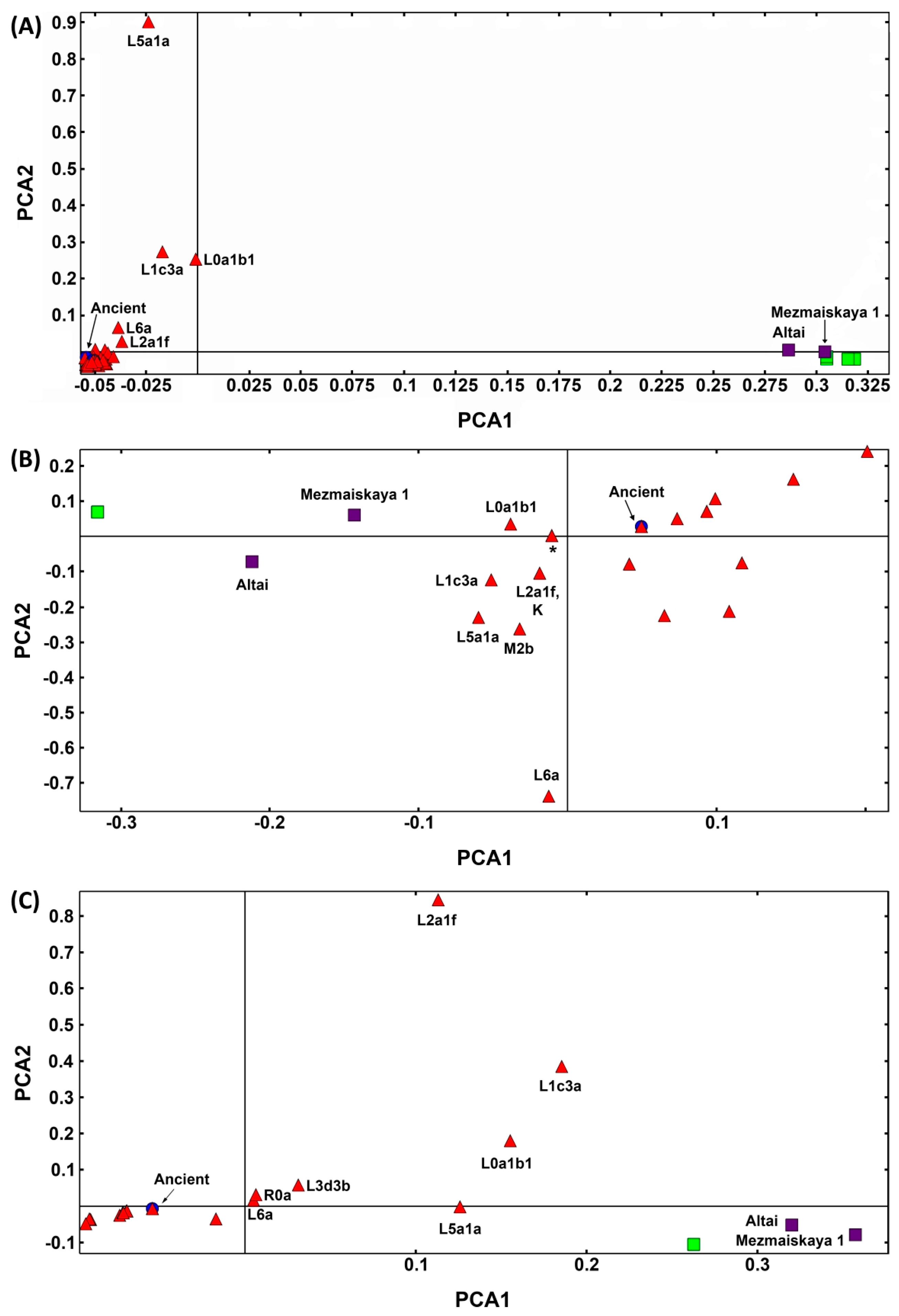
2.4. PCAs of Neanderthal and Human Mitogenomes
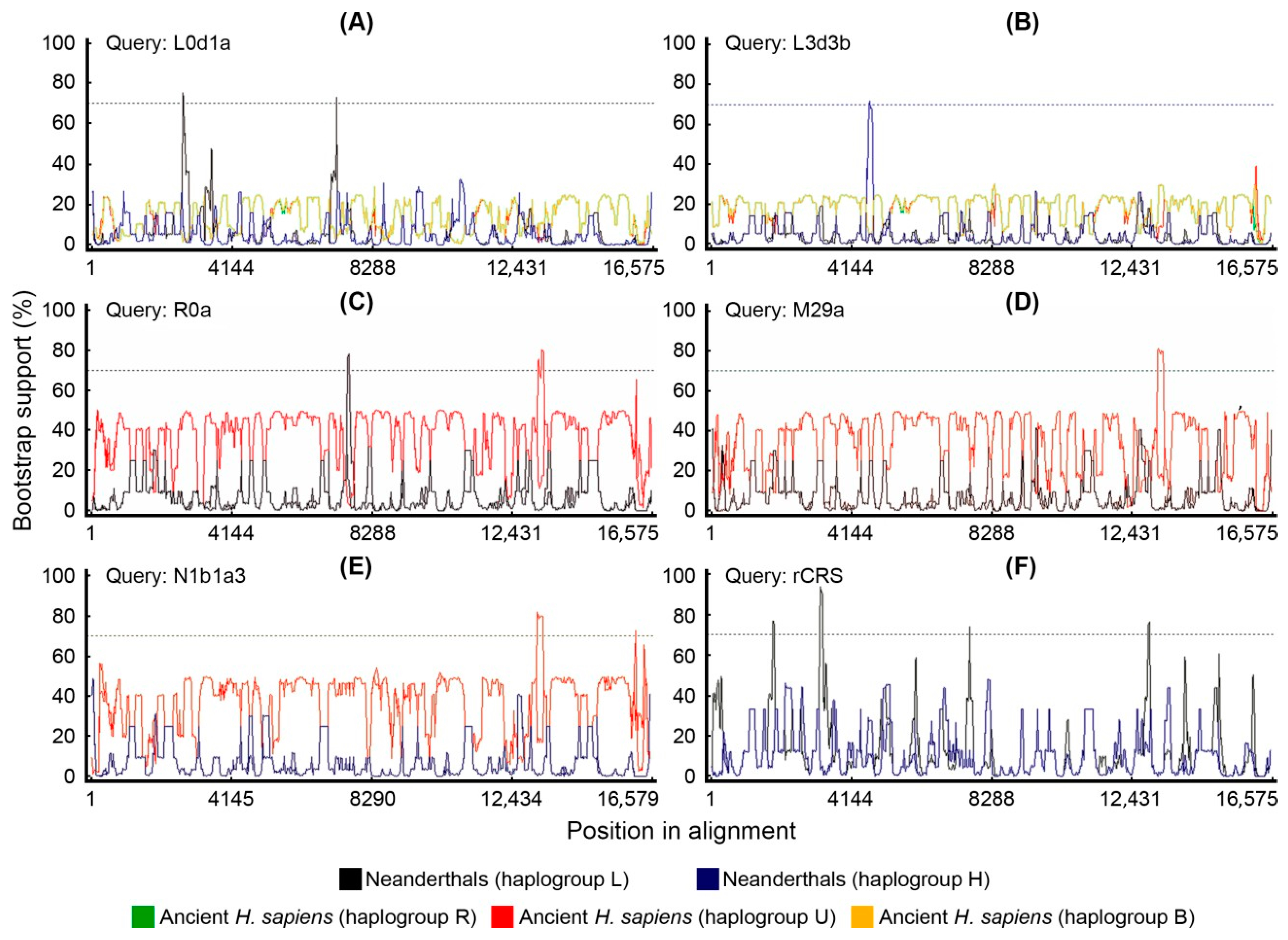
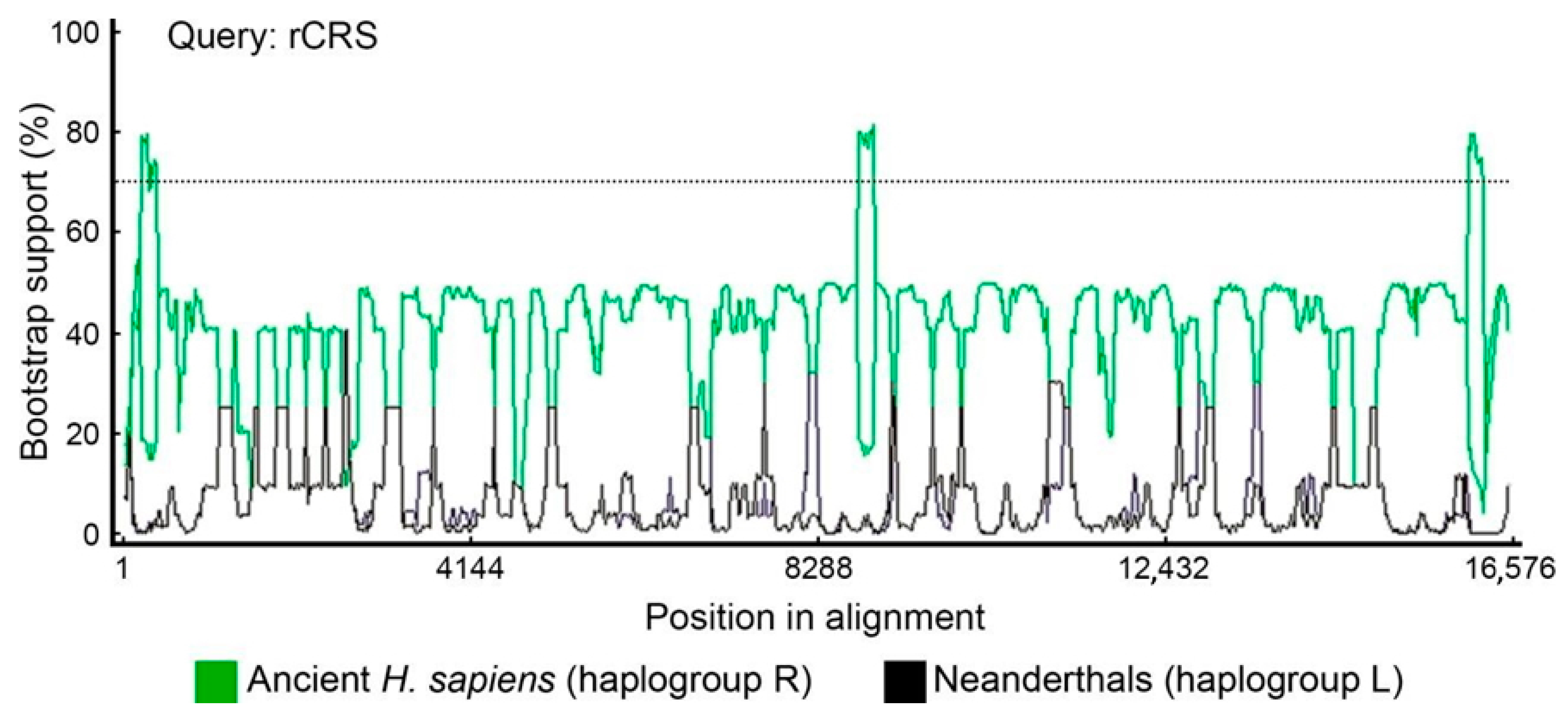
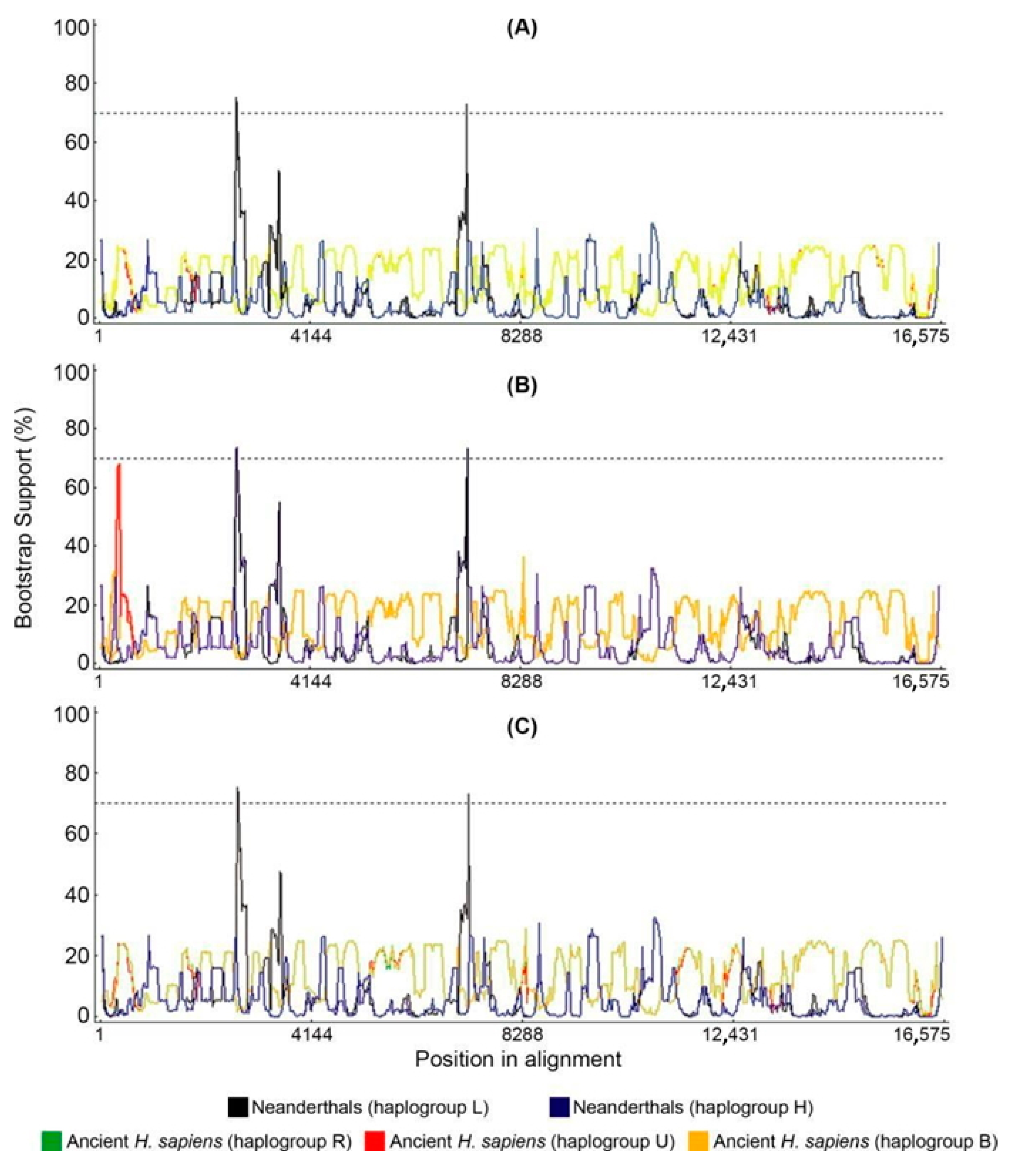
2.5. Bootscan Analysis of Mitogenomes
3. Discussion
4. Material and Methods
4.1. Mitochondrial Genome Sequences
4.2. Sequence Alignments
4.3. Phylogenetic Inference
4.4. Recombination Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M.; Patterson, N.; Li, H.; Zhai, W.; Fritz, M.H.-Y.; et al. A Draft Sequence of the Neandertal Genome. Science 2010, 328, 710–722. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.H. Hominin interbreeding and the evolution of human variation. J. Biol. Res.-Thessalon. 2016, 23, 17. [Google Scholar] [CrossRef] [PubMed]
- PPrüfer, K.; Racimo, F.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S.; Heinze, A.; Renaud, G.; Sudmant, P.H.; De Filippo, C.; et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 2014, 505, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Simonti, C.N.; Vernot, B.; Bastarache, L.; Bottinger, E.; Carrell, D.S.; Chisholm, R.L.; Crosslin, D.R.; Hebbring, S.J.; Jarvik, G.P.; Kullo, I.J.; et al. The phenotypic legacy of admixture between modern humans and Neandertals. Science 2016, 351, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, A. Close Encounters of the Prehistoric Kind. Science 2010, 328, 680–684. [Google Scholar] [CrossRef] [PubMed]
- Higham, T.; Douka, K.; Wood, R.; Ramsey, C.B.; Brock, F.; Basell, L.; Camps, M.; Arrizabalaga, A.; Baena, J.; Barroso-Ruíz, C.; et al. The timing and spatiotemporal patterning of Neanderthal disappearance. Nature 2014, 512, 306–309. [Google Scholar] [CrossRef]
- Serre, D.; Langaney, A.; Chech, M.; Teschler-Nicola, M.; Paunovic, M.; Mennecier, P.; Hofreiter, M.; Possnert, G.; Pääbo, S. No Evidence of Neandertal mtDNA Contribution to Early Modern Humans. PLoS Biol. 2004, 2, e57. [Google Scholar] [CrossRef] [PubMed]
- Kivisild, T. Maternal ancestry and population history from whole mitochondrial genomes. Investig. Genet. 2015, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Hill, G.E.; Johnson, J.D. The mitonuclear compatibility hypothesis of sexual selection. Proc. R. Soc. B. 2013, 280, 20131314. [Google Scholar] [CrossRef]
- Kraytsberg, Y.; Schwartz, M.; Brown, T.A.; Ebralidse, K.; Kunz, W.S.; Clayton, D.A.; Vissing, J.; Khrapko, K. Recombination of Human Mitochondrial DNA. Science 2004, 304, 981. [Google Scholar] [CrossRef]
- Piganeau, G.; Eyre-Walker, A. A reanalysis of the indirect evidence for recombination in human mitochondrial DNA. Heredity 2004, 92, 282–288. [Google Scholar] [CrossRef]
- Muller, H.J. Some Genetic Aspects of Sex. Am. Nat. 1932, 66, 118–138. [Google Scholar] [CrossRef]
- Loewe, L. Quantifying the genomic decay paradox due to Muller’s ratchet in human mitochondrial DNA. Genet. Res. 2006, 87, 133–159. [Google Scholar] [CrossRef]
- Felsenstein, J. The evolutionary advantage of recombination. Genetics 1974, 78, 737–756. [Google Scholar] [CrossRef]
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999, 23, 147. [Google Scholar] [CrossRef]
- Racimo, F.; Marnetto, D.; Huerta-Sánchez, E. Signatures of Archaic Adaptive Introgression in Present-Day Human Populations. Mol. Biol. Evol. 2017, 34, 296–317. [Google Scholar] [CrossRef]
- Lin, F.-H.; Lin, R.; Wisniewski, H.M.; Hwang, Y.-W.; Grundke-Iqbal, I.; Healy-Louie, G.; Iqbal, K. Detection of point mutations in codon 331 of mitochondrial NADH dehydrogenase subunit 2 in alzheimer’s brains. Biochem. Biophys. Res. Commun. 1992, 182, 238–246. [Google Scholar] [CrossRef]
- Petruzzella, V.; Chen, X.; Schon, E.A. Is a point mutation in the mitochondrial ND2 gene associated with alzheimer’s disease? Biochem. Biophys. Res. Commun. 1992, 186, 491–497. [Google Scholar] [CrossRef]
- Schnopp, N.M.; Kösel, S.; Egensperger, R.; Graeber, M.B. Regional heterogeneity of mtDNA heteroplasmy in parkinsonian brain. Clin. Neuropathol. 1996, 15, 348–352. [Google Scholar]
- Fitzpatrick, E.; Bourke, B.; Drumm, B.; Rowland, M. The incidence of cyclic vomiting syndrome in children: Population-based study. Am. J. Gastroenterol. 2008, 103, 991–995. [Google Scholar] [CrossRef]
- Lancet, T. Hearing loss: An important global health concern. Lancet 2016, 387, 2351. [Google Scholar] [CrossRef]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [CrossRef]
- Lott, M.T.; Leipzig, J.N.; Derbeneva, O.; Xie, H.M.; Chalkia, D.; Sarmady, M.; Procaccio, V.; Wallace, D.C. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr. Protoc. Bioinform. 2013, 44, 1.23.1–1.23.26. [Google Scholar] [CrossRef]
- Thomas, M.G.; Miller, K.W.P.; Mascie-Taylor, C.G.N. Mitochondrial DNA and IQ in Europe. Intelligence 1998, 26, 167–173. [Google Scholar] [CrossRef][Green Version]
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstätter, A.; Forer, L.; Specht, G.; Bandelt, H.-J.; Kronenberg, F.; Salas, A.; Schönherr, S. HaploGrep 2: Mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef]
- Posth, C.; Renaud, G.; Mittnik, A.; Drucker, D.G.; Rougier, H.; Cupillard, C.; Valentin, F.; Thevenet, C.; Furtwängler, A.; Wißing, C.; et al. Pleistocene Mitochondrial Genomes Suggest a Single Major Dispersal of Non-Africans and a Late Glacial Population Turnover in Europe. Curr. Biol. 2016, 26, 827–833. [Google Scholar] [CrossRef]
- Hervella, M.; Svensson, E.M.; Alberdi, A.; Günther, T.; Izagirre, N.; Munters, A.R.; Alonso, S.; Ioana, M.; Ridiche, F.; Soficaru, A.; et al. The mitogenome of a 35,000-year-old Homo sapiens from Europe supports a Palaeolithic back-migration to Africa. Sci. Rep. 2016, 6, 25501. [Google Scholar] [CrossRef]
- Formicola, V.; Pontrandolfi, A.; Svoboda, J. The Upper Paleolithic triple burial of Dolní Věstonice: Pathology and funerary behavior. Am. J. Phys. Anthropol. 2001, 115, 372–379. [Google Scholar] [CrossRef]
- Fu, Q.; Mittnik, A.; Johnson, P.L.F.; Bos, K.; Lari, M.; Bollongino, R.; Sun, C.; Giemsch, L.; Schmitz, R.; Burger, J.; et al. A Revised Timescale for Human Evolution Based on Ancient Mitochondrial Genomes. Curr. Biol. 2013, 23, 553–559. [Google Scholar] [CrossRef]
- Green, R.E.; Malaspinas, A.-S.; Krause, J.; Briggs, A.W.; Johnson, P.L.F.; Uhler, C.; Meyer, M.; Good, J.M.; Maricic, T.; Stenzel, U.; et al. A Complete Neandertal Mitochondrial Genome Sequence Determined by High-Throughput Sequencing. Cell 2008, 134, 416–426. [Google Scholar] [CrossRef]
- Gansauge, M.-T.; Meyer, M. Selective enrichment of damaged DNA molecules for ancient genome sequencing. Genome Res. 2014, 24, 1543–1549. [Google Scholar] [CrossRef]
- Briggs, A.W.; Good, J.M.; Green, R.E.; Krause, J.; Maricic, T.; Stenzel, U.; Lalueza-Fox, C.; Rudan, P.; Brajković, D.; Kućan, Ž. Targeted Retrieval and Analysis of Five Neandertal mtDNA Genomes. Science 2009, 325, 318–321. [Google Scholar] [CrossRef]
- Fu, Q.; Li, H.; Moorjani, P.; Jay, F.; Slepchenko, S.M.; Bondarev, A.A.; Johnson, P.L.F.; Aximu-Petri, A.; Prüfer, K.; De Filippo, C.; et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 2014, 514, 445–449. [Google Scholar] [CrossRef]
- Rieux, A.; Eriksson, A.; Li, M.; Sobkowiak, B.; Weinert, L.A.; Warmuth, V.; Ruiz-Linares, A.; Manica, A.; Balloux, F. Improved Calibration of the Human Mitochondrial Clock Using Ancient Genomes. Mol. Biol. Evol. 2014, 31, 2780–2792. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef]
- Steiper, M.E.; Young, N.M. Primate molecular divergence dates. Mol. Phylogenet Evol. 2006, 41, 384–394. [Google Scholar] [CrossRef]
- Bergsten, J. A review of long-branch attraction. Cladistics 2005, 21, 163–193. [Google Scholar] [CrossRef]
- Henn, B.M.; Botigué, L.R.; Gravel, S.; Wang, W.; Brisbin, A.; Byrnes, J.K.; Fadhlaoui-Zid, K.; Zalloua, P.A.; Moreno-Estrada, A.; Bertranpetit, J.; et al. Genomic Ancestry of North Africans Supports Back-to-Africa Migrations. PLoS Genet. 2012, 8, e1002397. [Google Scholar] [CrossRef]
- D’Aurelio, M.; Gajewski, C.D.; Lin, M.T.; Mauck, W.M.; Shao, L.Z.; Lenaz, G.; Moraes, C.T.; Manfredi, G. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 2004, 13, 3171–3179. [Google Scholar] [CrossRef]
- Mita, S.; Rizzuto, R.; Moraes, C.T.; Shanske, S.; Arnaudo, E.; Fabrizi, G.M.; Koga, Y.; DiMauro, S.; Schon, E.A. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 1990, 18, 561–567. [Google Scholar] [CrossRef]
- Schwartz, M.; Vissing, J. Paternal Inheritance of Mitochondrial DNA. N. Engl. J. Med. 2002, 347, 576–580. [Google Scholar] [CrossRef]
- Dokianakis, E.; Ladoukakis, E.D. Different degree of paternal mtDNA leakage between male and female progeny in interspecific Drosophila crosses. Ecol. Evol. 2014, 4, 2633–2641. [Google Scholar] [CrossRef]
- Neiman, M.; Taylor, D.R. The causes of mutation accumulation in mitochondrial genomes. Proc. R. Soc. B Biol. Sci. 2009, 276, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wolf, A.B.; Fu, W.; Li, L.; Akey, J.M. Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals. Cell 2020, 180, 677–687. [Google Scholar] [CrossRef]
- Horai, S.; Hayasaka, K.; Kondo, R.; Tsugane, K.; Takahata, N. Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc. Natl. Acad. Sci. USA 1995, 92, 532–536. [Google Scholar] [CrossRef]
- Van Oven, M.; Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009, 30, E386–E394. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0.b5; Sinauer Associates: Sunderland, UK, 2001. [Google Scholar]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Lindenbaum, P. JVarkit: Java-Based Utilities for Bioinformatics. Figshare 2015, 10, m9. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]

| Position | DbSNP | Change | Change | Change | Type | Prot. Effect | Alignment. | 1K Gen. | Gene | Codon |
|---|---|---|---|---|---|---|---|---|---|---|
| rs | AA | Nt | Codon | Freq. (%) | Freq. (%) | Position | ||||
| 146 | rs370482130 | - | T>C | - | Ts | - | 28.40% | 21.40% | - | - |
| 150 | rs62581312 | - | C>T | - | Ts | - | 20.90% | 19.10% | - | - |
| 152 | rs117135796 | - | T>C | - | Ts | - | 38.80% | 33% | - | - |
| 185 | rs879015046 | - | G>A | - | Ts | - | 4.50% | 3.70% | - | - |
| 189 | rs371543232 | - | A>G | - | Ts | - | 19.40% | 7.70% | - | - |
| 195 | rs2857291 | - | T>C | - | Ts | - | 13.40% | 24.70% | - | - |
| 247 | rs41334645 | - | G>A | - | Ts | - | 17.90% | 7.00% | - | - |
| 709 | rs2853517 | - | G>A | - | Ts | - | 26.90% | 16.70% | - | - |
| 769 | rs2853519 | - | G>A | - | Ts | - | 22.40% | 16.40% | - | - |
| 825 | rs2853520 | - | T>A | - | Tv | - | 17.90% | 7.10% | - | - |
| 827 | rs28358569 | - | A>G | - | Ts | - | 16.40% | 4.30% | - | - |
| 1018 | rs2856982 | - | G>A | - | Ts | - | 22.40% | 16.50% | - | - |
| 2706 | rs2854128 | - | A>G | - | Ts | - | 80.60% | 89.00% | - | - |
| 2758 | rs2856980 | - | G>A | - | Ts | - | 6.00% | 6.60% | - | - |
| 2885 | rs2854130 | - | T>C | - | Ts | - | 16.40% | 6.80% | - | - |
| 3010 | rs3928306 | - | G>A | - | Ts | - | 17.90% | 11.20% | - | - |
| 3594 | rs193303025 | - | C>T | GTC>GTT | Ts | No change | 20.90% | 15.90% | ND1 | 3 |
| 4104 | rs1117205 | - | A>G | CTA>CTG | Ts | No change | 19.40% | 15.80% | ND1 | 3 |
| 4312 | rs193303033 | - | C>T | - | Ts | - | 14.90% | 1.80% | tRNA | - |
| 4688 | rs878853056 | - | T>C | GCT>GCC | Ts | No change | 4.50% | 0.40% | ND2 | 3 |
| 5460 | rs3021088 | A>T | G>A | GCC>ACC | Ts | AA change | 19.40% | 7.50% | ND2 | 1 |
| 5471 | rs879108598 | - | G>A | ACG>ACA | Ts | No change | 16.40% | 0.50% | ND2 | 3 |
| 5821 | rs56133209 | - | G>A | - | Ts | - | 14.90% | 0.50% | tRNA | - |
| 6962 | rs1970771 | - | G>A | CTG>CTA | Ts | No change | 3.00% | 3.00% | COX1 | 3 |
| 7146 | rs372136420 | T>A | A>G | ACT>GCT | Ts | AA change | 6.00% | 6.10% | COX1 | 1 |
| 7256 | - | - | C>T | AAC>AAT | Ts | No change | 20.90% | 15.80% | COX1 | 3 |
| 7424 | - | - | A>G | GAA>GAG | Ts | No change | 16.40% | 2.30% | COX1 | 3 |
| 7521 | rs199474817 | - | G>A | - | Ts | - | 20.90% | 16.00% | tRNA | - |
| 7650 | - | T>I | C>T | ACC>ATC | Ts | AA change | 13.40% | 0.00% | COX2 | 2 |
| 8386 | - | - | C>T | ACC>ACT | Ts | No change | 14.90% | 0.00% | ATP8 | 3 |
| 8468 | rs1116907 | - | C>T | CTA>TTA | Ts | No change | 16.40% | 6.70% | ATP8 | 1 |
| 8655 | - | - | C>T | ATC>ATT | Ts | No change | 17.90% | 7.10% | ATP6 | 3 |
| 9053 | rs199646902 | S>N | G>A | AGC>AAC | Ts | AA change | 14.90% | 2.60% | ATP6 | 2 |
| 9090 | rs386829064 | - | T>C | TCT>TCC | Ts | No change | 3.00% | 0.30% | ATP6 | 3 |
| 9755 | rs2856985 | - | G>A | GAG>GAA | Ts | No change | 14.90% | 2.00% | COX3 | 3 |
| 10,310 | rs41467651 | - | G>A | CTG>CTA | Ts | No change | 16.40% | 4.00% | ND3 | 3 |
| 10,373 | rs28358277 | - | G>A | GAG>GAA | Ts | No change | 14.90% | 3.00% | ND3 | 3 |
| 10,586 | rs28358281 | - | G>A | TCG>TCA | Ts | No change | 3.00% | 2.60% | ND4L | 3 |
| 10,664 | - | - | C>T | GTC>GTT | Ts | No change | 14.90% | 1.80% | ND4L | 3 |
| 10,688 | rs2853488 | - | G>A | GTG>GTA | Ts | No change | 17.90% | 7.00% | ND4L | 3 |
| 10,810 | rs28358282 | - | T>C | CTT>CTC | Ts | No change | 17.9 | 7.10% | ND4 | 3 |
| 10,915 | rs2857285 | - | T>C | TGT>TGC | Ts | No change | 14.90% | 2.30% | ND4 | 3 |
| 11,914 | rs2853496 | - | G>A | ACG>ACA | Ts | No change | 23.90% | 12.70% | ND4 | 3 |
| 12,366 | - | - | A>G | CTA>CTG | Ts | No change | 14.90% | 0.20% | ND5 | 3 |
| 12,810 | rs28359174 | - | A>G | TGA>TGG | Ts | No change | 4.50% | 2.40% | ND5 | 3 |
| 13,105 | rs2853501 | I>V | A>G | ATC>GTC | Ts | AA change | 20.90% | 13.70% | ND5 | 1 |
| 13,276 | rs2853502 | M>V | A>G | ATA>GTA | Ts | AA change | 14.90% | 1.80% | ND5 | 1 |
| 14,178 | rs28357671 | I>V | T>C | ATT>GTT | Ts | AA change | 16.40% | 5.20% | ND6 | 1 |
| 14,560 | rs28357676 | - | G>A | GTC>GTT | Ts | No change | 16.40% | 5.40% | ND6 | 3 |
| 15,148 | rs527236206 | - | G>A | CCG>CCA | Ts | No change | 16.40% | 1.10% | CYTB | 3 |
| 15,244 | rs28357369 | - | A>G | GGA>GGG | Ts | No change | 4.50% | 2.10% | CYTB | 3 |
| 15,355 | rs527236181 | - | G>A | ACG>ACA | Ts | No change | 16.40% | 0.60% | CYTB | 3 |
| 16,086 | rs386420030 | - | T>C | - | Ts | - | 4.50% | 2.10% | - | - |
| 16,093 | rs2853511 | - | T>C | - | Ts | - | 13.40% | 5.60% | - | - |
| 16,148 | rs201893071 | - | C>T | - | Ts | - | 17.90% | 3.10% | - | - |
| 16,169 | rs879121566 | - | C>T | - | Ts | - | 1.50% | 0.80% | - | - |
| 16,182 | rs879044186 | - | A>C | - | Tv | - | 15.20% | 2.70% | - | - |
| 16,183 | rs28671493 | - | A>G | - | Ts | - | 1.50% | 0.40% | - | - |
| 16,209 | rs386829278 | - | T>C | - | Ts | - | 17.90% | 3.00% | - | - |
| 16,230 | rs2853514 | - | A>G | - | Ts | - | 14.90% | 2.10% | - | - |
| 16,234 | rs368259300 | - | T>C | - | Ts | - | 16.40% | 1.90% | - | - |
| 16,278 | rs41458645 | - | C>T | - | Ts | - | 25.40% | 20.80% | - | - |
| 16,298 | rs148377232 | - | T>C | - | Ts | - | 11.90% | 5.00% | - | - |
| 16,311 | rs34799580 | - | T>C | - | Ts | - | 34.30% | 18.70% | - | - |
| 16,320 | rs62581338 | - | C>T | - | Ts | - | 16.40% | 5.20% | - | - |
| 16,519 | rs3937033 | - | T>C | - | Ts | - | 61.20% | 62.30% | - | - |
| Position | 195 | 3010 | 5460 | 5821 | 16,093 | 16,183 | 16,519 |
|---|---|---|---|---|---|---|---|
| Region/gene | D-loop | 16S rRNA | ND2 | tRNA-Cys | D-loop | D-loop | D-loop |
| dbSNP rs | rs2857291 | rs3928306 | rs3021088 | rs56133209 | rs2853511 | rs28671493 | rs3937033 |
| DNA change | T>C | G>A | G>A | G>A | T>C | A>C | T>C |
| Type | Transition | Transition | Transition | Transition | Transition | Transversion | Transition |
| Codon position | - | - | 1 | - | - | - | - |
| Codon effect | - | - | Non-Syn. | - | - | - | - |
| Codon change | - | - | GCC>ACC | - | - | - | - |
| Protein change | - | - | Ala>Thr | - | - | - | - |
| Disease | Bipolar | Cyclic | Alzheimer’s | Deafness | Cyclic | Melanoma | Cyclic vomiting |
| associations | disorder/ | vomiting | disease/ | helper | vomiting | patients | syndrome with |
| melanoma | syndrome with | Parkinson’s | mutation | syndrome | migraine/metastasis/ | ||
| patients | migraine | disease | glioblastoma, gastric, | ||||
| lung, ovarian, prostate | |||||||
| tumors | |||||||
| Modern | L2a1f, | H1a1, J1c, D4 | L0a1b1, L4a1, | C7 | L4a1, R0a, | X1a, X3, | L1c3a, L2a1f, L6a, |
| haplogroups | L4a1, | W1, Q1 | K1, H3, T1, | U1a1d, B2, | H15, H1a1, H3, I1, K, | ||
| L5a1a, L6a, | A, C1a | M2b, M29a, O | K1, K2a2a, R0a1a3, | ||||
| W1, K, | R1a, U2c, V1a, W1, | ||||||
| J2a2a, M2b | X1a, X3, B2, C4, C7, | ||||||
| E1, F1a1, G1a1, M20, | |||||||
| M29a, M2b, M3b, | |||||||
| N1b1a3, N2a, O, P2 | |||||||
| Neanderthal | Altai | Altai | Altai | Altai | Feldhofer1 | El Sidron 1253 | Altai |
| El Sidron 1253 | El Sidron 1253 | El Sidron 1253 | Vin. 33.25 | Feldhofer 1 | El Sidron 1253 | ||
| Feldhofer 1 | Feldhofer 1 | Feldhofer 1 | Feldhofer 2 | Feldhofer 1 | |||
| Feldhofer 2 | Feldhofer 2 | Feldhofer 2 | Mezmaiskaya1 | Feldhofer 2 | |||
| Mezmaiskaya1 | Mezmaiskaya1 | Mezmaiskaya1 | Vindija 33.16 | Mezmaiskaya1 | |||
| Vindija 33.16 | Vindija 33.16 | Vindija 33.16 | Vindija 33.17 | Vindija 33.16 | |||
| Vindija 33.17 | Vindija 33.17 | Vindija 33.17 | Vindija 33.19 | Vindija 33.17 | |||
| Vindija 33.19 | Vindija 33.19 | Vindija 33.19 | Vindija 33.25 | Vindija 33.19 | |||
| Vindija 33.25 | Vindija 33.25 | Vindija 33.25 | Vindija 33.25 |
| Archaic AMH | Haplogroup | Quality ** (%) | Age (Years) | Location | Reference | GenBank/ENA * | |
|---|---|---|---|---|---|---|---|
| 1 | Berry Au Bac 1 | U5b1a | 98.78 | 7160–7319 | France | [26] | KU534977 |
| 2 | Bockstein | U5b1d1 | 98.72 | 8016–8329 | Germany | [26] | KU534973 |
| 3 | Cuiry Les Chaudardes 1 | U5b1b | 97.01 | 8050–8360 | France | [26] | KU534975 |
| 4 | Ofnet | U5b1d1 | 98.72 | 8159–8424 | Germany | [26] | KU534974 |
| 5 | Felsdach | U5a2c | 97.43 | 8380–8980 | Germany | [26] | KU534954 |
| 6 | Hohlenstein Stadel | U5b2c1 | 94.77 | 8446–8809 | Germany | [26] | KU534979 |
| 7 | Falkenstein | U5b2a | 92.19 | 8993–9409 | Germany | [26] | KU534980 |
| 8 | Mareuil Les Meaux 1 | U5a2 + 16362 | 100 | 9080–9500 | France | [26] | KU534959 |
| 9 | Les Closeaux 3 | U5a2 | 98.27 | 9580–10,230 | France | [26] | KU534958 |
| 10 | Ranchot 88 | U5b1 | 95.95 | 9933–10,235 | France | [26] | KU534978 |
| 11 | Iboussieres 31-2 | U5b1 + 16189 | 99.01 | 10,140 | France | [26] | KU534976 |
| 12 | Iboussieres 25-1 | U5b2a | 93.92 | 10,140 | France | [26] | KU534981 |
| 13 | Iboussieres 39 | U5b2b | 92.40 | 11,600–12,040 | France | [26] | KU534972 |
| 14 | Hohle Fels 10 | U8a | 96.92 | 12,700 | Germany | [26] | KU534961 |
| 15 | Rochedane | U5b2b | 97.01 | 12,830–13,090 | France | [26] | KU534971 |
| 16 | Burkhardtshohle | U8a | 96.92 | 14,150–15,080 | Germany | [26] | KU534960 |
| 17 | Hohle Fels 79 | U8a | 96.92 | 14,270–15,070 | Germany | [26] | KU534962 |
| 18 | Brillenhohle | U8a | 97.66 | 14,400–15,120 | Germany | [26] | KU534947 |
| 19 | Oberkassel 998 | U5b1+ | 99.45 | 14,000 | Germany | [27] | KC521457 |
| 20 | Goyet Q-2 | U8a | 96.92 | 14,780–15,230 | Belgium | [26] | KU534963 |
| 21 | Rigney 1 | U2′3′4′7′8′9 | 87.68 | 15,240–15,690 | France | [26] | KU534957 |
| 22 | Hohle Fels 49 | U8a | 96.92 | 15,568–16,250 | Germany | [26] | KU534964 |
| 23 | Paglicci 71 | U5b2b | 97.97 | 18,197–18,973 | Italy | [26] | KU534950 |
| 24 | Dolni Vestonice 43 | U5 | 97.44 | 25,000 | Czech Rep. | [26] | KU534970 |
| 25 | Goyet Q56-16 | U2 | 91.40 | 26,040–26,600 | Belgium | [26] | KU534965 |
| 26 | Goyet 2878-21 | U5 | 92.05 | 26,269–27,055 | Belgium | [26] | KU534955 |
| 27 | Goyet Q376-19 | U2 | 89.43 | 27,310–27,720 | Belgium | [26] | KU534967 |
| 28 | Goyet Q55-2 | U2 | 85.86 | 27,310–27,730 | Belgium | [26] | KU534948 |
| 29 | La Rochette | M | 92.38 | 27,400–27,784 | France | [26] | KU534951 |
| 30 | Goyet Q53-1 | U2 | 93.23 | 27,720–28,230 | Belgium | [26] | KU534966 |
| 31 | Paglicci 108 | U2′3′4′7′8′9 | 92.40 | 27,831–28,961 | Italy | [26] | KU534968 |
| 32 | Paglicci 133 | U8c | 99.41 | 28,000–29,000 | Italy | [26] | KU534956 |
| 33 | Dolni Vestonice 16 | U5 | 97.94 | 29,386–30,567 | Czech Rep. | [26] | KU534949 |
| 34 | Doni Vestonice 14 | U5 | 100 | 31,000 | Czech Rep. | [28] | KC521458 |
| 35 | Cioclovina 1 | U | 96.72 | 32,519–33,905 | Romania | [26] | KU534969 |
| 36 | Goyet Q376-3 | M | 88.67 | 33,140–33,940 | Belgium | [26] | KU534953 |
| 37 | Goyet Q116-1 | M | 93.64 | 34,430–35,160 | Belgium | [26] | KU534952 |
| 38 | Kostenki 14 | U2 | 92.69 | 36–39,000 | Russia | [27] | FN600416 * |
| 39 | Fumane 2 | R | 94.43 | 39–41,000 | Italy | [27] | KP718913 |
| 40 | Tianyuan | B4′5 | 91.17 | 40,000 | China | [29] | KC417443 |
| 41 | Ust-Ishim | R | 87.89 | 45,000 | West Siberia | [27] | PRJEB6622 * |
| Neanderthals | |||||||
| 1 | Vindija 33.16 | H1e | 52.74 | 38,000 | Croatia | [30] | AM948965 |
| 2 | Vindija 33.17 | H1e | 52.75 | Not dated | Croatia | [31] | KJ533544 |
| 3 | Vindija 33.19 | H1e | 52.74 | Not dated | Croatia | [31] | KJ533545 |
| 4 | Vindija 33.25 | H1as | 52.84 | Not dated | Croatia | [32] | FM865410 |
| 5 | El Sidron 1253 | H1e | 52.81 | 39,000 | Spain | [32] | FM865409 |
| 6 | Feldhofer 1 | H1as | 52.84 | 40,000 | Germany | [32] | FM865407 |
| 7 | Feldhofer 2 | H1e | 52.81 | 40,000 | Germany | [32] | FM865408 |
| 8 | Altai Neanderthal | L1′2′3′4′5′6 | 55.96 | 50,000 | Siberia | [3] | KC879692 |
| 9 | Mezmaskaya 1 | L1′2′3′4′5′6 | 52.12 | 65,000 | Russia | [32] | FM865411 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, R.C.; Rodrigues, C.R.; Broach, J.R.; Briones, M.R.S. Convergent Mutations and Single Nucleotide Variants in Mitochondrial Genomes of Modern Humans and Neanderthals. Int. J. Mol. Sci. 2024, 25, 3785. https://doi.org/10.3390/ijms25073785
Ferreira RC, Rodrigues CR, Broach JR, Briones MRS. Convergent Mutations and Single Nucleotide Variants in Mitochondrial Genomes of Modern Humans and Neanderthals. International Journal of Molecular Sciences. 2024; 25(7):3785. https://doi.org/10.3390/ijms25073785
Chicago/Turabian StyleFerreira, Renata C., Camila R. Rodrigues, James R. Broach, and Marcelo R. S. Briones. 2024. "Convergent Mutations and Single Nucleotide Variants in Mitochondrial Genomes of Modern Humans and Neanderthals" International Journal of Molecular Sciences 25, no. 7: 3785. https://doi.org/10.3390/ijms25073785
APA StyleFerreira, R. C., Rodrigues, C. R., Broach, J. R., & Briones, M. R. S. (2024). Convergent Mutations and Single Nucleotide Variants in Mitochondrial Genomes of Modern Humans and Neanderthals. International Journal of Molecular Sciences, 25(7), 3785. https://doi.org/10.3390/ijms25073785







