Grape SnRK2.7 Positively Regulates Drought Tolerance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results Analysis
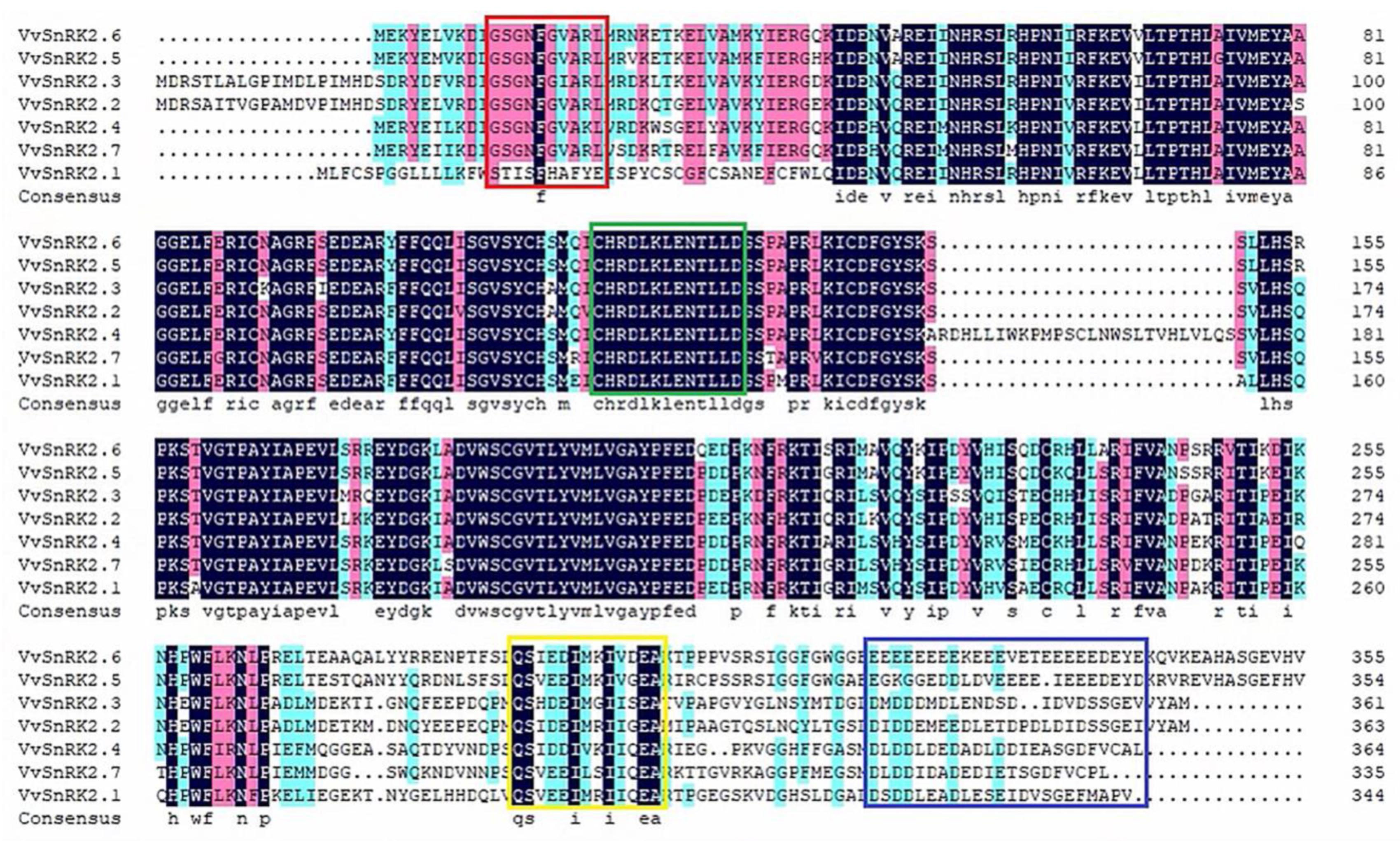
2.1. Evolutionary Analysis of VvSNRK2 Genes
2.2. Analysis of Physicochemical Properties of VvSnRK2.7
2.3. Subcellular Localization of VvSNRK2.7
2.4. Yeast Library Screening and PCR Assay
2.5. Validation of VvSnRK2.7 Interactions with bZIP Protein
2.6. pCAMBIA1300-VvSnRK2.7 Overexpression Vector Construction and Characterization of Transgenic Arabidopsis
2.7. Morphological Observation and Expression Analysis of Transgenic Arabidopsis after Drought Stress
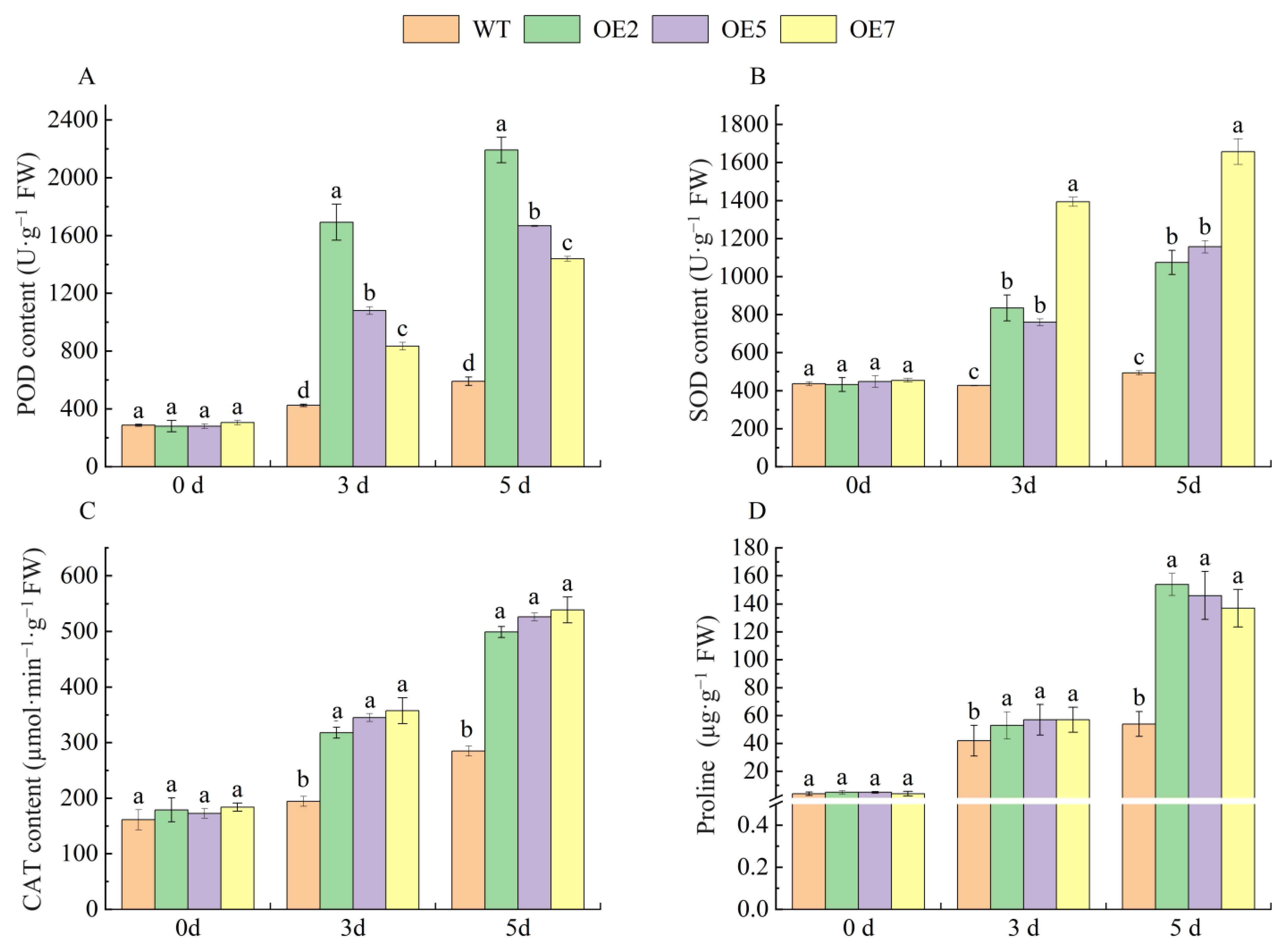
2.8. VvSnRK2.7 Increased Antioxidant Enzyme Activities and Proline Content under Drought Stress
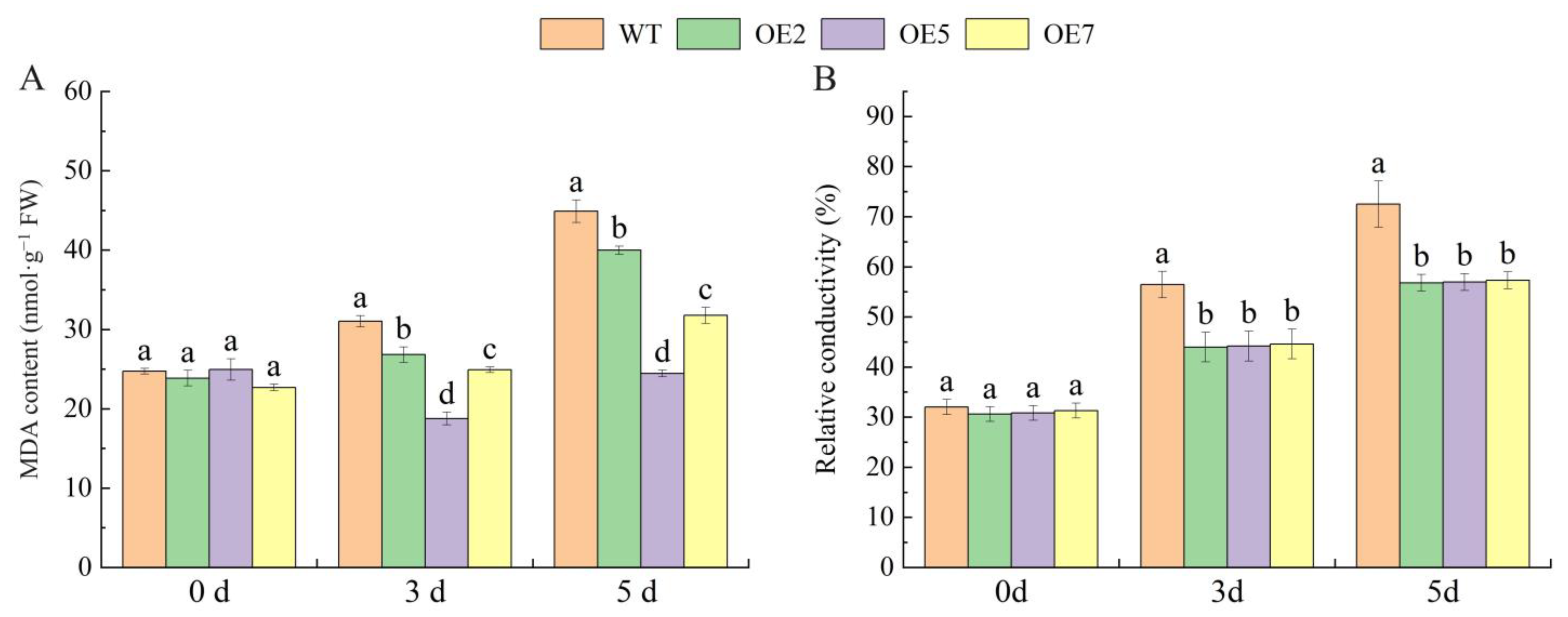
2.9. Overexpression of the VvSnRK2.7 Alters Arabidopsis Leaf MDA Content and Relative Conductance
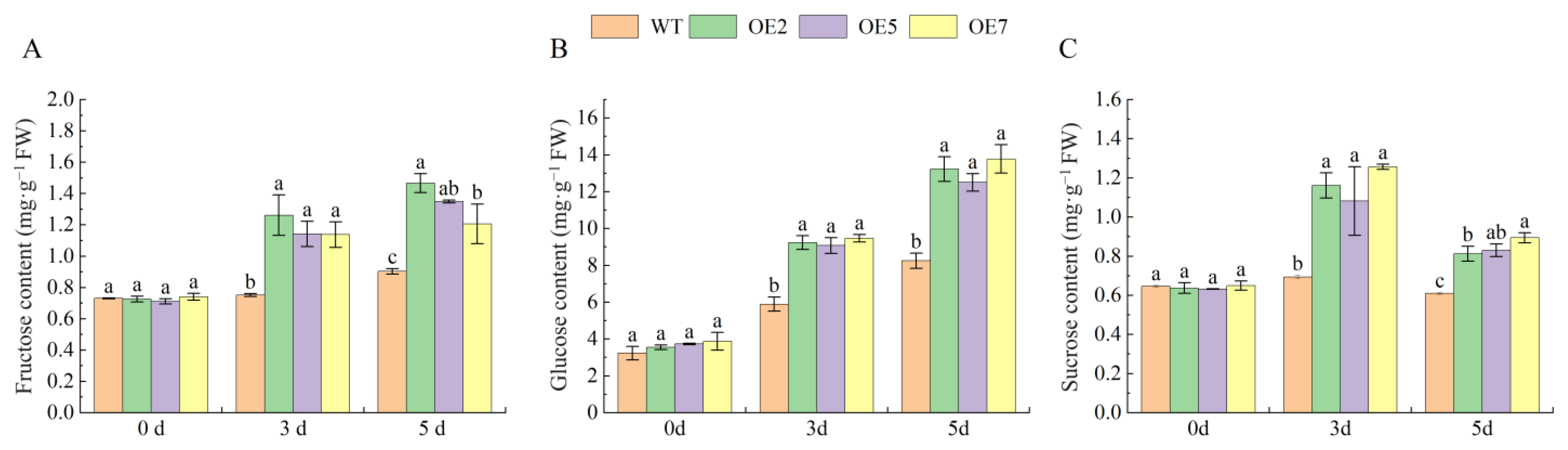
2.10. Overexpression of VvSnRK2.7 Increases Leaf Sugar Content in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Test Materials
4.2. Obtaining and Cloning of Grape VvSnRK2.7
4.3. Overexpression and Subcellular Localization of Recombinant Plasmid Transformation
4.4. Functional Analysis of Drought Resistance in Transgenic Arabidopsis
4.5. Measurement of Relevant Indicators after Stress
4.6. Screening of VvSnRK2.7-Interacting Proteins with a Mating Assay
4.7. Validation of the VvSnRK2.7 with VvbZIP
4.8. qRT-PCR Analysis
4.9. Data Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Song, J.; Li, C.; Ma, J.; Liu, J.; Zhu, X.; Li, J.; He, F.; Yang, C. Genome-Wide Identification and Expression Profile Analysis of the SnRK2 Gene Family in Nicotiana tabacum. Biochem. Genet. 2022, 60, 1511–1526. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.J.; Man, L.L.; Cao, S.; Liu, P.; Li, Z.G.; Wang, X.D. Heterologous expression of an Agropyron cristatum SnRK2 protein kinase gene (AcSnRK2.11) increases freezing tolerance in transgenic yeast and tobacco. 3 Biotech 2020, 10, 209. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; An, L.; Li, W. The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014, 33, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017, 40, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Li, Y.; Rehman, S.U.; Miao, L.; Zhang, Y.; Chen, X.; Yu, C.; Wang, J.; Li, C.; Jing, R. The Sucrose Non-Fermenting 1-Related Protein Kinase 2 (SnRK2) Genes Are Multifaceted Players in Plant Growth, Development and Response to Environmental Stimuli. Plant Cell Physiol. 2020, 61, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.J.; Sun, M.H.; Lu, J.; Kang, H.; You, C.X.; Hao, Y.J. An apple sucrose transporter MdSUT2.2 is a phosphorylation target for protein kinase MdCIPK22 in response to drought. Plant Biotechnol. J. 2019, 17, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.F.; Li, Y.B.; Nai, G.J.; Liang, G.P.; Ma, Z.H.; Chen, B.H.; Mao, J. Changes and response mechanism of sugar and organic acids in fruits under water deficit stress. PeerJ 2022, 10, e13691. [Google Scholar] [CrossRef] [PubMed]
- Diédhiou, C.J.; Popova, O.V.; Dietz, K.J.; Golldack, D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zhang, D.F.; Li, H.Y.; Liu, Y.H.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize SnRK2 protein kinase gene confers enhanced salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2011, 30, 1683–1699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mao, X.; Wang, C.; Jing, R. Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS ONE 2010, 5, e16041. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e5. [Google Scholar] [CrossRef] [PubMed]
- Giribaldi, M.; Gény, L.; Delrot, S.; Schubert, A. Proteomic analysis of the effects of ABA treatments on ripening Vitis vinifera berries. J. Exp. Bot. 2010, 61, 2447–2458. [Google Scholar] [CrossRef] [PubMed]
- Rook, F.; Hadingham, S.A.; Li, Y.; Bevan, M.W. Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ. 2006, 29, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.R.; Shin, M.; Shen, J.Q. The wheat PKABA1-interacting factor TaABF1 mediates both abscisic acid-suppressed and abscisic acid-induced gene expression in bombarded aleurone cells. Plant Mol. Biol. 2008, 68, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005, 44, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein kinases—Key regulators of plant response to abiotic stresses. OMICS 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Murcia, G.N.; Pontin, M.; Piccoli, P. Role of ABA and Gibberellin A3 on gene expression pattern of sugar transporters and invertases in Vitis vinifera cv. Malbec during berry ripening. Plant Growth Regul. 2018, 84, 275–283. [Google Scholar] [CrossRef]
- Xiang, D.J.; Man, L.L.; Cao, S.; Liu, P.; Li, Z.G.; Wang, X.D. Ectopic expression of an oat SnRK2 gene, AsSnRK2D, enhances dehydration and salinity tolerance in tobacco by modulating the expression of stress-related genes. Braz. J. Bot. 2020, 43, 429–446. [Google Scholar] [CrossRef]
- Tak, H.; Mhatre, M. Cloning and molecular characterization of a putative bZIP transcription factor VvbZIP23 from Vitis vinifera. Protoplasma 2013, 250, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, H.; Tian, S.; Chang, X.; Jing, R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 2010, 61, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Yoshida, R.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 17306–17311. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Mao, X.; Zhang, H.; Chen, S.; Zhai, C.; Yang, S.; Jing, R. Cloning and characterization of TaSnRK2.3, a novel SnRK2 gene in common wheat. J. Exp. Bot. 2013, 64, 2063–2080. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Jossier, M.; Bouly, J.P.; Meimoun, P.; Arjmand, A.; Lessard, P.; Hawley, S.; Grahame Hardie, D.; Thomas, M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 2009, 59, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Lu, S.; Li, W.; Nai, G.; Ma, Z.; Li, Y.; Chen, B.; Mao, J. Transcriptome and metabolites analysis of water-stressed grape berries at different growth stages. Physiol. Plant. 2023, 175, e13910. [Google Scholar] [CrossRef]
- Lu, S.; Wang, P.; Nai, G.; Li, Y.; Su, Y.; Liang, G.; Chen, B.; Mao, J. Insight into VvGH3 genes evolutional relationship from monocotyledons and dicotyledons reveals that VvGH3-9 negatively regulates the drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2022, 172, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Fang, W.; Shi, S.; Zhao, Y.; Li, X.; Xiao, K. Wheat WRKY Type Transcription Factor Gene TaWRKY1 is Essential in Mediating Drought Tolerance Associated with an ABA-Dependent Pathway. Plant Mol. Biol. Report. 2016, 34, 1111–1126. [Google Scholar] [CrossRef]
- Rohde, P.; Hincha, D.K.; Heyer, A.G. Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J. 2004, 38, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, P.; Dai, Y.; Liu, X.; Lu, S.; Guo, L.; Gou, H.; Mao, J. Evolution of the 14-3-3 gene family in monocotyledons and dicotyledons and validation of MdGRF13 function in transgenic Arabidopsis thaliana. Plant Cell Rep. 2023, 42, 1345–1364. [Google Scholar] [CrossRef] [PubMed]




| Gene | Amino Acid Number | Molecular Weight | Isoelectric Point | Full Length/bp | Instability Index |
|---|---|---|---|---|---|
| PNVvSnRK2.7 | 355 | 40.61 | 6.05 | 5680 | 45.07 |
| RGVvSnRK2.1 | 355 | 40.62 | 6.1 | 5683 | 47.12 |
| ChVvSnRk2.1 | 355 | 40.68 | 6.05 | 5690 | 47.12 |
| Gene | Primer Sequence (5′-3′) | Purpose |
|---|---|---|
| VvSnRK2.7-EGFP | 5′-gagctcggtacccggggatccATGGAGAAGTATGAAATGGTGAAGG | Transient expression |
| 3′-ggtgtcgactctagaggatccACTAACGTGAAATTCTCCGCTTG | Transient expression | |
| pCAMBIA1300-VvSnRK2.7 | 5′-gagctcggtacccggggatccATGGAGAAGTATGAAATGGTGAAGG | Overexpression |
| 3′-ggtgtcgactctagaggatccTTAACTAACGTGAAATTCTCCGCT | Overexpression | |
| pGBKT7-VvSnRK2.7 | 5′-aggccgaattcccggggatccttATGGAGAAGTATGAAATGGTGAAGG | BD vector primers |
| 3′-ccgctgcaggtcgacggatccTTAACTAACGTGAAATTCTCCGCT | BD vector primers | |
| pGADT7-VvbZIP | 5′-gtgggcatcgatacgggatccatATGTTGTCATCAACAGGTGGCG | AD vector primers |
| 3′-cagctcgagctcgatggatccTCAAAATGGGGCTGTTGAAGTT | AD vector primers | |
| qRT-AtActin | 5′-CTTGCACCAAGCAGCATGAA | qRT-PCR primers |
| 3′-CCGATCCAGACACTGTACTTCCTT | qRT-PCR primers | |
| qRT-VvSnRK2.7 | 5′-AGAAGAGGGTAAAGGCGGTGAGG | qRT-PCR primers |
| 3′-R-CGCTTGCATGGACTTCCCTAACC | qRT-PCR primers |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, G.; Ma, W.; Nai, G.; Liang, G.; Lu, S.; Ma, Z.; Mao, J.; Chen, B. Grape SnRK2.7 Positively Regulates Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2024, 25, 4473. https://doi.org/10.3390/ijms25084473
Lan G, Ma W, Nai G, Liang G, Lu S, Ma Z, Mao J, Chen B. Grape SnRK2.7 Positively Regulates Drought Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2024; 25(8):4473. https://doi.org/10.3390/ijms25084473
Chicago/Turabian StyleLan, Guanquecailang, Weifeng Ma, Guojie Nai, Guoping Liang, Shixiong Lu, Zonghuan Ma, Juan Mao, and Baihong Chen. 2024. "Grape SnRK2.7 Positively Regulates Drought Tolerance in Transgenic Arabidopsis" International Journal of Molecular Sciences 25, no. 8: 4473. https://doi.org/10.3390/ijms25084473





