Development of Detection Antibody Targeting the Linear Epitope in SARS-CoV-2 Nucleocapsid Protein with Ultra-High Sensitivity
Abstract
:1. Introduction
2. Results
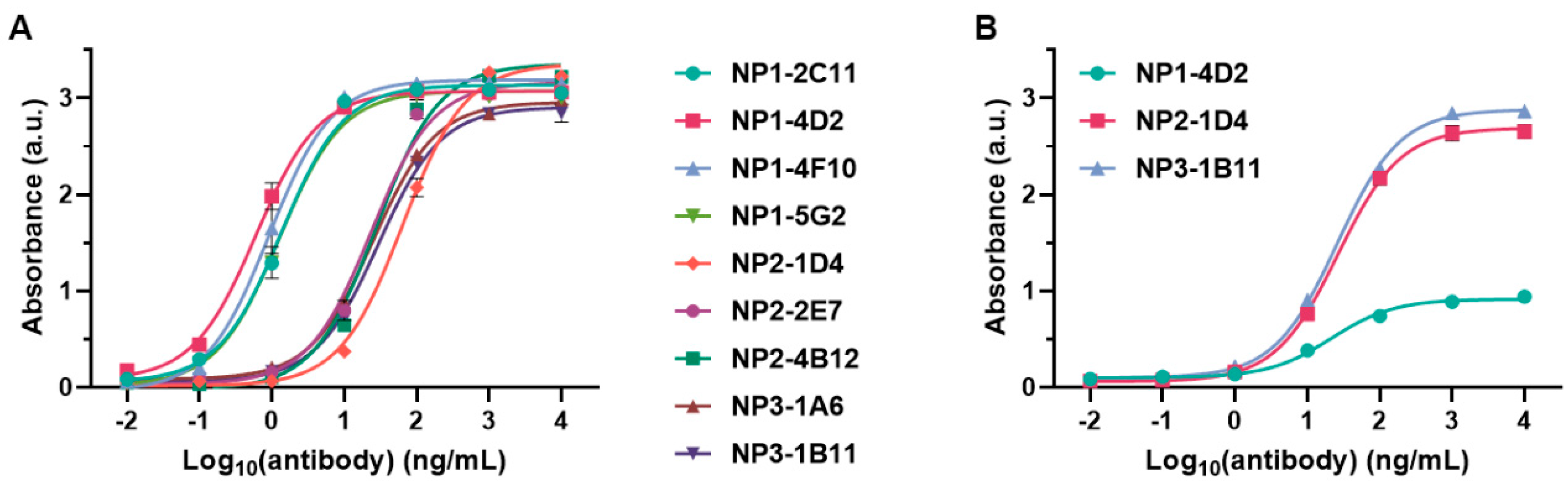
2.1. Preparation of Antibodies Targeting Peptide Antigens with Conserved Sequences
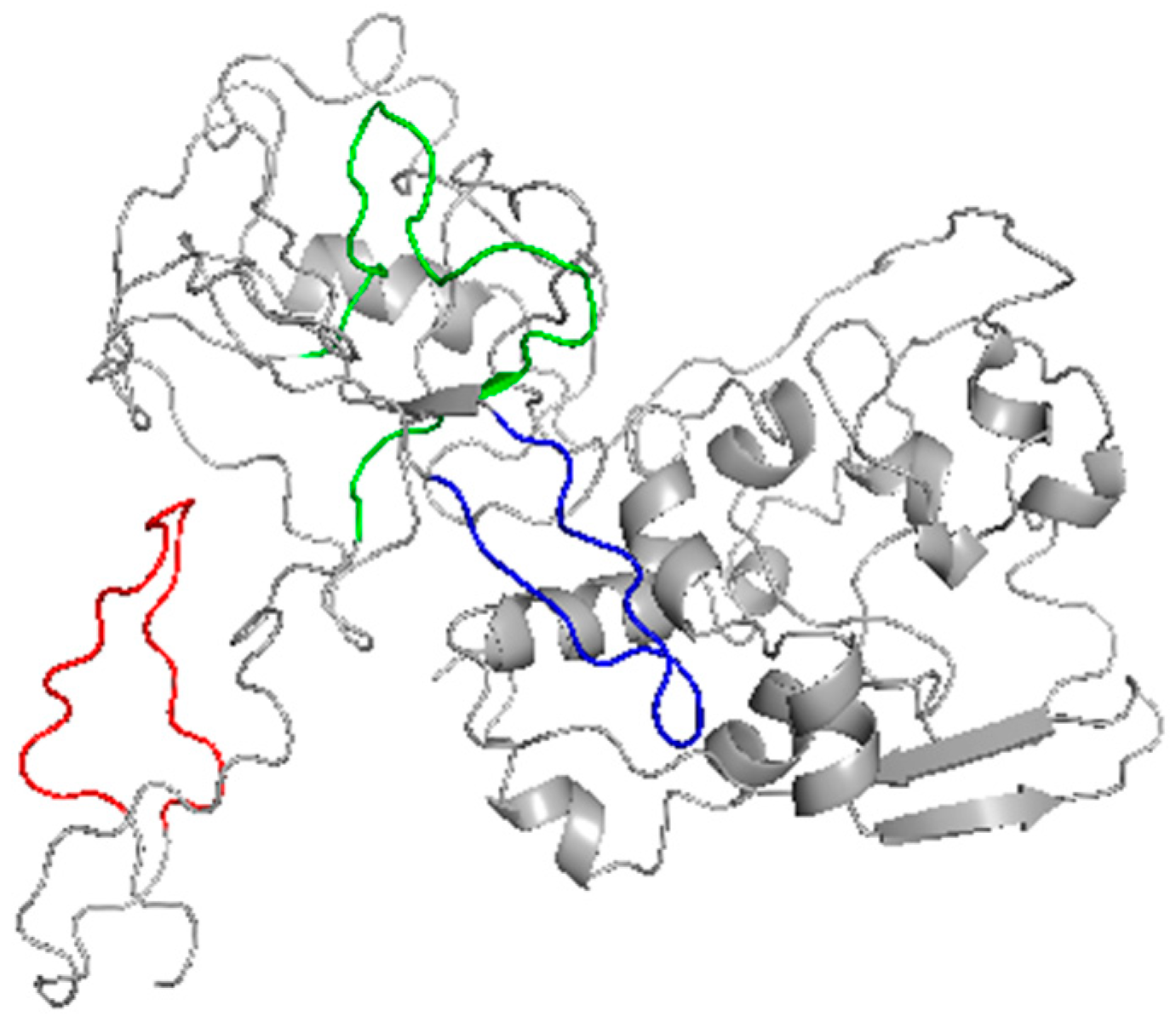
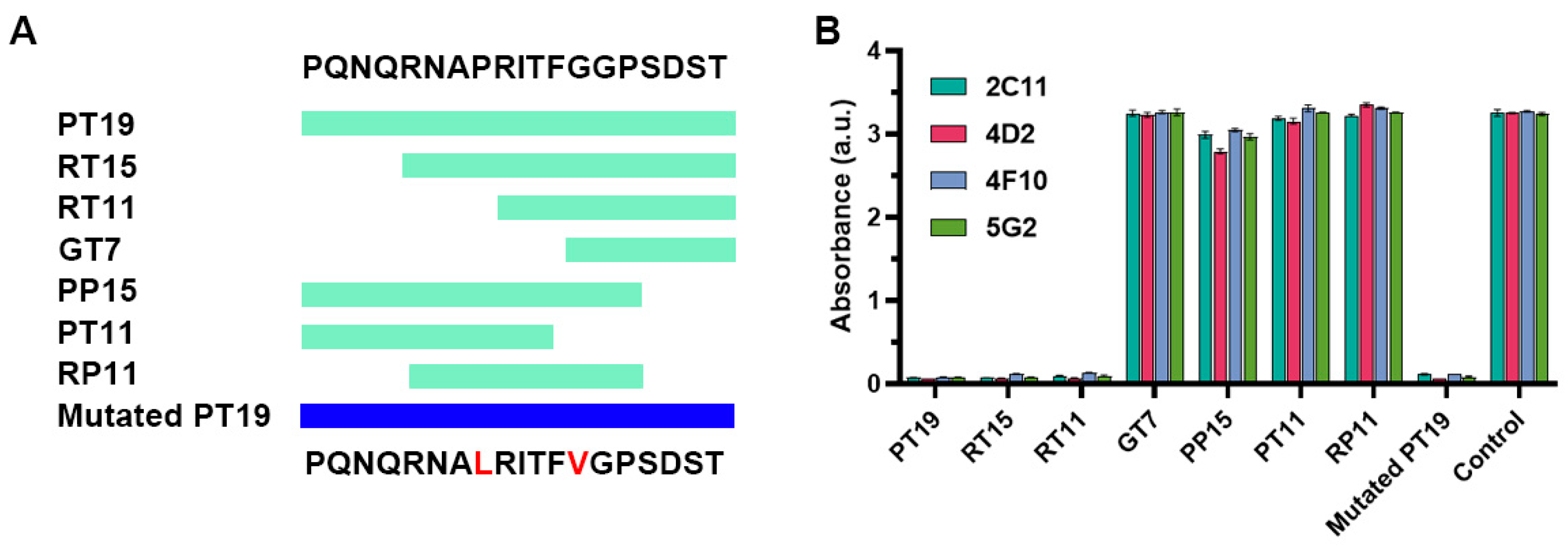
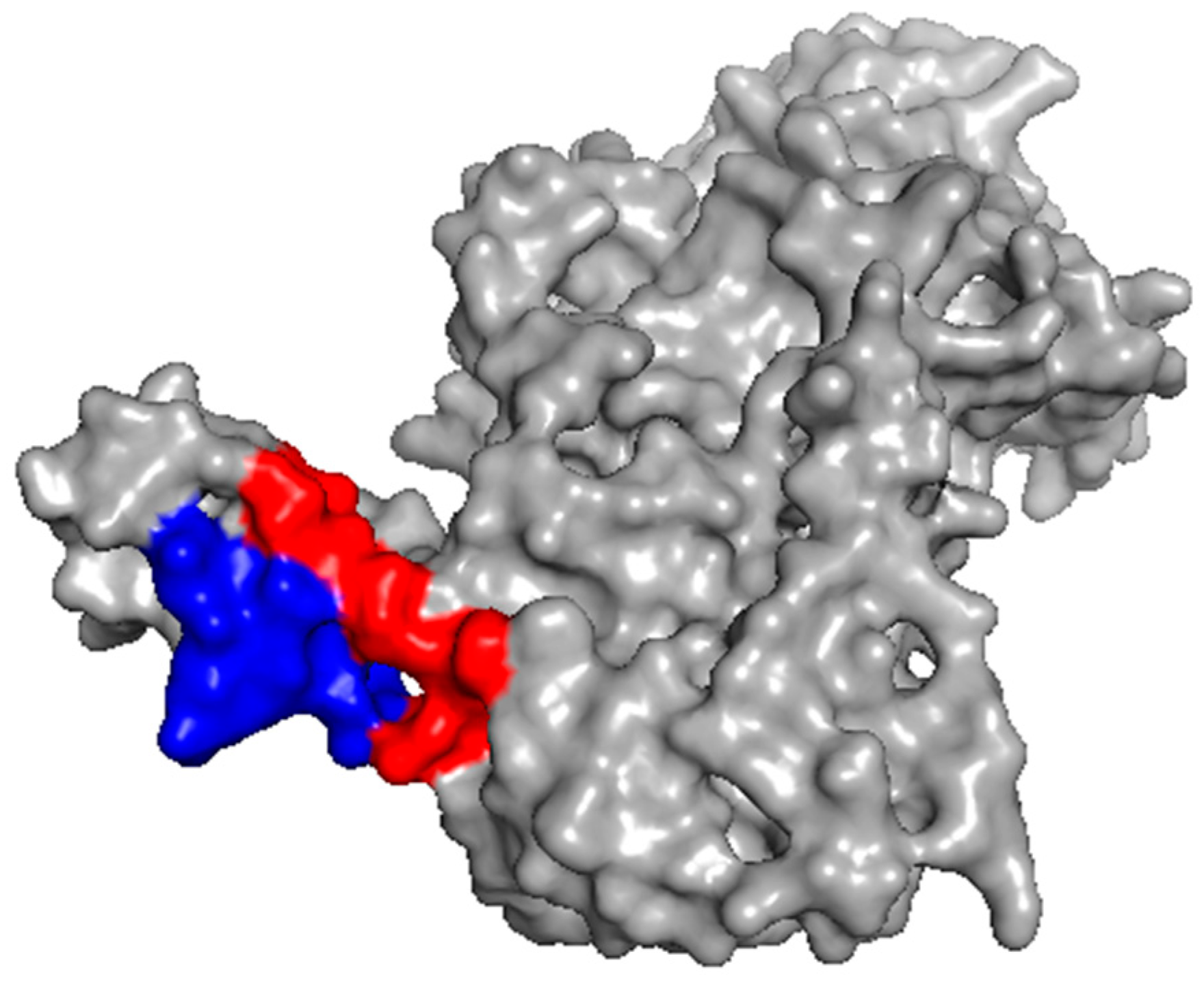
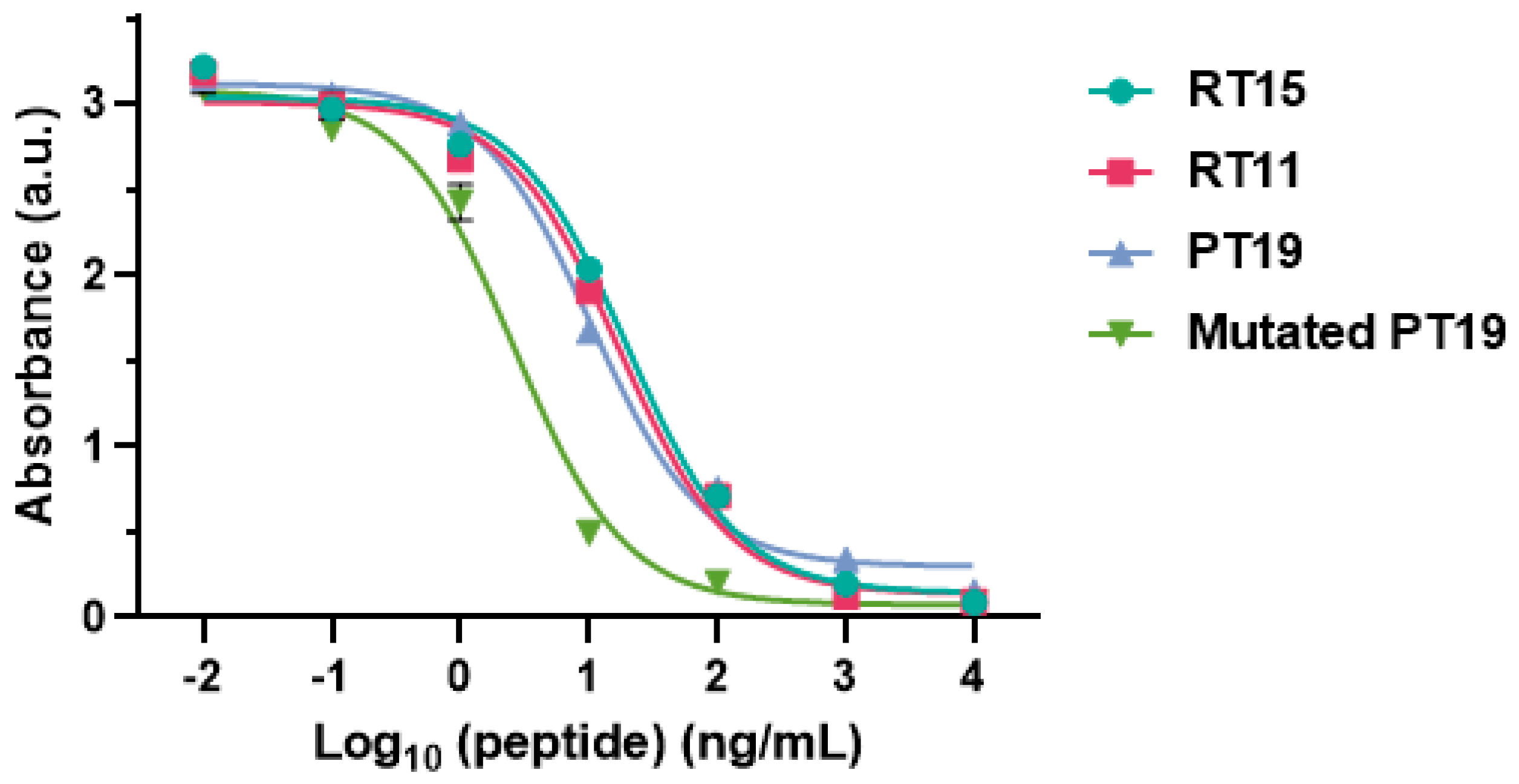
2.2. Investigation of the Epitope in CoV-NP1
2.3. Recognition of Mutated Peptide Antigen by CoV-NP1-Targeting Antibodies
2.4. Detection of the SARS-CoV-2 N Protein Using Colloidal Gold Immunochromatography
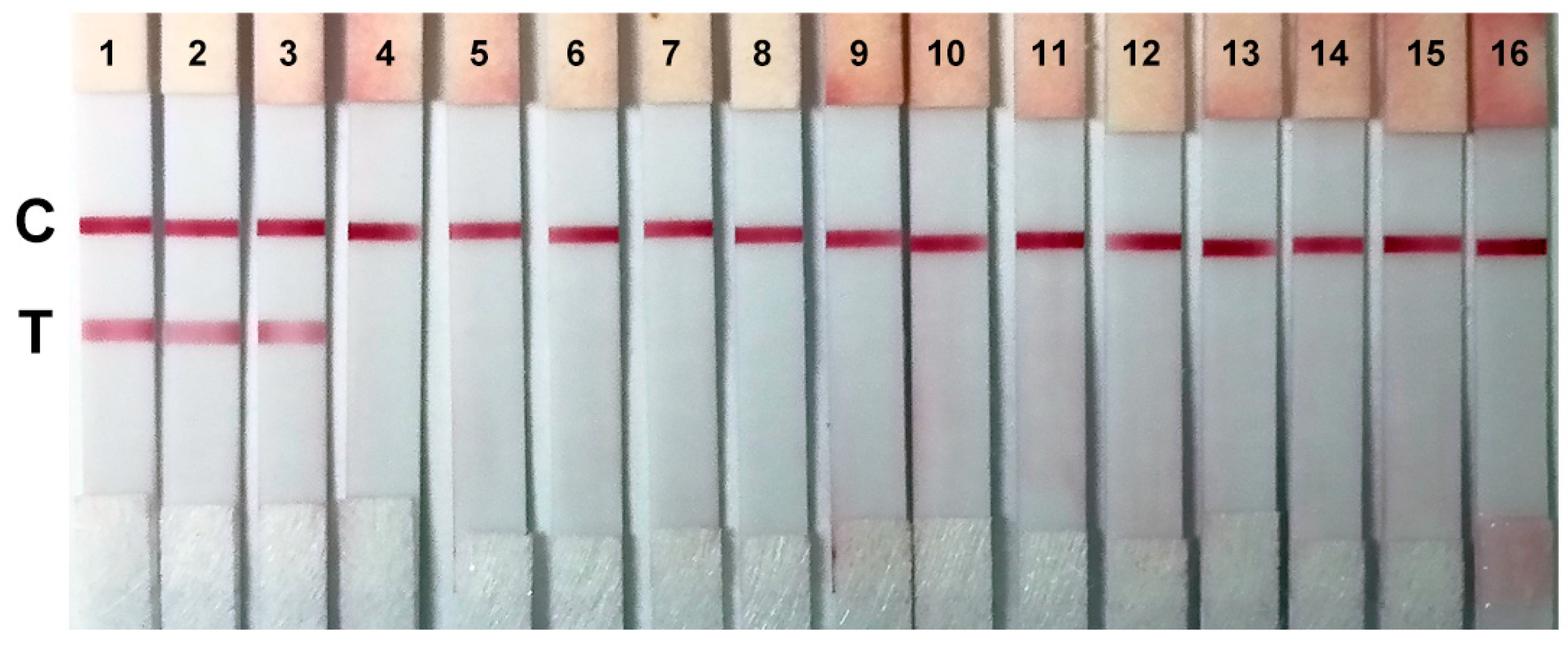
2.5. Sensitivity and Specificity of the Colloidal Gold Immunochromatographic Strips
3. Discussion
4. Materials and Methods
4.1. Screening of Peptides with Conserved Sequences as Antigens
4.2. Preparation of Monoclonal Antibodies
4.3. Evaluation of Binding Affinities
4.4. Investigation of the Inhibition of Binding between Antibody and N Protein by Different Peptides
4.5. Preparation of Colloidal Gold Immunochromatographic Strips
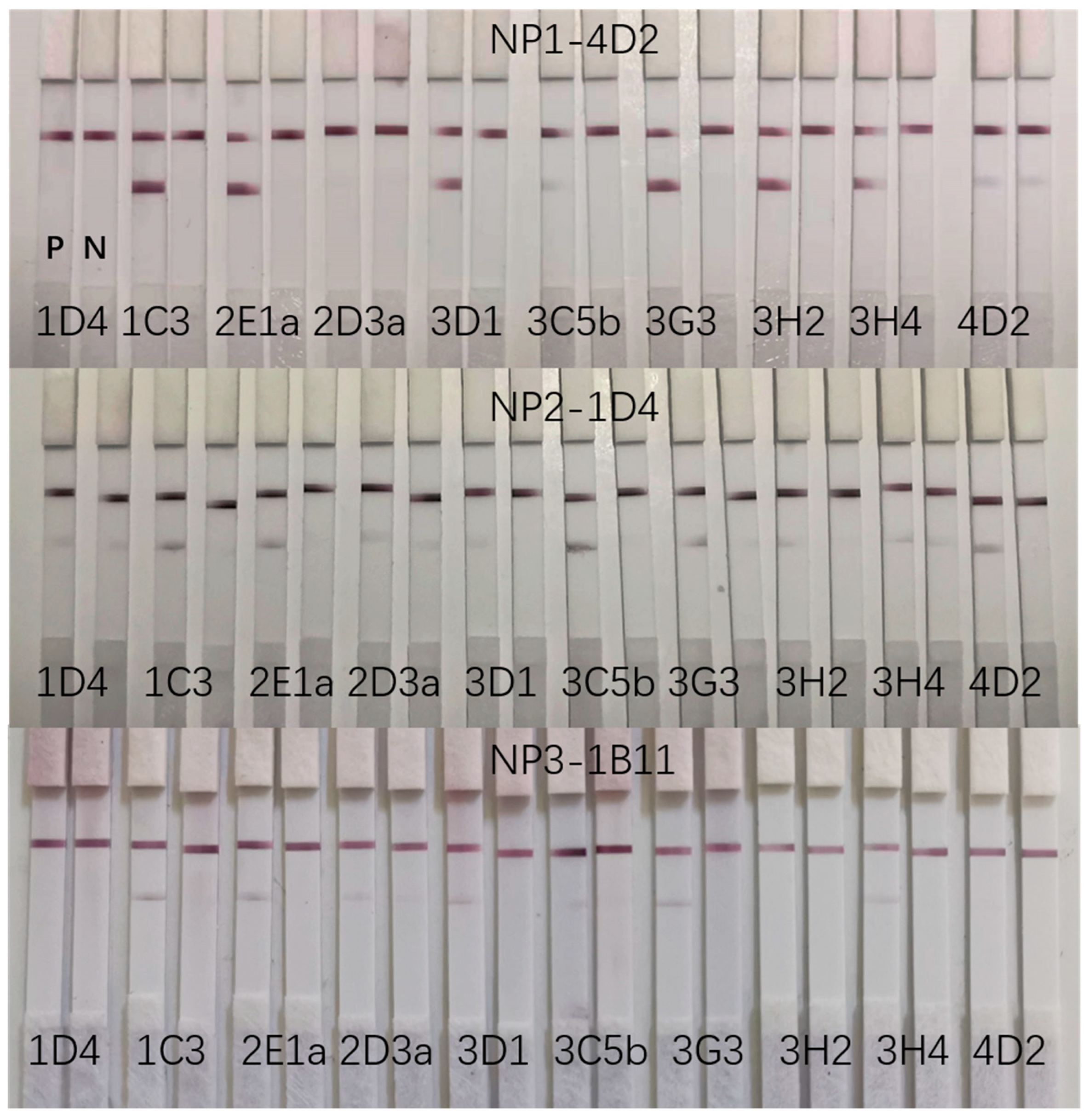
4.6. Screening of Matched Antibody Pairs
4.7. Evaluation of the Sensitivity and Specificity of Colloidal Gold Immunochromatographic Strips
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Fang, Z.; Ma, F.; Li, J.; Huang, Z.; Zhang, Y.; Li, J.; Chen, K. The role of SARS-CoV-2 N protein in diagnosis and vaccination in the context of emerging variants: Present status and prospects. Front. Microbiol. 2023, 14, 1217567. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Zhong, Q.; Gao, G.F. Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 2022, 65, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wu, F.; Cen, Y.; Ye, L.; Shi, X.; Huang, Y.; Fang, S.; Ma, L. Comparative research on nucleocapsid and spike glycoprotein as the rapid immunodetection targets of COVID-19 and establishment of immunoassay strips. Mol. Immunol. 2021, 131, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wu, Q.; Chen, J.; Ni, X.; Dai, J. A DNA Aptamer Based Method for Detection of SARS-CoV-2 Nucleocapsid Protein. Virol. Sin. 2020, 35, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Samper, I.C.; McMahon, C.J.; Schenkel, M.S.; Clark, K.M.; Khamcharoen, W.; Anderson, L.B.R.; Terry, J.S.; Gallichotte, E.N.; Ebel, G.D.; Geiss, B.J.; et al. Electrochemical Immunoassay for the Detection of SARS-CoV-2 Nucleocapsid Protein in Nasopharyngeal Samples. Anal. Chem. 2022, 94, 4712–4719. [Google Scholar] [CrossRef] [PubMed]
- Brandmeier, J.C.; Jurga, N.; Grzyb, T.; Hlaváček, A.; Obořilová, R.; Skládal, P.; Farka, Z.; Gorris, H.H. Digital and Analog Detection of SARS-CoV-2 Nucleocapsid Protein via an Upconversion-Linked Immunosorbent Assay. Anal. Chem. 2023, 95, 4753–4759. [Google Scholar] [CrossRef]
- Ang, G.Y.; Chan, K.G.; Yean, C.Y.; Yu, C.Y. Lateral Flow Immunoassays for SARS-CoV-2. Diagnostics 2022, 12, 2854. [Google Scholar] [CrossRef]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Dhankhar, P.; Dalal, V.; Singh, V.; Tomar, S.; Kumar, P. Computational guided identification of novel potent inhibitors of N-terminal domain of nucleocapsid protein of severe acute respiratory syndrome coronavirus 2. J. Biomol. Struct. Dyn. 2022, 40, 4084–4099. [Google Scholar] [CrossRef]
- Shamsi, A.; Mohammad, T.; Anwar, S.; Amani, S.; Khan, M.S.; Husain, F.M.; Rehman, M.T.; Islam, A.; Hassan, M.I. Potential drug targets of SARS-CoV-2: From genomics to therapeutics. Int. J. Biol. Macromol. 2021, 177, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Cheng, Y.; Zhou, H.; Sun, C.; Zhang, S. The SARS-CoV-2 nucleocapsid protein: Its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virol. J. 2023, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, L.; Xie, Y.-H.; Wu, J. Advancements in detection of SARS-CoV-2 infection for confronting COVID-19 pandemics. Lab. Investig. 2022, 102, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Calorenni, P.; Leonardi, A.A.; Sciuto, E.L.; Rizzo, M.G.; Faro, M.J.L.; Fazio, B.; Irrera, A.; Conoci, S. PCR-Free Innovative Strategies for SARS-CoV-2 Detection. Adv. Healthc. Mater. 2023, 12, 2300512. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Du, N.; Lei, Y.; Dorje, S.; Qi, J.; Luo, T.; Gao, G.F.; Song, H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020, 39, e105938. [Google Scholar] [CrossRef]
- Nagler, A.; Kalaora, S.; Barbolin, C.; Gangaev, A.; Ketelaars, S.L.C.; Alon, M.; Pai, J.; Benedek, G.; Yahalom-Ronen, Y.; Erez, N.; et al. Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA peptidomics. Cell Rep. 2021, 35, 109305. [Google Scholar] [CrossRef]
- Drain, P.K. Rapid Diagnostic Testing for SARS-CoV-2. N. Engl. J. Med. 2022, 386, 264–272. [Google Scholar] [CrossRef]
- Abavisani, M.; Rahimian, K.; Mahdavi, B.; Tokhanbigli, S.; Mollapour Siasakht, M.; Farhadi, A.; Kodori, M.; Mahmanzar, M.; Meshkat, Z. Mutations in SARS-CoV-2 structural proteins: A global analysis. Virol. J. 2022, 19, 220. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, C.; Cracknell Daniels, B.; Brancaccio, G.; Brazzale, A.R.; Lavezzo, E.; Ciavarella, C.; Onelia, F.; Franchin, E.; Manuto, L.; Bianca, F.; et al. Impact of antigen test target failure and testing strategies on the transmission of SARS-CoV-2 variants. Nat. Commun. 2022, 13, 5870. [Google Scholar] [CrossRef] [PubMed]
- Hilti, D.; Wehrli, F.; Roditscheff, A.; Risch, M.; Risch, L.; Egli, A.; Bodmer, T.; Wohlwend, N. SARS-CoV-2 Nucleocapsid Protein Mutations Found in Switzerland Disrupt N-Gene Amplification in Commonly Used Multiplex RT-PCR Assay. Pathogens 2023, 12, 1383. [Google Scholar] [CrossRef] [PubMed]
- Laine, P.; Nihtilä, H.; Mustanoja, E.; Lyyski, A.; Ylinen, A.; Hurme, J.; Paulin, L.; Jokiranta, S.; Auvinen, P.; Meri, T. SARS-CoV-2 variant with mutations in N gene affecting detection by widely used PCR primers. J. Med. Virol. 2022, 94, 1227–1231. [Google Scholar] [CrossRef] [PubMed]
- de Mello Malta, F.; Amgarten, D.; Marra, A.R.; Petroni, R.C.; da Silva Nali, L.H.; Siqueira, R.A.; Neto, M.C.; Oler, S.C.; Pinho, J.R.R. Nucleocapsid single point-mutation associated with drop-out on RT-PCR assay for SARS-CoV-2 detection. BMC Infect. Dis. 2023, 23, 714. [Google Scholar] [CrossRef] [PubMed]
- Budd, J.; Miller, B.S.; Weckman, N.E.; Cherkaoui, D.; Huang, D.; Decruz, A.T.; Fongwen, N.; Han, G.-R.; Broto, M.; Estcourt, C.S.; et al. Lateral flow test engineering and lessons learned from COVID-19. Nat. Rev. Bioeng. 2023, 1, 13–31. [Google Scholar] [CrossRef]
- Mak, G.C.K.; Cheng, P.K.C.; Lau, S.S.Y.; Wong, K.K.Y.; Lau, C.S.; Lam, E.T.K.; Chan, R.C.W.; Tsang, D.N.C. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 129, 104500. [Google Scholar] [CrossRef] [PubMed]
- Mak, G.C.K.; Lau, S.S.Y.; Wong, K.K.Y.; Chow, N.L.S.; Lau, C.S.; Lam, E.T.K.; Chan, R.C.W.; Tsang, D.N.C. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 133, 104684. [Google Scholar] [CrossRef] [PubMed]
- Mak, G.C.K.; Lau, S.S.Y.; Wong, K.K.Y.; Chow, N.L.S.; Lau, C.S.; Lam, E.T.K.; Chan, R.C.W.; Tsang, D.N.C. Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS-CoV-2. J. Clin. Virol. 2021, 134, 104712. [Google Scholar] [CrossRef]
- Sun, Q.; Ning, Q.; Li, T.; Jiang, Q.; Feng, S.; Tang, N.; Cui, D.; Wang, K. Immunochromatographic enhancement strategy for SARS-CoV-2 detection based on nanotechnology. Nanoscale 2023, 15, 15092–15107. [Google Scholar] [CrossRef]
- Ruantip, S.; Pimpitak, U.; Rengpipat, S.; Pasomsub, E.; Seepiban, C.; Gajanandana, O.; Torvorapanit, P.; Hirankarn, N.; Jaru-ampornpan, P.; Siwamogsatham, S.; et al. Self-enhancement lateral flow immunoassay for COVID-19 diagnosis. Sens. Actuators B Chem. 2023, 389, 133898. [Google Scholar] [CrossRef]
- Kim, J.; Kim, C.; Park, J.S.; Lee, N.E.; Lee, S.; Cho, S.-Y.; Park, C.; Yoon, D.S.; Yoo, Y.K.; Lee, J.H. Affordable on-site COVID-19 test using non-powered preconcentrator. Biosens. Bioelectron. 2023, 222, 114965. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.-K.; Kim, K.; Park, J.; Im, H.; Maher, S.; Kim, M.-G. Plasmon color-preserved gold nanoparticle clusters for high sensitivity detection of SARS-CoV-2 based on lateral flow immunoassay. Biosens. Bioelectron. 2022, 205, 114094. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Jo, E.-J.; Jung, C.; Kim, M.-G. Absorption-Modulated SiO2@Au Core–Satellite Nanoparticles for Highly Sensitive Detection of SARS-CoV-2 Nucleocapsid Protein in Lateral Flow Immunosensors. ACS Appl. Mater. Interfaces 2022, 14, 45189–45200. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Jiao, X.; Liang, Z.; Zhao, H.; Zhao, Y.; Xie, J.; Jiang, Y.; Yu, X.; Fang, X.; Dai, X. Lateral Flow Immunoassay Coupled with Copper Enhancement for Rapid and Sensitive SARS-CoV-2 Nucleocapsid Protein Detection. Biosensors 2022, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Zhang, W.; Wang, S.-a.; Li, C.; Li, W.; Liu, J.; Yu, F.; Tao, Y.; Cheng, S.; Xie, H.; et al. A semi-quantitative upconversion nanoparticle-based immunochromatographic assay for SARS-CoV-2 antigen detection. Front. Microbiol. 2023, 14, 1289682. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Liu, Z.; Fang, F.; Zhao, S.; Li, Y.; Xu, M.; Peng, Y.; Chen, H.; Yuan, F.; Zhang, W.; et al. Next-Generation Rapid and Ultrasensitive Lateral Flow Immunoassay for Detection of SARS-CoV-2 Variants. ACS Sens. 2023, 8, 3733–3743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, L.; Du, K.; Zhang, Y.; Wang, J.; Chen, L.; Lyu, Y.; Li, J.; Liu, H.; Huo, J.; et al. Foundation and Clinical Evaluation of a New Method for Detecting SARS-CoV-2 Antigen by Fluorescent Microsphere Immunochromatography. Front. Cell. Infect. Microbiol. 2020, 10, 553837. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Ding, H.; Ding, J.; Xue, Y.; Lu, S.; Lv, H. Preparation of highly specific monoclonal antibodies against SARS-CoV-2 nucleocapsid protein and the preliminary development of antigen detection test strips. J. Med. Virol. 2022, 94, 1633–1640. [Google Scholar] [CrossRef]
- Zhu, J.; Guo, G.; Liu, J.; Li, X.; Yang, X.; Liu, M.; Fu, C.; Zeng, J.; Li, J. One-pot synthesized Au@Pt nanostars-based lateral flow immunoassay for colorimetric and photothermal dual-mode detection of SARS-CoV-2 nucleocapsid antibody. Anal. Chim. Acta 2024, 1292, 342241. [Google Scholar] [CrossRef]
- Xie, Z.; Feng, S.; Pei, F.; Xia, M.; Hao, Q.; Liu, B.; Tong, Z.; Wang, J.; Lei, W.; Mu, X. Magnetic/fluorescent dual-modal lateral flow immunoassay based on multifunctional nanobeads for rapid and accurate SARS-CoV-2 nucleocapsid protein detection. Anal. Chim. Acta 2022, 1233, 340486. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Dong, L.; Feng, X.; Yang, Y.; Wang, X.; Niu, C.; Liang, Z.; Qu, W.; Zou, Q.; Dai, X.; et al. Relationship between SARS-CoV-2 nucleocapsid protein and N gene and its application in antigen testing kits evaluation. Talanta 2023, 258, 124462. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Huang, Y.; Jiang, Z.; Wu, F.; Ma, L. Deciphering the Subtype Differentiation History of SARS-CoV-2 Based on a New Breadth-First Searching Optimized Alignment Method Over a Global Data Set of 24,768 Sequences. Front. Genet. 2021, 11, 591833. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]


| Peptide | Sequence |
|---|---|
| CoV-NP1 | PQNQRNAPRITFGGPSDST |
| CoV-NP2 | SWFTALTQHGKEDLKFPRGQGVPINT |
| CoV-NP3 | TRRIRGGDGKMKDLSP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.; Jiang, Y.; Yang, H.; Ma, L. Development of Detection Antibody Targeting the Linear Epitope in SARS-CoV-2 Nucleocapsid Protein with Ultra-High Sensitivity. Int. J. Mol. Sci. 2024, 25, 4436. https://doi.org/10.3390/ijms25084436
Wu F, Jiang Y, Yang H, Ma L. Development of Detection Antibody Targeting the Linear Epitope in SARS-CoV-2 Nucleocapsid Protein with Ultra-High Sensitivity. International Journal of Molecular Sciences. 2024; 25(8):4436. https://doi.org/10.3390/ijms25084436
Chicago/Turabian StyleWu, Feng, Yike Jiang, Hongtian Yang, and Lan Ma. 2024. "Development of Detection Antibody Targeting the Linear Epitope in SARS-CoV-2 Nucleocapsid Protein with Ultra-High Sensitivity" International Journal of Molecular Sciences 25, no. 8: 4436. https://doi.org/10.3390/ijms25084436





