Exosomal miRNA Changes Associated with Restoration to Sinus Rhythm in Atrial Fibrillation Patients
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
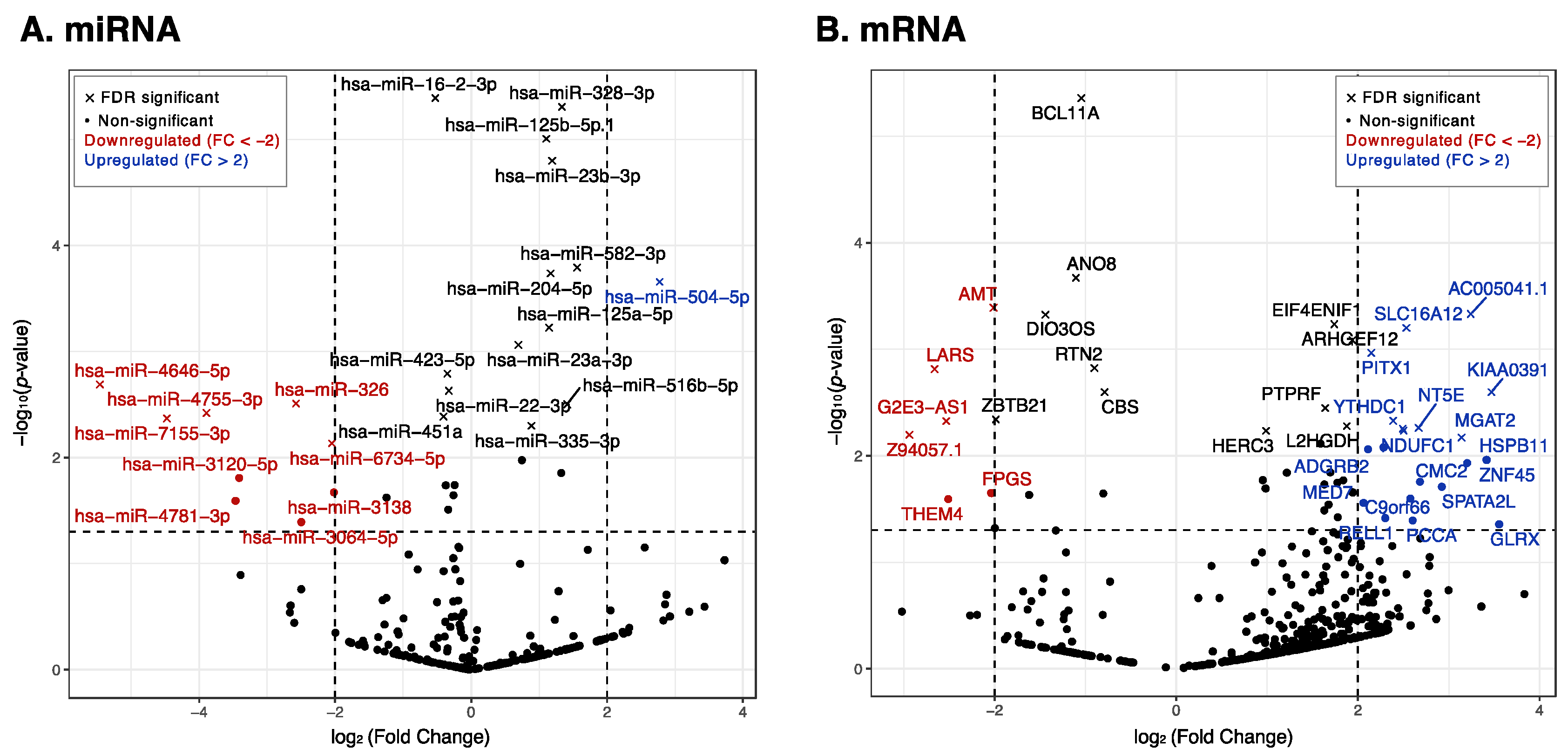
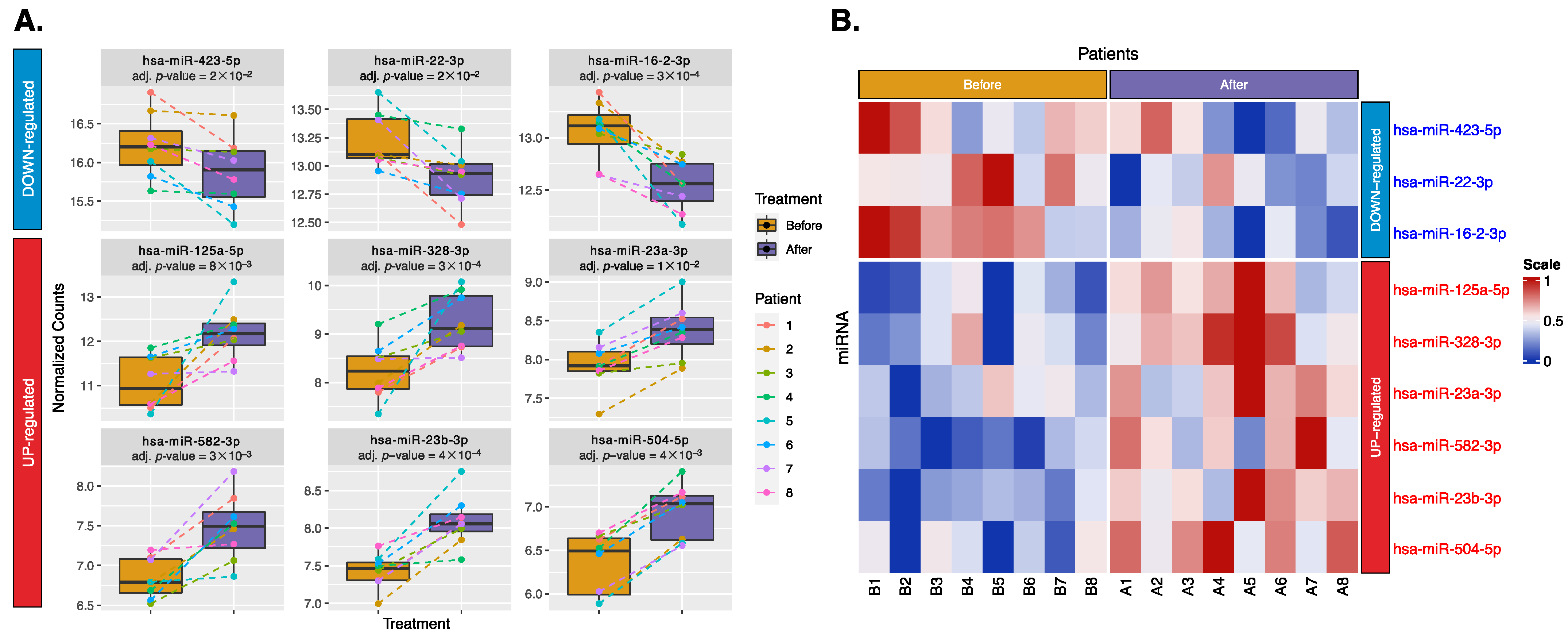
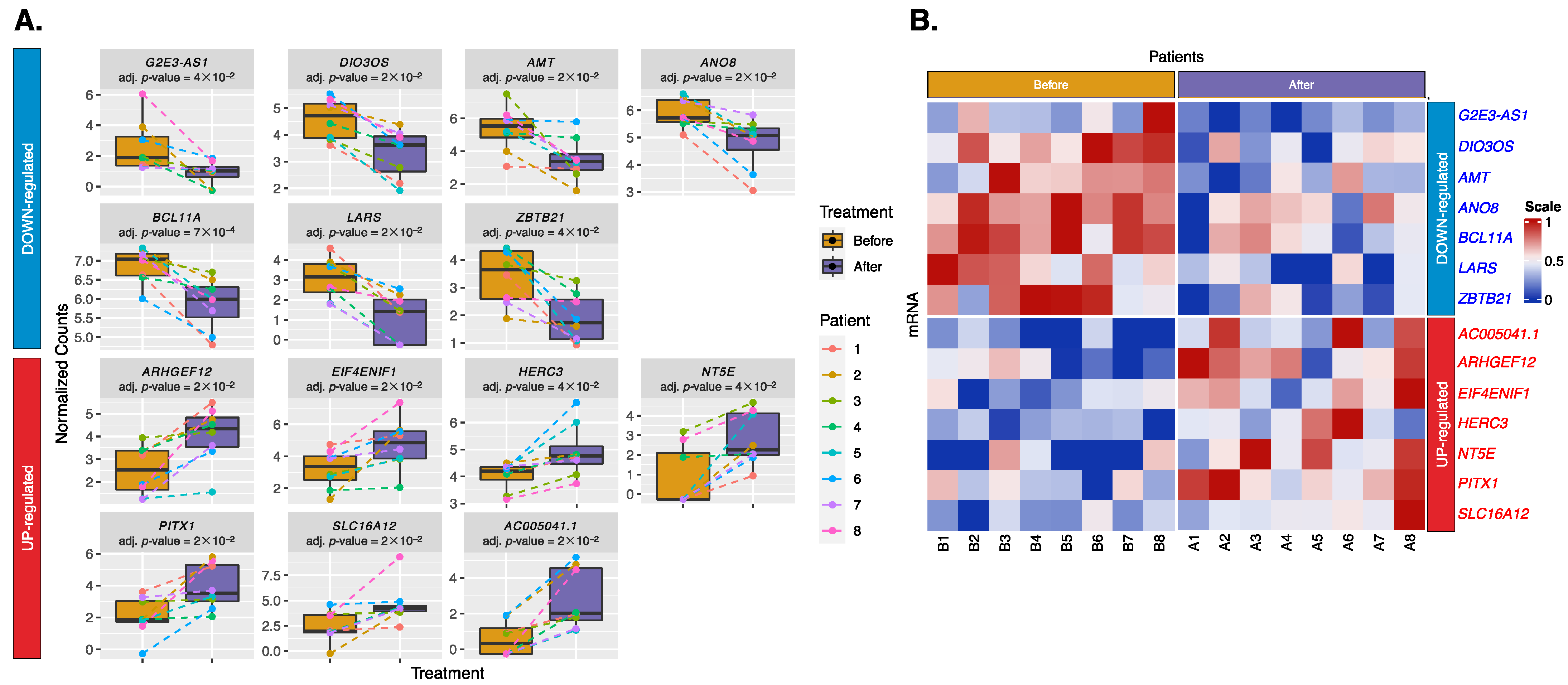
2.2. Differential Expression of Exosomal miRNAs and mRNAs
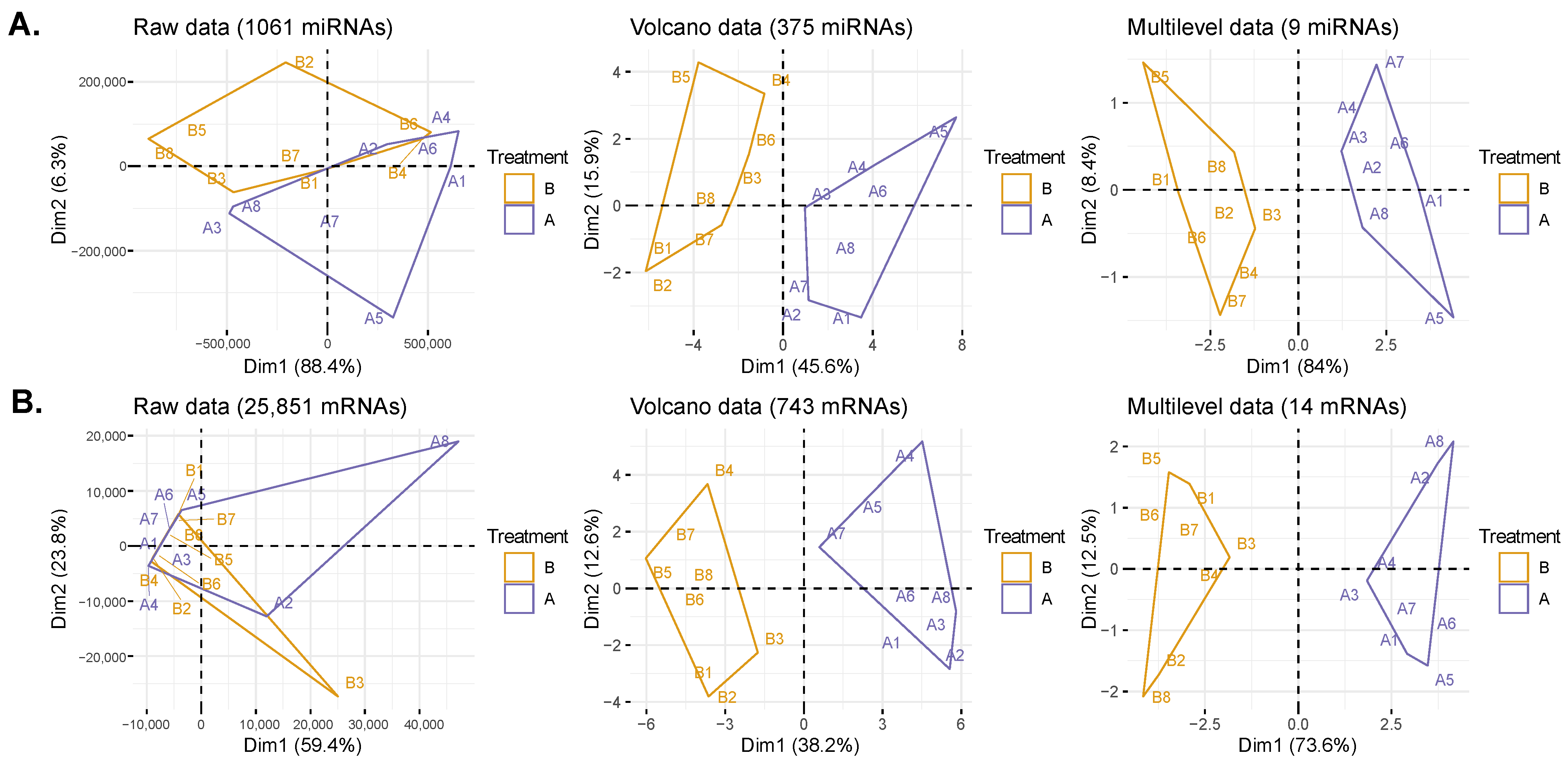
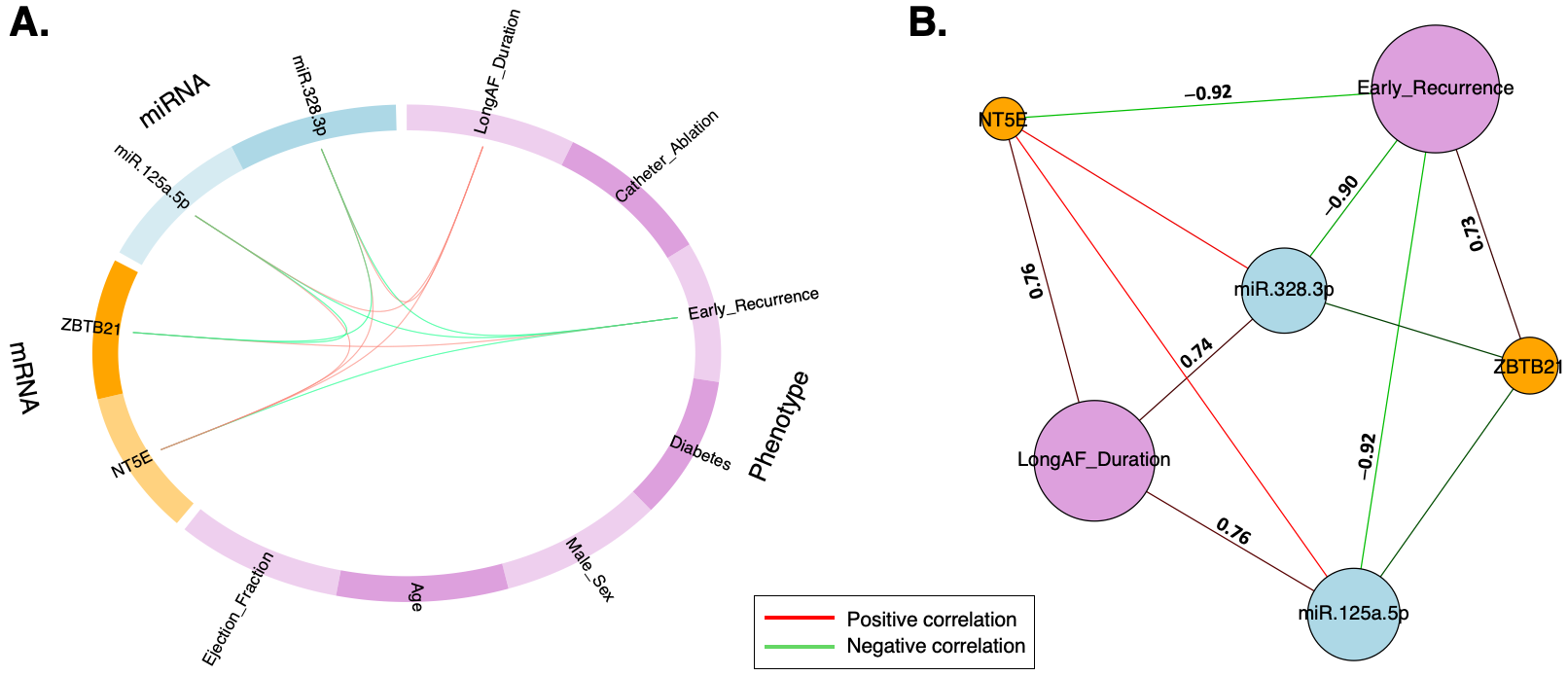
2.3. Data Transformation and DIABLO Model
3. Discussion
3.1. Part 1: The Role of miRNAs in SR Restoration
3.1.1. Downregulated miRNAs:
3.1.2. Upregulated miRNAs:
3.2. Part 2: The Role of mRNAs in SR Restoration
3.2.1. Downregulated mRNAs:
3.2.2. Upregulated mRNAs:
3.3. Part 3: Early Recurrence of AF and Underlying Cardiac Remodeling
3.4. Part 4: Strengths and Limitations of this Study
4. Materials and Methods
4.1. Participant Recruitment and Sample Collection
4.2. RNA Purification
4.3. Small RNA Library Preparation and Sequencing
4.4. Exosomal microRNA and mRNA Analysis
4.5. DIABLO Analysis for Multiomics Integration and Prediction
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AF | Atrial fibrillation |
| SR | Sinus rhythm |
| RFCA | Radiofrequency catheter ablation |
| EC | Electrical cardioversion |
| miRNA | microRNA |
| PCA | Principal component analysis |
References
- Zheng, D.; Huo, M.; Li, B.; Wang, W.; Piao, H.; Wang, Y.; Zhu, Z.; Li, D.; Wang, T.; Liu, K. The Role of Exosomes and Exosomal MicroRNA in Cardiovascular Disease. Front. Cell Dev. Biol. 2021, 8, 616161. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Deng, Y.; Xu, J.; Liu, J.; Liu, L.; Fan, C. The Role of Exosomes and Their Cargos in the Mechanism, Diagnosis, and Treatment of Atrial Fibrillation. Front. Cardiovasc. Med. 2021, 8, 712828. [Google Scholar] [CrossRef]
- Aliot, E.; Botto, G.L.; Crijns, H.J.; Kirchhof, P. Quality of life in patients with atrial fibrillation: How to assess it and how to improve it. EP Eur. 2014, 16, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Geng, M.; Lin, A.; Nguyen, T.P. Revisiting Antiarrhythmic Drug Therapy for Atrial Fibrillation: Reviewing Lessons Learned and Redefining Therapeutic Paradigms. Front. Pharmacol. 2020, 11, 581837. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.T.; Belley-Côté, E.P.; Ibrahim, O.; Um, K.J.; Lengyel, A.; Adli, T.; Qiu, Y.; Wong, M.; Sibilio, S.; Benz, A.P.; et al. Techniques improving electrical cardioversion success for patients with atrial fibrillation: A systematic review and meta-analysis. EP Eur. 2023, 25, 318–330. [Google Scholar] [CrossRef] [PubMed]
- AlTurki, A.; Proietti, R.; Dawas, A.; Alturki, H.; Huynh, T.; Essebag, V. Catheter ablation for atrial fibrillation in heart failure with reduced ejection fraction: A systematic review and meta-analysis of randomized controlled trials. BMC Cardiovasc. Disord. 2019, 19, 18. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Velasco, E.; Inácio, J.M.; Sousa, I.; Guimarães, A.R.; Franco, D.; Moura, G.; Belo, J.A. miRNAs in Heart Development and Disease. Int. J. Mol. Sci. 2024, 25, 1673. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Yan, S.; Zhao, X.; Han, X.; Fang, N.; Zhang, Y.; Dai, C.; Li, W.; Yu, H.; Gao, Y.; et al. Atrial myocyte-derived exosomal microRNA contributes to atrial fibrosis in atrial fibrillation. J. Transl. Med. 2022, 20, 407. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Li, H.; Zhang, A.; Li, Z.; Zhang, Y.; Ren, M.; Zhang, Y.; Hou, Y. MicroRNAs sequencing of plasma exosomes derived from patients with atrial fibrillation: miR-124-3p promotes cardiac fibroblast activation and proliferation by regulating AXIN1. J. Physiol. Biochem. 2022, 78, 85–98. [Google Scholar] [CrossRef]
- Li, Z.; Wang, S.; Hidru, T.H.; Sun, Y.; Gao, L.; Yang, X.; Xia, Y. Long Atrial Fibrillation Duration and Early Recurrence Are Reliable Predictors of Late Recurrence after Radiofrequency Catheter Ablation. Front. Cardiovasc. Med. 2022, 9, 864417. [Google Scholar] [CrossRef]
- Ortega-Hernandez, J.A.; Escobar-Alvarado, J.; Sanchez-Munoz, F.; Amezcua-Guerra, L.M.; Sanchez-Gloria, J.L.; Toledo-Lugo, A.; Gonzalez-Pacheco, H.; Marquez-Murillo, M.; Springall, R. Utility of miR-155, miR-21 and miR-16 in plasma as biomarkers of atrial fibrillation. Eur. Heart J. 2021, 42, ehab724.0323. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, L.; Li, H.; Han, L.; Yu, X. The Relation between microRNA-16 and Cardiac Remodelling Induced by Acute Myocardial Infarction: A Case Control Study. J. Biol. Regul. Homeost. Agents 2022, 36, 1379–1385. [Google Scholar] [CrossRef]
- Liu, Y.; Zhong, C.; Chen, S.; Xue, Y.; Wei, Z.; Dong, L.; Kang, L. Circulating exosomal mir-16-2-3p is associated with coronary microvascular dysfunction in diabetes through regulating the fatty acid degradation of endothelial cells. Cardiovasc. Diabetol. 2024, 23, 60. [Google Scholar] [CrossRef] [PubMed]
- van Boven, N.; Akkerhuis, K.M.; Anroedh, S.S.; Rizopoulos, D.; Pinto, Y.; Battes, L.C.; Hillege, H.L.; Caliskan, K.C.; Germans, T.; Manintveld, O.C.; et al. Serially measured circulating miR-22-3p is a biomarker for adverse clinical outcome in patients with chronic heart failure: The Bio-SHiFT study. Int. J. Cardiol. 2017, 235, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Park, H.; Park, J. Circulating microRNA-423 attenuates the phosphorylation of calcium handling proteins in atrial fibrillation. Mol. Med. Rep. 2022, 25, 186. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, M.; Yao, Y.; He, S.; Wang, Y.; Cao, Z.; Chen, H.; Fu, Y.; Liu, H.; Zhao, Q. Cardiac Fibroblasts Promote Ferroptosis in Atrial Fibrillation by Secreting Exo-miR-23a-3p Targeting SLC7A11. Oxidative Med. Cell. Longev. 2022, 2022, 3961495. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xiao, Z.; Guo, H.; Fang, X.; Liang, J.; Zhu, J.; Yang, J.; Li, H.; Pan, R.; Yuan, S.; et al. Novel role of the clustered miR-23b-3p and miR-27b-3p in enhanced expression of fibrosis-associated genes by targeting TGFBR3 in atrial fibroblasts. J. Cell. Mol. Med. 2019, 23, 3246–3256. [Google Scholar] [CrossRef]
- Li, S.; Ren, J.; Sun, Q. The expression of microRNA-23a regulates acute myocardial infarction in patients and in vitro through targeting PTEN. Mol. Med. Rep. 2018, 17, 6866–6872. [Google Scholar] [CrossRef]
- Zhang, H.; Caudle, Y.; Shaikh, A.; Yao, B.; Yin, D. Inhibition of microRNA-23b prevents polymicrobial sepsis-induced cardiac dysfunction by modulating TGIF1 and PTEN. Biomed. Pharmacother. 2018, 103, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.B.; Zhang, S.H.; Li, H.Y.; Chi, X.D.; Jiang, L.; Huang, Q.L.; Xu, S.H. Rs12976445 Polymorphism Is Associated with Post-Ablation Recurrence of Atrial Fibrillation by Modulating the Expression of MicroRNA-125a and Interleukin-6R. Med. Sci. Monit. 2018, 24, 6349–6358. [Google Scholar] [CrossRef]
- Vaze, A.; Donahue, K.; Spring, M.; Sardana, M.; Tanriverdi, K.; Freedman, J.; Keaney, J.; Benjamin, E.; Mph, S.; Rosenthal, L.; et al. Plasma MicroRNAs Relate to Atrial Fibrillation Recurrence after Catheter Ablation: Longitudinal Findings from the MiRhythm Study. J. Clin. Exp. Cardiol. 2017, 8, 3. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, Y.; Wang, N.; Pan, Z.; Gao, X.; Zhang, F.; Zhang, Y.; Shan, H.; Luo, X.; Bai, Y.; et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 2010, 122, 2378–2387. [Google Scholar] [CrossRef] [PubMed]
- Soeki, T.; Matsuura, T.; Bando, S.; Tobiume, T.; Uematsu, E.; Ise, T.; Kusunose, K.; Yamaguchi, K.; Yagi, S.; Fukuda, D.; et al. Relationship between local production of microRNA-328 and atrial substrate remodeling in atrial fibrillation. J. Cardiol. 2016, 68, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Adamcova, M.; Parova, H.; Lencova-Popelova, O.; Kollarova-Brazdova, P.; Baranova, I.; Slavickova, M.; Stverakova, T.; Mikyskova, P.S.; Mazurova, Y.; Sterba, M. Cardiac miRNA expression during the development of chronic anthracycline-induced cardiomyopathy using an experimental rabbit model. Front. Pharmacol. 2023, 14, 1298172. [Google Scholar] [CrossRef] [PubMed]
- Linna-Kuosmanen, S.; Tomas Bosch, V.; Moreau, P.R.; Bouvy-Liivrand, M.; Niskanen, H.; Kansanen, E.; Kivelä, A.; Hartikainen, J.; Hippeläinen, M.; Kokki, H.; et al. NRF2 is a key regulator of endothelial microRNA expression under proatherogenic stimuli. Cardiovasc. Res. 2021, 117, 1339–1357. [Google Scholar] [CrossRef] [PubMed]
- Goto, J.; Otaki, Y.; Watanabe, T.; Watanabe, M. The Role of HECT-Type E3 Ligase in the Development of Cardiac Disease. Int. J. Mol. Sci. 2021, 22, 6065. [Google Scholar] [CrossRef] [PubMed]
- Kuś, A.; Chaker, L.; Teumer, A.; Peeters, R.P.; Medici, M. The Genetic Basis of Thyroid Function: Novel Findings and New Approaches. J. Clin. Endocrinol. Metab. 2020, 105, 1707–1721. [Google Scholar] [CrossRef] [PubMed]
- Arslan, S.; Şahin, N.Ö.; Bayyurt, B.; Berkan, Ö.; Yılmaz, M.B.; Aşam, M.; Ayaz, F. Role of lncRNAs in Remodeling of the Coronary Artery Plaques in Patients with Atherosclerosis. Mol. Diagn. Ther. 2023, 27, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.B.; Hui, L.; Zhang, Q.H.; Chen, X.; Zhang, C.; Zheng, L.; Feng, X.; Wang, Y.P.; Ding, Z.J.; Chen, R.R.; et al. The Mutation Analysis of the AMT Gene in a Chinese Family with Nonketotic Hyperglycinemia. Front. Genet. 2022, 13, 854712. [Google Scholar] [CrossRef]
- Jha, A.; Chung, W.Y.; Vachel, L.; Maleth, J.; Lake, S.; Zhang, G.; Ahuja, M.; Muallem, S. Anoctamin 8 tethers endoplasmic reticulum and plasma membrane for assembly of Ca<sup>2+</sup> signaling complexes at the ER/PM compartment. EMBO J. 2019, 38, e101452. [Google Scholar] [CrossRef]
- van de Vegte, Y.J.; Tegegne, B.S.; Verweij, N.; Snieder, H.; van der Harst, P. Genetics and the heart rate response to exercise. Cell. Mol. Life Sci. 2019, 76, 2391–2409. [Google Scholar] [CrossRef]
- Chen, W.; Lin, Y.; Jiang, M.; Wang, Q.; Shu, Q. Identification of LARS as an essential gene for osteosarcoma proliferation through large-Scale CRISPR-Cas9 screening database and experimental verification. J. Transl. Med. 2022, 20, 355. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yang, J.; Lv, W.; Lu, Y.; Zhang, L.; Zhang, Y.; Musha, Z.; Fan, P.; Yang, B.; Zhou, X.; et al. Screening of differentially expressed microRNAs of essential hypertension in Uyghur population. Lipids Health Dis. 2019, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Wass, S.Y.; Offerman, E.J.; Sun, H.; Hsu, J.; Rennison, J.H.; Cantlay, C.C.; McHale, M.L.; Gillinov, A.M.; Moravec, C.; Smith, J.D.; et al. Novel functional atrial fibrillation risk genes and pathways identified from coexpression analyses in human left atria. Heart Rhythm 2023, 20, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Kasippillai, T.; MacArthur, D.G.; Kirby, A.; Thomas, B.; Lambalk, C.B.; Daly, M.J.; Welt, C.K. Mutations in eIF4ENIF1 Are Associated With Primary Ovarian Insufficiency. J. Clin. Endocrinol. Metab. 2013, 98, E1534–E1539. [Google Scholar] [CrossRef] [PubMed]
- Minor, M.; Alcedo, K.P.; Battaglia, R.A.; Snider, N.T. Cell type- and tissue-specific functions of ecto-5′-nucleotidase (CD73). Am. J. Physiol. Cell Physiol. 2019, 317, C1079–C1092. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Qin, X.; Wen, P.; Wu, Y.; Zhuang, J. Systematic analysis of molecular mechanisms of heart failure through the pathway and network-based approach. Life Sci. 2021, 265, 118830. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.C.; Choi, S.H.; Klarin, D.; Smith, J.G.; Loh, P.R.; Chaffin, M.; Roselli, C.; Hulme, O.L.; Lunetta, K.L.; Dupuis, J.; et al. Heritability of Atrial Fibrillation. Circ. Cardiovasc. Genet. 2017, 10, e001838. [Google Scholar] [CrossRef]
- Miyazawa, K.; Ito, K.; Ito, M.; Zou, Z.; Kubota, M.; Nomura, S.; Matsunaga, H.; Koyama, S.; Ieki, H.; Akiyama, M.; et al. Cross-ancestry genome-wide analysis of atrial fibrillation unveils disease biology and enables cardioembolic risk prediction. Nat. Genet. 2023, 55, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ponnusamy, M.; Liu, C.; Gao, J.; Wang, K.; Li, P. MicroRNA as a Therapeutic Target in Cardiac Remodeling. BioMed Res. Int. 2017, 2017, 1278436. [Google Scholar] [CrossRef]
- Wang, Y.; Tan, J.; Wang, L.; Pei, G.; Cheng, H.; Zhang, Q.; Wang, S.; He, C.; Fu, C.; Wei, Q. MiR-125 Family in Cardiovascular and Cerebrovascular Diseases. Front. Cell Dev. Biol. 2021, 9, 799049. [Google Scholar] [CrossRef]
- Zhao, D.; Li, C.; Yan, H.; Li, T.; Qian, M.; Zheng, N.; Jiang, H.; Liu, L.; Xu, B.; Wu, Q.; et al. Cardiomyocyte Derived miR-328 Promotes Cardiac Fibrosis by Paracrinely Regulating Adjacent Fibroblasts. Cell. Physiol. Biochem. 2018, 46, 1555–1565. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.K.; Hsu, H.C.; Liu, Y.H.; Tsai, W.S.; Ma, C.P.; Chen, Y.T.; Tan, B.C.; Lai, Y.Y.; Chang, I.Y.; Yang, C.; et al. EV-miRome-wide profiling uncovers miR-320c for detecting metastatic colorectal cancer and monitoring the therapeutic response. Cell. Oncol. 2022, 45, 621–638. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Shannon, C.P.; Gautier, B.; Rohart, F.; Vacher, M.; Tebbutt, S.J.; Le Cao, K.A. DIABLO: An integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 2019, 35, 3055–3062. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
| Patient | Sex | Age (Years) | AF Duration (Months) | Recurrence | Diabetes | LVEF (%) | Treatment Type |
|---|---|---|---|---|---|---|---|
| 1 | Female | 65 | 9 | Late | No | 68 | RFCA |
| 2 | Female | 61 | 15 | Late | No | 65 | EC |
| 3 | Male | 39 | 33 | Late | Yes | 50 | RFCA |
| 4 | Male | 66 | 89 | Late | No | 53 | EC |
| 5 | Male | 70 | 35 | Late | Yes | 50 | RFCA |
| 6 | Male | 74 | 9 | Early | No | 78 | EC |
| 7 | Male | 56 | 5 | Early | No | 50 | EC |
| 8 | Male | 62 | 22 | Late | No | 65 | EC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, P.-C.; Ko, A.M.-S.; Chen, Y.-L.; Chiu, C.-H.; Yeh, Y.-H.; Tsai, F.-C. Exosomal miRNA Changes Associated with Restoration to Sinus Rhythm in Atrial Fibrillation Patients. Int. J. Mol. Sci. 2024, 25, 3861. https://doi.org/10.3390/ijms25073861
Tsai P-C, Ko AM-S, Chen Y-L, Chiu C-H, Yeh Y-H, Tsai F-C. Exosomal miRNA Changes Associated with Restoration to Sinus Rhythm in Atrial Fibrillation Patients. International Journal of Molecular Sciences. 2024; 25(7):3861. https://doi.org/10.3390/ijms25073861
Chicago/Turabian StyleTsai, Pei-Chien, Albert Min-Shan Ko, Yu-Lin Chen, Cheng-Hsun Chiu, Yung-Hsin Yeh, and Feng-Chun Tsai. 2024. "Exosomal miRNA Changes Associated with Restoration to Sinus Rhythm in Atrial Fibrillation Patients" International Journal of Molecular Sciences 25, no. 7: 3861. https://doi.org/10.3390/ijms25073861




