Serotonin Transporter Deficiency Induces Metabolic Alterations in the Ileal Mucosa
Abstract
:1. Introduction
2. Results
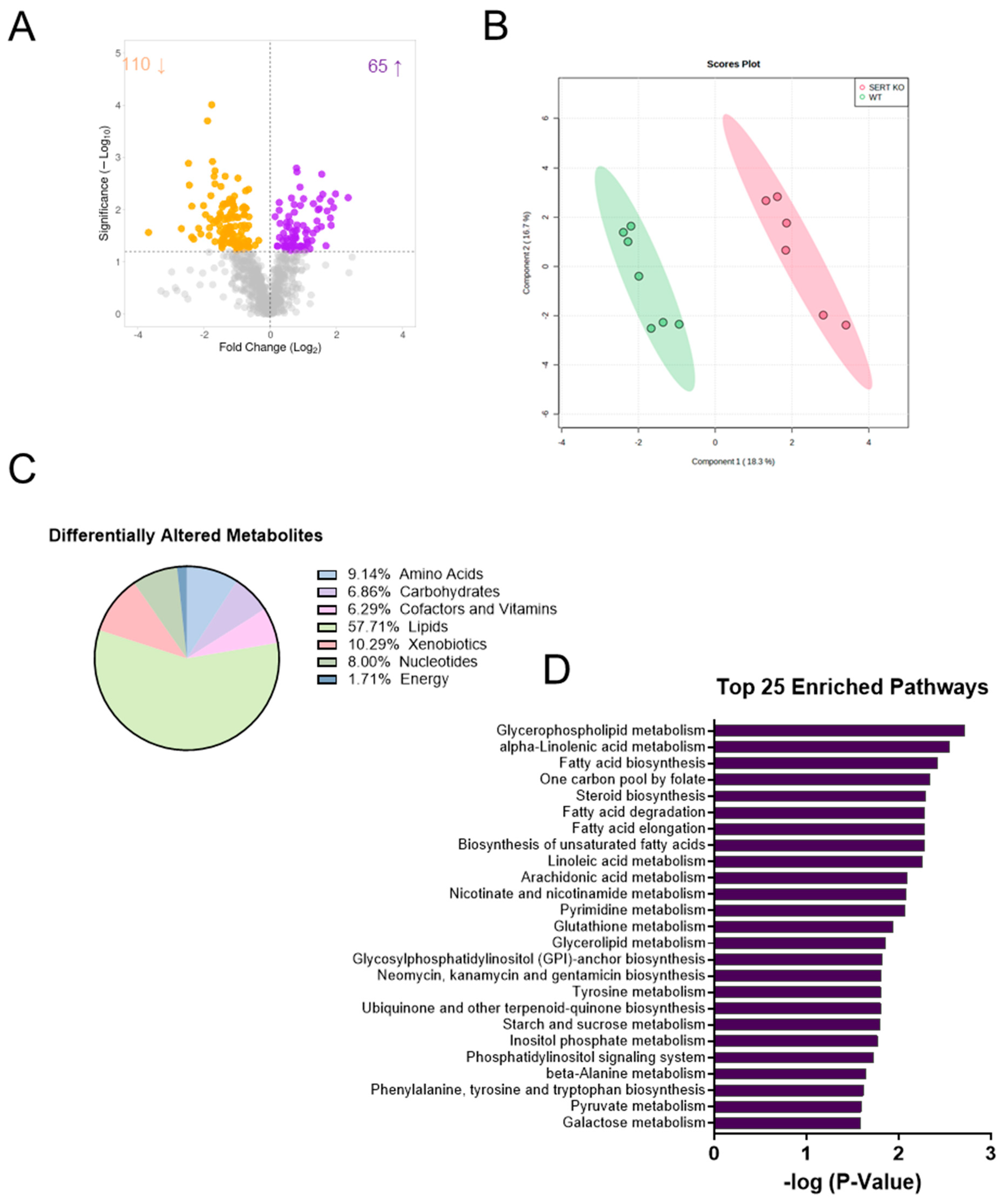
2.1. SERT Deficiency Leads to Distinct Metabolomic Profile in the Ileal Mucosa Characterized by Alterations in Lipid Metabolism
2.2. Lack of SERT Results in Altered Mucosal Glycerols and Long Chain Monounsaturated Fatty Acids
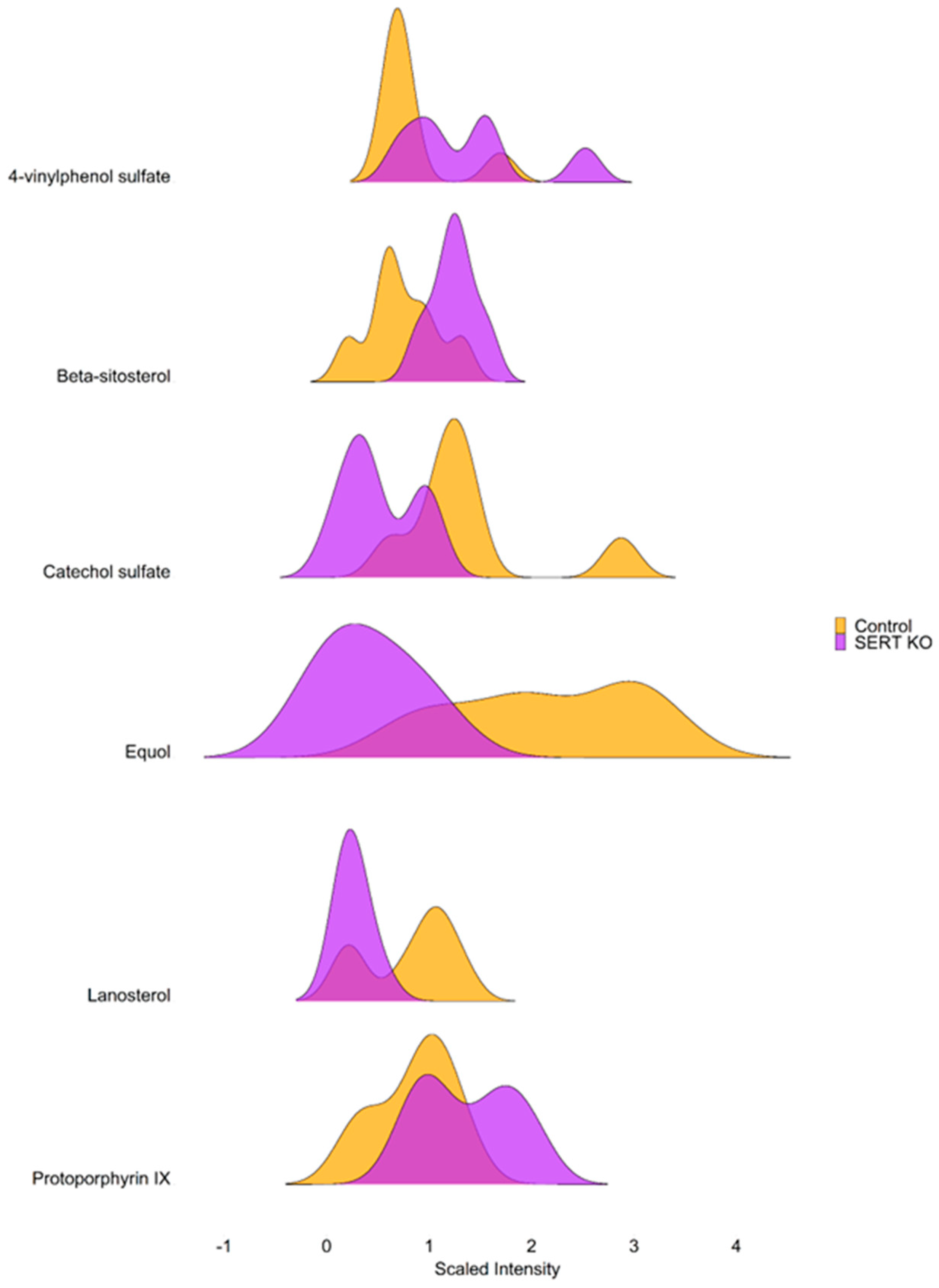
2.3. SERT KO Mice Exhibit Broad Changes in Metabolite Abundances
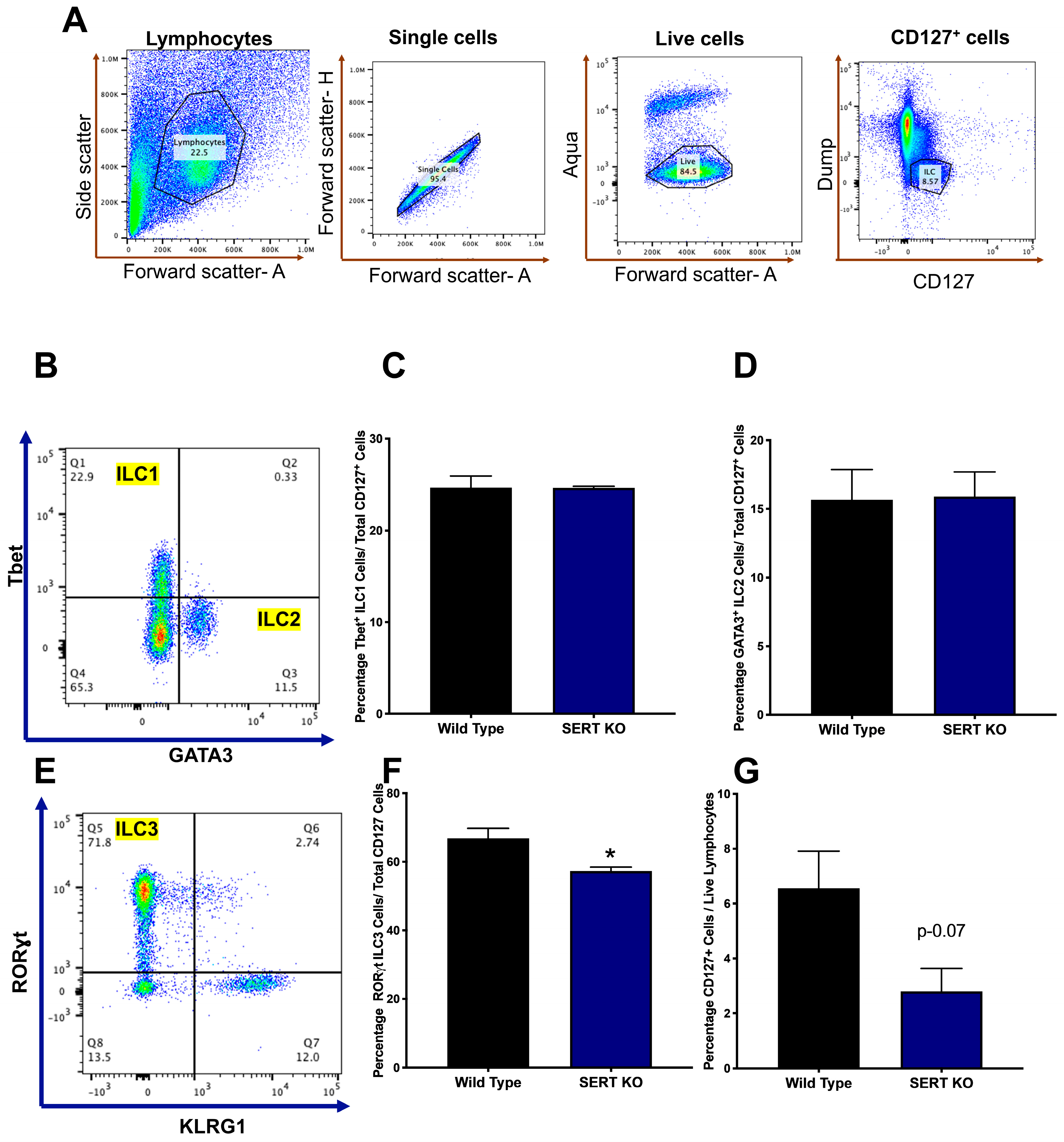
2.4. SERT KO Mice Exhibit Distinct Innate Lymphoid Cell Populations
3. Discussion
3.1. Facts and Perspectives
3.2. Strengths of the Study
4. Materials and Methods
4.1. Animals
4.2. Metabolomic Analysis
4.2.1. Sample Preparation
4.2.2. Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS)
4.2.3. Compound Identification and Metabolite Quantification
4.3. Lamina Propria Lymphocyte Isolation
4.4. Flow Cytometry
4.5. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Terry, N.; Margolis, K.G. Serotonergic Mechanisms Regulating the GI Tract: Experimental Evidence and Therapeutic Relevance. In Handbook of Experimental Pharmacology; Springer: Cham, Switzerland, 2017; Volume 239, pp. 319–342. [Google Scholar] [CrossRef]
- Gershon, M.D. 5-Hydroxytryptamine (Serotonin) in the Gastrointestinal Tract. Curr. Opin. Endocrinol. Diabetes Obes. 2013, 20, 14–21. [Google Scholar] [CrossRef]
- Koopman, N.; Katsavelis, D.; Hove, A.S.T.; Brul, S.; de Jonge, W.J.; Seppen, J. The Multifaceted Role of Serotonin in Intestinal Homeostasis. Int. J. Mol. Sci. 2021, 22, 9487. [Google Scholar] [CrossRef] [PubMed]
- Grondin, J.A.; Khan, W.I. Emerging Roles of Gut Serotonin in Regulation of Immune Response, Microbiota Composition and Intestinal Inflammation. J. Can. Assoc. Gastroenterol. 2024, 7, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Alcaino, C.; Knutson, K.R.; Treichel, A.J.; Yildiz, G.; Strege, P.R.; Linden, D.R.; Li, J.H.; Leiter, A.B.; Szurszewski, J.H.; Farrugia, G.; et al. A Population of Gut Epithelial Enterochromaffin Cells Is Mechanosensitive and Requires Piezo2 to Convert Force into Serotonin Release. Proc. Natl. Acad. Sci. USA 2018, 115, E7632–E7641. [Google Scholar] [CrossRef] [PubMed]
- Bayrer, J.R.; Castro, J.; Venkataraman, A.; Touhara, K.K.; Rossen, N.D.; Morrie, R.D.; Maddern, J.; Hendry, A.; Braverman, K.N.; Garcia-Caraballo, S.; et al. Gut Enterochromaffin Cells Drive Visceral Pain and Anxiety. Nature 2023, 616, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.A.; Sun, E.W.; Martin, A.M.; Keating, D.J. The Ever-Changing Roles of Serotonin. Int. J. Biochem. Cell Biol. 2020, 125, 105776. [Google Scholar] [CrossRef] [PubMed]
- Beyder, A. In Pursuit of the Epithelial Mechanosensitivity Mechanisms. Front. Endocrinol. 2018, 9, 804. [Google Scholar] [CrossRef]
- Wade, P.R.; Chen, J.; Jaffe, B.; Kassem, I.S.; Blakely, R.D.; Gershon, M.D. Localization and Function of a 5-HT Transporter in Crypt Epithelia of the Gastrointestinal Tract. J. Neurosci. 1996, 16, 2352–2364. [Google Scholar] [CrossRef] [PubMed]
- Gajeswski-Kurdziel, P.A.; Walsh, A.E.; Blakely, R.D. Functional and Pathological Consequences of Being Fast on the Uptake: Protein Kinase G and P38α MAPK Regulation of Serotonin Transporters. Curr. Res. Physiol. 2024, 7, 100117. [Google Scholar] [CrossRef] [PubMed]
- Mercado, C.P.; Kilic, F. Molecular Mechanisms of SERT in Platelets: Regulation of Plasma Serotonin Levels. Mol. Interv. 2010, 10, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Gill, R.K.; Pant, N.; Saksena, S.; Singla, A.; Nazir, T.M.; Vohwinkel, L.; Turner, J.R.; Goldstein, J.; Alrefai, W.A.; Dudeja, P.K. Function, Expression, and Characterization of the Serotonin Transporter in the Native Human Intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G254–G262. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.L.; Lerner, A.; Rudnick, G.; Lesch, K.-P. Serotonin Transporter: Gene, Genetic Disorders, and Pharmacogenetics. Mol. Interv. 2004, 4, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Lowy, M.T.; Meltzer, H.Y. 5-Hydroxytryptophan-Induced Cortisol Response and CSF 5-HIAA in Depressed Patients. Am. J. Psychiatry 1987, 144, 334–337. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, R.J.; Zweig, R.M.; Whitehouse, P.J.; Wenk, G.L.; Singer, H.S.; Mayeux, R.; Price, D.L.; Snyder, S.H. Aminergic Systems in Alzheimer’s Disease and Parkinson’s Disease. Ann. Neurol. 1987, 22, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Esmaili, A.; Nazir, S.F.; Borthakur, A.; Yu, D.; Turner, J.R.; Saksena, S.; Singla, A.; Hecht, G.A.; Alrefai, W.A.; Gill, R.K. Enteropathogenic Escherichia Coli Infection Inhibits Intestinal Serotonin Transporter Function and Expression. Gastroenterology 2009, 137, 2074–2083. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, C.; Matheus, N.; Iceta, R.; Mesonero, J.E.; Alcalde, A.I. Lipopolysaccharide Induces Alteration of Serotonin Transporter in Human Intestinal Epithelial Cells. Innate Immun. 2009, 15, 243–250. [Google Scholar] [CrossRef]
- Foley, K.F.; Pantano, C.; Ciolino, A.; Mawe, G.M. IFN-Gamma and TNF-Alpha Decrease Serotonin Transporter Function and Expression in Caco2 Cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G779–G784. [Google Scholar] [CrossRef] [PubMed]
- Matheus, N.; Mendoza, C.; Iceta, R.; Mesonero, J.E.; Alcalde, A.I. Regulation of Serotonin Transporter Activity by Adenosine in Intestinal Epithelial Cells. Biochem. Pharmacol. 2009, 78, 1198–1204. [Google Scholar] [CrossRef]
- Bengel, D.; Murphy, D.L.; Andrews, A.M.; Wichems, C.H.; Feltner, D.; Heils, A.; Mössner, R.; Westphal, H.; Lesch, K.P. Altered Brain Serotonin Homeostasis and Locomotor Insensitivity to 3, 4-Methylenedioxymethamphetamine (“Ecstasy”) in Serotonin Transporter-Deficient Mice. Mol. Pharmacol. 1998, 53, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.L.; Lesch, K.-P. Targeting the Murine Serotonin Transporter: Insights into Human Neurobiology. Nat. Rev. Neurosci. 2008, 9, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Zha, W.; Ho, H.T.B.; Hu, T.; Hebert, M.F.; Wang, J. Serotonin Transporter Deficiency Drives Estrogen-Dependent Obesity and Glucose Intolerance. Sci. Rep. 2017, 7, 1137. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Margolis, K.J.; Gershon, M.D.; Schwartz, G.J.; Sze, J.Y. Reduced Serotonin Reuptake Transporter (SERT) Function Causes Insulin Resistance and Hepatic Steatosis Independent of Food Intake. PLoS ONE 2012, 7, e32511. [Google Scholar] [CrossRef] [PubMed]
- Warden, S.J.; Haney, E.M. Skeletal Effects of Serotonin (5-Hydroxytryptamine) Transporter Inhibition: Evidence from in Vitro and Animal-Based Studies. J. Musculoskelet. Neuronal Interact. 2008, 8, 121–132. [Google Scholar] [PubMed]
- Jørandli, J.W.; Thorsvik, S.; Skovdahl, H.K.; Kornfeld, B.; Sæterstad, S.; Gustafsson, B.I.; Sandvik, A.K.; van Beelen Granlund, A. The Serotonin Reuptake Transporter Is Reduced in the Epithelium of Active Crohn’s Disease and Ulcerative Colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G761–G768. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, S.C.; Mailer, R.; Pabst, O.; Weier, G.; Sedlik, W.; Li, Z.; Chen, J.J.; Murphy, D.L.; Gershon, M.D. Role of Serotonin in Intestinal Inflammation: Knockout of Serotonin Reuptake Transporter Exacerbates 2,4,6-Trinitrobenzene Sulfonic Acid Colitis in Mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G685–G695. [Google Scholar] [CrossRef] [PubMed]
- Haub, S.; Ritze, Y.; Bergheim, I.; Pabst, O.; Gershon, M.D.; Bischoff, S.C. Enhancement of Intestinal Inflammation in Mice Lacking Interleukin 10 by Deletion of the Serotonin Reuptake Transporter. Neurogastroenterol. Motil. 2010, 22, 826-e229. [Google Scholar] [CrossRef] [PubMed]
- Singhal, M.; Turturice, B.A.; Manzella, C.R.; Ranjan, R.; Metwally, A.A.; Theorell, J.; Huang, Y.; Alrefai, W.A.; Dudeja, P.K.; Finn, P.W.; et al. Serotonin Transporter Deficiency Is Associated with Dysbiosis and Changes in Metabolic Function of the Mouse Intestinal Microbiome. Sci. Rep. 2019, 9, 2138. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut Microbiome Structure and Metabolic Activity in Inflammatory Bowel Disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wen, X.-L.; Duan, S.-L. Role of Metabolites Derived from Gut Microbiota in Inflammatory Bowel Disease. World J. Clin. Cases 2022, 10, 2660–2677. [Google Scholar] [CrossRef]
- Aldars-García, L.; Gisbert, J.P.; Chaparro, M. Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review. Pharmaceuticals 2021, 14, 1190. [Google Scholar] [CrossRef] [PubMed]
- Kolho, K.-L.; Pessia, A.; Jaakkola, T.; de Vos, W.M.; Velagapudi, V. Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J. Crohn’s Colitis 2017, 11, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Bjerrum, J.T.; Wang, Y.; Hao, F.; Coskun, M.; Ludwig, C.; Günther, U.; Nielsen, O.H. Metabonomics of Human Fecal Extracts Characterize Ulcerative Colitis, Crohn’s Disease and Healthy Individuals. Metabolomics 2015, 11, 122–133. [Google Scholar] [CrossRef] [PubMed]
- De Preter, V.; Machiels, K.; Joossens, M.; Arijs, I.; Matthys, C.; Vermeire, S.; Rutgeerts, P.; Verbeke, K. Faecal Metabolite Profiling Identifies Medium-Chain Fatty Acids as Discriminating Compounds in IBD. Gut 2015, 64, 447–458. [Google Scholar] [CrossRef]
- Jansson, J.; Willing, B.; Lucio, M.; Fekete, A.; Dicksved, J.; Halfvarson, J.; Tysk, C.; Schmitt-Kopplin, P. Metabolomics Reveals Metabolic Biomarkers of Crohn’s Disease. PLoS ONE 2009, 4, e6386. [Google Scholar] [CrossRef] [PubMed]
- Perez, K.; Ngollo, M.; Rabinowitz, K.; Hammoudi, N.; Seksik, P.; Xavier, R.J.; Daly, M.J.; Dotan, I.; Le Bourhis, L.; Allez, M. Meta-Analysis of IBD Gut Samples Gene Expression Identifies Specific Markers of Ileal and Colonic Diseases. Inflamm. Bowel Dis. 2022, 28, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Manzella, C.R.; Jayawardena, D.; Pagani, W.; Li, Y.; Alrefai, W.A.; Bauer, J.; Jung, B.; Weber, C.R.; Gill, R.K. Serum Serotonin Differentiates Between Disease Activity States in Crohn’s Patients. Inflamm. Bowel Dis. 2020, 26, 1607–1618. [Google Scholar] [CrossRef] [PubMed]
- Manzella, C.; Singhal, M.; Alrefai, W.A.; Saksena, S.; Dudeja, P.K.; Gill, R.K. Serotonin Is an Endogenous Regulator of Intestinal CYP1A1 via AhR. Sci. Rep. 2018, 8, 6103. [Google Scholar] [CrossRef] [PubMed]
- Fernandes Silva, L.; Hokkanen, J.; Vangipurapu, J.; Oravilahti, A.; Laakso, M. Metabolites as Risk Factors for Diabetic Retinopathy in Patients with Type 2 Diabetes: A 12-Year Follow-up Study. J. Clin. Endocrinol. Metab. 2023, 109, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Mayo, B.; Vázquez, L.; Flórez, A.B. Equol: A Bacterial Metabolite from The Daidzein Isoflavone and Its Presumed Beneficial Health Effects. Nutrients 2019, 11, 2231. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.A.; Jensen, C.L.; French, H.T.; Stein, A.R.; Huang, S.-J.; Tolliver, T.J.; Murphy, D.L. Neurochemical, Behavioral and Physiological Effects of Pharmacologically Enhanced Serotonin Levels in Serotonin Transporter (SERT)-Deficient Mice. Psychopharmacology 2008, 201, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-W.; Ho, P.-S.; Kuo, S.-C.; Chen, C.-Y.; Liang, C.-S.; Yen, C.-H.; Huang, C.-C.; Ma, K.-H.; Shiue, C.-Y.; Huang, W.-S.; et al. Disproportionate Reduction of Serotonin Transporter May Predict the Response and Adherence to Antidepressants in Patients with Major Depressive Disorder: A Positron Emission Tomography Study with 4-[18F]-ADAM. Int. J. Neuropsychopharmacol. 2015, 18, pyu120. [Google Scholar] [CrossRef] [PubMed]
- Veniaminova, E.; Cespuglio, R.; Chernukha, I.; Schmitt-Boehrer, A.G.; Morozov, S.; Kalueff, A.V.; Kuznetsova, O.; Anthony, D.C.; Lesch, K.-P.; Strekalova, T. Metabolic, Molecular, and Behavioral Effects of Western Diet in Serotonin Transporter-Deficient Mice: Rescue by Heterozygosity? Front. Neurosci. 2020, 14, 24. [Google Scholar] [CrossRef] [PubMed]
- El Aidy, S.; Ramsteijn, A.S.; Dini-Andreote, F.; van Eijk, R.; Houwing, D.J.; Salles, J.F.; Olivier, J.D.A. Serotonin Transporter Genotype Modulates the Gut Microbiota Composition in Young Rats, an Effect Augmented by Early Life Stress. Front. Cell. Neurosci. 2017, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- McFie, P.J.; Patel, A.; Stone, S.J. The Monoacylglycerol Acyltransferase Pathway Contributes to Triacylglycerol Synthesis in HepG2 Cells. Sci. Rep. 2022, 12, 4943. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Armenia, S.J.; Shaughnessy, M.P.; Greig, C.J.; Cowles, R.A. Potentiation of Serotonin Signaling Leads to Increased Carbohydrate and Lipid Absorption in the Murine Small Intestine. J. Pediatr. Surg. 2019, 54, 1245–1249. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Kambayashi, T. The Immunomodulatory Functions of Diacylglycerol Kinase ζ. Front. Cell Dev. Biol. 2016, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Erion, D.M.; Shulman, G.I. Diacylglycerol-Mediated Insulin Resistance. Nat. Med. 2010, 16, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Monnerie, S.; Comte, B.; Ziegler, D.; Morais, J.A.; Pujos-Guillot, E.; Gaudreau, P. Metabolomic and Lipidomic Signatures of Metabolic Syndrome and Its Physiological Components in Adults: A Systematic Review. Sci. Rep. 2020, 10, 669. [Google Scholar] [CrossRef]
- Meikle, P.J.; Wong, G.; Barlow, C.K.; Weir, J.M.; Greeve, M.A.; MacIntosh, G.L.; Almasy, L.; Comuzzie, A.G.; Mahaney, M.C.; Kowalczyk, A.; et al. Plasma Lipid Profiling Shows Similar Associations with Prediabetes and Type 2 Diabetes. PLoS ONE 2013, 8, e74341. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liao, Z.; Mo, Y.; Zhao, W.; Zhou, X.; Xiao, X.; Cui, W.; Feng, G.; Zhong, S.; Liang, Y.; et al. Inactivation of 3-Hydroxybutyrate Dehydrogenase Type 2 Promotes Proliferation and Metastasis of Nasopharyngeal Carcinoma by Iron Retention. Br. J. Cancer 2020, 122, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Tan, Y.-Y.; Ding, Y.; Fang, Y.; Yang, X.; Fang, J.; Xu, D.-C.; Zhang, H.; Lu, W.; Li, M.; et al. β-Sitosterol Improves Experimental Colitis in Mice with a Target against Pathogenic Bacteria. J. Cell. Biochem. 2019, 120, 5687–5694. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.; Devarajan, N.; Rajagopal, P.; Babu, S.; Ganesan, S.K.; Veeraraghavan, V.P.; Palanisamy, C.P.; Cui, B.; Periyasamy, V.; Chandrasekar, K. β-Sitosterol Circumvents Obesity Induced Inflammation and Insulin Resistance by down-Regulating IKKβ/NF-κB and JNK Signaling Pathway in Adipocytes of Type 2 Diabetic Rats. Molecules 2021, 26, 2101. [Google Scholar] [CrossRef] [PubMed]
- Kuryłowicz, A. The Role of Isoflavones in Type 2 Diabetes Prevention and Treatment—A Narrative Review. Int. J. Mol. Sci. 2021, 22, 218. [Google Scholar] [CrossRef] [PubMed]
- Hamaura, K.; Murakami, H.; Tamura, A.; Matsuki, K.; Sato, E.; Tanabe, J.; Yanagimachi, M.; Oishi, M.; Iino, K.; Okuyama, S.; et al. Association between Equol Producers and Type 2 Diabetes Mellitus among Japanese Older Adults. J. Diabetes Investig. 2023, 14, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Susanti, D.; Amiroudine, M.Z.A.M.; Rezali, M.F.; Taher, M. Friedelin and Lanosterol from Garcinia Prainiana Stimulated Glucose Uptake and Adipocytes Differentiation in 3T3-L1 Adipocytes. Nat. Prod. Res. 2013, 27, 417–424. [Google Scholar] [CrossRef]
- Kang, L.; Pang, J.; Zhang, X.; Liu, Y.; Wu, Y.; Wang, J.; Han, D. L-Arabinose Attenuates LPS-Induced Intestinal Inflammation and Injury through Reduced M1 Macrophage Polarization. J. Nutr. 2023, 153, 3327–3340. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pan, H.; Liu, J.-X.; Li, T.; Liu, S.; Shi, W.; Sun, C.; Fan, M.; Xue, L.; Wang, Y.; et al. L-Arabinose Inhibits Colitis by Modulating Gut Microbiota in Mice. J. Agric. Food Chem. 2019, 67, 13299–13306. [Google Scholar] [CrossRef] [PubMed]
- Bethlehem, L.; van Echten-Deckert, G. Ectoines as Novel Anti-Inflammatory and Tissue Protective Lead Compounds with Special Focus on Inflammatory Bowel Disease and Lung Inflammation. Pharmacol. Res. 2021, 164, 105389. [Google Scholar] [CrossRef] [PubMed]
- Hamaguchi, M.; Okamura, T.; Fukuda, T.; Nishida, K.; Yoshimura, Y.; Hashimoto, Y.; Ushigome, E.; Nakanishi, N.; Majima, S.; Asano, M.; et al. Group 3 Innate Lymphoid Cells Protect Steatohepatitis from High-Fat Diet Induced Toxicity. Front. Immunol. 2021, 12, 648754. [Google Scholar] [CrossRef] [PubMed]
- Jarade, A.; Di Santo, J.P.; Serafini, N. Group 3 Innate Lymphoid Cells Mediate Host Defense against Attaching and Effacing Pathogens. Curr. Opin. Microbiol. 2021, 63, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Withers, D.R.; Hepworth, M.R. Group 3 Innate Lymphoid Cells: Communications Hubs of the Intestinal Immune System. Front. Immunol. 2017, 8, 1298. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, A.; Gnafakis, S.; Shomrat, O. Innate Lymphoid Cell-Epithelial Cell Modules Sustain Intestinal Homeostasis. Immunity 2020, 52, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Bostick, J.W.; Zhou, L. Regulation of Innate Lymphoid Cells by Aryl Hydrocarbon Receptor. Front. Immunol. 2018, 8, 311218. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Milburn, M.V.; Ryals, J.A.; Lonergan, S.C.; Mitchell, M.W.; Wulff, J.E.; Alexander, D.C.; Evans, A.M.; Bridgewater, B.; Miller, L.; et al. Plasma Metabolomic Profiles Enhance Precision Medicine for Volunteers of Normal Health. Proc. Natl. Acad. Sci. USA 2015, 112, E4901–E4910. [Google Scholar] [CrossRef] [PubMed]
- Calzadilla, N.; Qazi, A.; Sharma, A.; Mongan, K.; Comiskey, S.; Manne, J.; Youkhana, A.G.; Khanna, S.; Saksena, S.; Dudeja, P.K.; et al. Mucosal Metabolomic Signatures in Chronic Colitis: Novel Insights into the Pathophysiology of Inflammatory Bowel Disease. Metabolites 2023, 13, 873. [Google Scholar] [CrossRef] [PubMed]
- Calzadilla, N.; Zilberstein, N.; Hanscom, M.; Al Rashdan, H.T.; Chacra, W.; Gill, R.K.; Alrefai, W.A. Serum Metabolomic Analysis in Cirrhotic Alcohol-Associated Liver Disease Patients Identified Differentially Altered Microbial Metabolites and Novel Potential Biomarkers for Disease Severity. Dig. Liver Dis. 2023, in press. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Yu, G.; Xu, C.; Zhang, D.; Ju, F.; Ni, Y. MetOrigin: Discriminating the Origins of Microbial Metabolites for Integrative Analysis of the Gut Microbiome and Metabolome. iMeta 2022, 1, e10. [Google Scholar] [CrossRef]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A Web Server for Metabolomic Data Analysis and Interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]


| Name | Fold Change Relative Abundance | p-Value | q-Value |
|---|---|---|---|
| Tryptophan | 0.78 | 0.1489 | 0.1998 |
| Serotonin | 3.15 | 0.1147 | 0.1833 |
| Indoleacetate | 0.69 | 0.0987 | 0.1671 |
| Xanthurenate | 0.53 * | 0.0406 | 0.1236 |
| Indoleacetylglycine | 0.4 ** | 0.0023 | 0.09 |
| Kynurenate | 0.6 ** | 0.0088 | 0.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calzadilla, N.; Jayawardena, D.; Qazi, A.; Sharma, A.; Mongan, K.; Comiskey, S.; Eathara, A.; Saksena, S.; Dudeja, P.K.; Alrefai, W.A.; et al. Serotonin Transporter Deficiency Induces Metabolic Alterations in the Ileal Mucosa. Int. J. Mol. Sci. 2024, 25, 4459. https://doi.org/10.3390/ijms25084459
Calzadilla N, Jayawardena D, Qazi A, Sharma A, Mongan K, Comiskey S, Eathara A, Saksena S, Dudeja PK, Alrefai WA, et al. Serotonin Transporter Deficiency Induces Metabolic Alterations in the Ileal Mucosa. International Journal of Molecular Sciences. 2024; 25(8):4459. https://doi.org/10.3390/ijms25084459
Chicago/Turabian StyleCalzadilla, Nathan, Dulari Jayawardena, Aisha Qazi, Anchal Sharma, Kai Mongan, Shane Comiskey, Abhijith Eathara, Seema Saksena, Pradeep K. Dudeja, Waddah A. Alrefai, and et al. 2024. "Serotonin Transporter Deficiency Induces Metabolic Alterations in the Ileal Mucosa" International Journal of Molecular Sciences 25, no. 8: 4459. https://doi.org/10.3390/ijms25084459






