Analysis and Characterization of the Extracellular Vesicles Released in Non-Cancer Diseases Using Matrix-Assisted Laser Desorption Ionization/Mass Spectrometry
Abstract
:1. Introduction
2. EV Isolation Techniques
2.1. Size-Based Methods
2.2. Novel Techniques
3. MALDI/MS-Based Biomarker Analysis of EVs from Non-Cancer Cells
3.1. Applications
3.1.1. Various Applications
3.1.2. Hormonal and Metabolic Disorders
3.1.3. Neurodegenerative Diseases
3.1.4. Immune System, Infectious Disease, and Vaccines
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Jabeen, S.; Thirumalai, V. The interplay between electrical and chemical synaptogenesis. J. Neurophysiol. 2018, 120, 1914–1922. [Google Scholar] [CrossRef]
- Dang, V.D.; Jella, K.K.; Ragheb, R.R.T.; Denslow, N.D.; Alli, A.A. Lipidomic and proteomic analysis of exosomes from mouse cortical collecting duct cells. FASEB J. 2017, 31, 5399–5408. [Google Scholar] [CrossRef]
- Ridder, K.; Keller, S.; Dams, M.; Rupp, A.K.; Schlaudraff, J.; Del Turco, D.; Starmann, J.; Macas, J.; Karpova, D.; Devraj, K.; et al. Extracellular Vesicle-Mediated Transfer of Genetic Information between the Hematopoietic System and the Brain in Response to Inflammation. PLoS Biol. 2014, 12, e1001874. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- EL Andaloussi, S.; Mager, I.; Breakefield, X.; Wood, M. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Medeirosa, T.; Myettea, R.L.; Almeida, J.R.; Silva, A.A.; Burger, D. Extracellular Vesicles: Cell-Derived Biomarkers of Glomerular and Tubular Injury. Cell Physiol. Biochem. 2020, 54, 88–109. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Huang, G.; Lin, G.; Zhu, Y.; Duand, W.; Jin, D. Emerging technologies for profiling extracellular vesicle heterogeneity. Lab Chip 2020, 20, 2423–2437. [Google Scholar] [CrossRef]
- Yokoi, A.; Ochiya, T. Exosomes and extracellular vesicles: Rethinking the essential values in cancer biology. Semin. Cancer Biol. 2021, 74, 79–91. [Google Scholar] [CrossRef]
- Thakur, A.; Parra, D.C.; Motallebnejad, P.; Brocchi, M.; Chen, H.J. Exosomes: Small vesicles with big roles in cancer, vaccine development, and therapeutics. Bioact. Mater. 2022, 10, 281–294. [Google Scholar] [CrossRef]
- Szwedowicz, U.; Lapinska, Z.; Gajewska-Naryniecka, A.; Choromanska, A. Exosomes and Other Extracellular Vesicles with High Therapeutic Potential: Their Applications in Oncology, Neurology, and Dermatology. Molecules 2022, 27, 1303. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Sidhom, K.; Obi, P.O.; Saleem, A. A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option? Int. J. Mol. Sci. 2020, 21, 6466. [Google Scholar] [CrossRef]
- Martins, T.S.; Vaz, M.; Henriques, A.G. A review on comparative studies addressing exosome isolation methods from body fuids. Anal. Bioanal. Chem. 2023, 415, 1239–1263. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Ehab, S.; Cho, J. Relevance of multilamellar and multicompartmental vesicles in biological fluids: Understanding the significance of proportional variations and disease correlation. Biomark. Res. 2023, 11, 77. [Google Scholar] [CrossRef]
- Cho, Y.T.; Su, H.; Huang, T.L.; Chen, H.C.; Wu, P.W.J.; Wu, C.; Wu, D.C.; Shiea, J. Matrix-assisted laser desorption ionization/time-of-flight mass spectrometry for clinical diagnosis. Clin. Chim. Acta 2013, 415, 266–275. [Google Scholar] [CrossRef]
- Zambonin, C.; Aresta, A. MALDI-TOF/MS Analysis of Non-Invasive Human Urine and Saliva Samples for the Identification of New Cancer Biomarkers. Molecules 2022, 27, 1925. [Google Scholar] [CrossRef]
- Jalaludin, I.; Lubman, D.M.; Kim, J. MALDI/MS: A Powerful but Underutilized Mass Spectrometric Technique for Exosome Research. Mass Spectrom. Lett. 2021, 12, 93–105. [Google Scholar]
- Zambonin, C. MALDI-TOF/MS analysis of extracellular vesicles released by cancer cells. Appl. Sci. 2022, 12, 6149. [Google Scholar] [CrossRef]
- Cvjetkovic, A.; Lötvall, J.; Lässer, C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J. Extracell. Vesicles 2014, 3, 23111. [Google Scholar] [CrossRef]
- Kuo, W.P.; Jia, S. Extracellular Vesicles: Methods and Protocols; Humana Press: New York, NY, USA, 2017; Volume 1660. [Google Scholar]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Rood, I.M.; Deegens, J.K.J.; Merchant, M.L.; Tamboer, W.P.M.; Wilkey, D.W.; Wetzels, J.F.M.; Klein, J.B. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010, 78, 810–816. [Google Scholar] [CrossRef]
- Piotrowska, M.; Ciura, K.; Zalewska, M.; Dawid, M.; Correia, B.; Sawicka, P.; Lewczuk, B.; Kasprzyk, J.; Sola, L.; Piekoszewski, W.; et al. Capillary zone electrophoresis of bacterial extracellular vesicles: A proof of concept. J. Chromatogr. A 2020, 1621, 461047. [Google Scholar] [CrossRef]
- Vergauwen, G.; Dhondt, B.; Van Deun, J.; De Smedt, E.; Berx, G.; Timmerman, E.; Gevaert, K.; Miinalainen, I.; Cocquyt, V.; Braems, G.; et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci. Rep. 2017, 7, 2704. [Google Scholar] [CrossRef]
- Cheruvanky, A.; Zhou, H.; Pisitkun, T.; Kopp, J.B.; Knepper, M.A.; Yuen, P.S.; Star, R.A. Rapid isolation of urinary exosome biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Renal Physiol. 2007, 292, F1657–F1661. [Google Scholar] [CrossRef]
- Brezgin, S.; Parodi, A.; Kostyusheva, A.; Ponomareva, N.; Lukashev, A.; Sokolova, D.; Pokrovsky, V.S.; Slatinskaya, O.; Maksimov, G.; Zamyatnin, A.A.; et al. Technological aspects of manufacturing and analytical control of biological nanoparticles. Biotechnol. Adv. 2023, 64, 108122–108148. [Google Scholar] [CrossRef]
- Han, Z.; Peng, C.; Yi, J.; Zhang, D.; Xiang, X.; Peng, X.; Su, B.; Liu, B.; Shen, Y.; Qiao, L. Highly efficient exosome purification from human plasma by tangential flow filtration based microfluidic chip. Sens. Actuators B Chem. 2021, 333, 129563. [Google Scholar] [CrossRef]
- Hua, X.; Zhu, Q.; Liu, Y.; Zhou, S.; Huang, P.; Li, Q.; Liu, S. A double tangential flow filtration-based microfluidic device for highly efficient separation and enrichment of exosomes. Anal. Chim. Acta 2023, 1258, 341160. [Google Scholar] [CrossRef]
- Ye, W.; Pan, R.; Shi, K.Q.; Li, H.P.; Lee, L.P.; Liu, F. Isolation of small extracellular vesicles from a drop of plasma via EXODUS and their fingerprint proteomics profiling by MALDI-TOF MS. Biosens. Bioelectron. X 2022, 10, 100099. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, Q.; Cheng, L.; Wang, Y.; Li, M.; Yang, Q.; Hu, L.; Lou, D.; Li, J.; Dong, X.; et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods 2021, 18, 212–218. [Google Scholar] [CrossRef]
- Liu, W.; Ma, Z.; Kang, X. Current status and outlook of advances in exosome isolation. Anal. Bioanal. Chem. 2022, 414, 7123–7141. [Google Scholar] [CrossRef]
- Royo, F.; Théry, C.; Falcón-Pérez, J.M.; Nieuwland, R.; Witwer, K.W. Methods for Separation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey Performed by the ISEV Rigor and Standardization Subcommittee. Cells 2020, 9, 1955. [Google Scholar] [CrossRef]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef]
- Castelli, F.A.; Rosati, G.; Moguet, C.; Fuentes, C.; Marrugo-Ramírez, J.; Lefebvre, T.; Volland, H.; Merkoçi, A.; Simon, S.; Fenaille, F.; et al. Metabolomics for personalized medicine: The input of analytical chemistry from biomarker discovery to point-of-care tests. Anal. Bioanal. Chem. 2022, 414, 759–789. [Google Scholar] [CrossRef]
- Navajas, R.; Corrales, F.J.; Paradela, A. Serum Exosome Isolation by Size-Exclusion Chromatography for the Discovery and Validation of Preeclampsia-Associated Biomarkers BT—Proteomics for Biomarker Discovery: Methods and Protocols; Brun, V., Couté, Y., Eds.; Springer: New York, NY, USA, 2019; pp. 39–50. [Google Scholar]
- Darie-Ion, L.; Whitham, D.; Jayathirtha, M.; Rai, Y.; Neagu, A.N.; Darie, C.C.; Petre, B.A. Applications of MALDI/MS/MS-Based Proteomics in Biomedical Research. Molecules 2022, 27, 6196. [Google Scholar] [CrossRef]
- Chen, Y.; Leach III, F.E.; Kaiser, N.K.; Dang, X.; Ibrahim, Y.M.; Norheim, R.V.; Gordon, A.A.; Richard, D.S.; Alan, G.M. Improved ion optics for introduction of ions into a 9.4-T Fourier transform ion cyclotron resonance mass spectrometer. J. Mass Spectrom. 2015, 50, 280–284. [Google Scholar] [CrossRef]
- Guilhaus, M.; Selby, D.; Mlynski, V. Orthogonal acceleration time-of-flight mass spectrometry. Mass Spectrom. Rev. 2000, 19, 65–107. [Google Scholar] [CrossRef]
- Zubarev, R.A.; Makarov, A. Orbitrap Mass Spectrometry. Anal. Chem. 2013, 85, 5288–5296. [Google Scholar] [CrossRef]
- Liu, X.R.; Zhang, M.M.; Gross, M.L. Mass Spectrometry-Based Protein Footprinting for High Order Structure Analysis: Fundamentals and Applications. Chem. Rev. 2022, 120, 4355–4454. [Google Scholar] [CrossRef]
- Peterka, O.; Jirasko, R.; Chocholouskova, M.; Kuchar, L.; Wolrab, D.; Hajek, R.; Vrana, D.; Strouhal, O.; Melichar, B.; Holcapek, M. Lipidomic characterization of exosomes isolated from human plasma using various mass spectrometry techniques. BBA Mol. Cell Biol. Lipids 2020, 1865, 158634. [Google Scholar] [CrossRef]
- Banliat, C.; Le Bourhis, D.; Bernardi, O.; Tomas, D.; Labas, V.; Salvetti, P.; Guyonnet, B.; Mermillod, P.; Saint-Dizier, M. Oviduct Fluid Extracellular Vesicles Change the Phospholipid Composition of Bovine Embryos Developed In Vitro. Int. J. Mol. Sci. 2020, 21, 5326. [Google Scholar] [CrossRef]
- Madonna, R.; Angelucci, S.; Di Giuseppe, F.; Doria, V.; Giricz, Z.; Gorbe, A.; Ferdinandy, P.; De Caterina, R. Proteomic analysis of the secretome of adipose tissue-derived murine mesenchymal cells overexpressing telomerase and myocardin. J. Mol. Cell. Cardiol. 2019, 131, 171–186. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Lee, D.; Kim, Y.; Paek, M.; Kim, M.; Jang, K.S.; Oh, J.; Lee, Y.S.; Yeon, J.E.; Lubman, D.M.; et al. Platelet Factor 4 as a Novel Exosome Marker in MALDI/MS Analysis of Exosomes from Human Serum. Anal. Chem. 2019, 91, 13297–13305. [Google Scholar] [CrossRef]
- Burkova, E.E.; Dmitrenok, P.S.; Bulgakov, D.V.; Vlassov, V.V.; Ryabchikova, E.I.; Nevinsky, G.A. Exosomes from Human Placenta Purified by Affinity Chromatography on Sepharose Bearing Immobilized Antibodies against CD81 Tetraspanin Contain Many Peptides and Small Proteins. IUBMB Life 2018, 70, 1144–1155. [Google Scholar] [CrossRef]
- Burkova, E.E.; Grigoreva, A.E.; Bulgakov, D.V.; Dmitrenok, P.S.; Vlassov, V.V.; Ryabchikova, E.I.; Sedykh, S.E.; Nevinsky, G.A. Extra Purified Exosomes from Human Placenta Contain an Unpredictable Small Number of Different Major Proteins. Int. J. Mol. Sci. 2019, 20, 2434. [Google Scholar] [CrossRef]
- Saraswat, M.; Joenvaara, S.; Musante, L.; Peltoniemi, H.; Holthofer, H.; Renkonen, R. N-linked (N-) Glycoproteomics of Urimary Exosomes. Mol. Cell. Proteom. 2015, 14, 263–276. [Google Scholar] [CrossRef]
- Zhao, Z.; Du, S.; Shen, S.; Luo, P.; Ding, S.; Wang, G.; Wang, L. Biomarkers screening for viral myocarditis through proteomics analysis of plasma exosomes. Natl. Med. J. China 2019, 99, 343–348. [Google Scholar]
- Xie, X.F.; Chu, H.J.; Xu, Y.F.; Hua, L.; Wang, Z.P.; Huang, P.; Jia, H.L.; Zhang, L. Proteomics study of serum exosomes in Kawasaki disease patients with coronary artery aneurysms. Cardiol. J. 2019, 26, 584–593. [Google Scholar] [CrossRef]
- Song, W.; Zhou, X.; Benktander, J.D.; Gaunitz, S.; Zou, G.; Wang, Z.; Novotny, M.V.; Jacobson, S.C. In-Depth Compositional and Structural Characterization of N-Glycans Derived from Human Urinary Exosomes. Anal. Chem. 2019, 91, 13528–13537. [Google Scholar] [CrossRef]
- Korenevsky, A.V.; Milyutina, Y.P.; Berezkina, M.E.; Alexandrova, E.P.; Balabas, O.A.; Markova, K.L.; Selkov, S.A.; Sokolov, D.I. MALDI-TOF mass spectrometric protein profiling of THP-1 cells and their microvesicles. Med. Immunol. 2021, 23, 275–292. [Google Scholar] [CrossRef]
- Sedykh, S.E.; Purvinish, L.V.; Monogarov, A.S.; Burkova, E.; Grigoreva, A.E.; Bulgakov, D.V.; Dmitrenok, P.S.; Vlassov, V.V.; Ryabchikova, E.I.; Nevinsky, G.A. Purified horse milk exosomes contain an unpredictable small number of major proteins. Biochim. Open 2017, 4, 61–72. [Google Scholar] [CrossRef]
- Sedykh, S.E.; Purvinish, L.V.; Burkova, E.; Dmitrenok, P.S.; Vlassov, V.V.; Ryabchikova, E.I.; Nevinsky, G.A. Analysis of peptides and small proteins in preparations of horse milk exosomes, purified on anti-CD81-Sepharose. Int. Dairy J. 2021, 117, 104994. [Google Scholar] [CrossRef]
- Kaminska, A.; Platt, M.; Kasprzyk, J.; Kusnierz-Cabala, B.; Gala-Bladzinska, A.; Woznicka, O.; Jany, B.R.; Krok, F.; Piekoszewski, W.; Kuzniewski, M.; et al. Urinary Extracellular Vesicles: Potential Biomarkers of Renal Function in Diabetic Patients. J. Diabetes Res. 2016, 2016, 5741518. [Google Scholar] [CrossRef]
- Gu, D.; Chen, Y.; Masucci, M.; Xiong, C.; Zou, H.; Holthofer, H. Potential urine biomarkers for the diagnosis of prediabetes and early diabetic nephropathy based on ISN CKHDP program. Clin. Nephrol. 2020, 193, S129–S133. [Google Scholar] [CrossRef]
- Wu, G.; Geng, H.; Xu, R.; Deng, M.; Yang, C.; Xun, C.; Wang, Y.; Cai, Q.; Chen, P. Preparation of a CaTiO3/Al3+/Pr3+/Sm3+ nanocomposite for enrichment of exosomes in human serum. Talanta 2021, 226, 122186. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, L.; Ma, H.; Ni, F.; Liu, F.; Chen, H. Detection of Tear Components Using Matrix-Assisted Laser Desorption Ionization/Time-of-Flight Mass Spectrometry for Rapid Dry Eye Diagnosis. J. Proteome Res. 2020, 19, 3644–3651. [Google Scholar] [CrossRef]
- Paingankar, M.S.; Arankalle, V.A. Identification and characterization of cellular proteins interacting with Hepatitis E virus untranslated regions. Virus Res. 2015, 208, 98–109. [Google Scholar] [CrossRef]
- Medvedeva, E.S.; Mouzykantov, A.A.; Baranova, N.B.; Dramchini, M.A.; Chernova, O.A.; Chernov, V.M. Data on proteomic profiling of cells and extracellular vesicles of the melittin-resistant Acholeplasma laidlawii strain. Data Brief 2019, 25, 104169. [Google Scholar] [CrossRef]
- Mouzykantov, A.A.; Medvedeva, E.; Baranova, N.; Lopuhov, V.; Usachev, K.; Chernova, O.A.; Chernov, V.M. Data on the genome and proteome profiles of ciprofloxacin-resistant Acholeplasma laidlawii strains selected under different conditions in vitro. Data Brief 2020, 33, 106412. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C.; Xie, P.; Pan, Y.; Tan, Y.; Tang, L. Changes of cytokines and protein expression of macrophages stimulated with exosome from macrophages by Mycobacterium avium infection. Chin. J. Cell. Mol. Immunol. 2013, 29, 123–1261. [Google Scholar]
- Wang, J.; Chen, C.; Xie, P.; Pan, Y.; Tan, Y.; Tang, L. Proteomic analysis and immune properties of exosomes released by macrophages infected with Mycobacterium avium. Microbes Infect. 2014, 16, 283–291. [Google Scholar] [CrossRef]
- Sviben, D.; Forcic, D.; Halassy, B.; Allmaier, G.; Marchetti-Deschmann, M.; Brgles, M. Mass spectrometry-based investigation of measles and mumps virus proteome. Virol. J. 2018, 15, 160. [Google Scholar] [CrossRef] [PubMed]
- Korenevsky, A.V.; Shcherbitskaia, A.D.; Berezkina, M.E.; Markova, K.L.; Alexandrova, E.P.; Balabas, O.A.; Selkov, S.A.; Sokolov, D.I. MALDI-TOF mass spectrometric protein profiling of microvesicles produced by the NK-92 natural killer cell line. Med. Immunol. 2020, 22, 633–646. [Google Scholar] [CrossRef]
- Castaño, C.; Novials, A.; Párrizas, M. Exosomes and diabetes. Diabetes Metab. Res. Rev. 2019, 35, e3107. [Google Scholar] [CrossRef] [PubMed]
- Mohsenipour, R.; Mahdieh, N.; Rostami, P. Pancytopenia, a rare case report of epimerase deficiency galactosemia. J. Clin. Images Med. Case Rep. 2023, 4, 2588. [Google Scholar]
- Staubach, S.; Schadewaldt, P.; Wendel, U.; Nohroudi, K.; Hanisch, F.G. Differential Glycomics of Epithelial Membrane Glycoproteins from Urinary Exovesicles Reveals Shifts toward Complex-Type N-Glycosylation in Classical Galactosemia. J. Proteome Res. 2012, 11, 906–916. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Hornung, S.; Taha, H.B.; Bitan, G. Biomarkers for parkinsonian disorders in CNS-originating EVs: Promise and challenges. Acta Neuropathol. 2023, 145, 515–540. [Google Scholar] [CrossRef] [PubMed]
- Bitan, G. Disease-modifying therapy for proteinopathies: Can the exception become the rule? Prog. Mol. Biol. Transl. Sci. 2019, 168, 277–287. [Google Scholar] [PubMed]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef]
- Shipley, S.M.; Frederick, M.C.; Filley, C.M.; Kluger, B.M. Potential for misdiagnosis in community-acquired PET scans for dementia. Neurol. Clin. Pract. 2013, 3, 305–312. [Google Scholar] [CrossRef]
- Skinner, T.R.; Scott, I.A.; Martin, J.H. Diagnostic errors in older patients: A systematic review of incidence and potential causes in seven prevalent diseases. Int. J. Gen. Med. 2016, 9, 137–146. [Google Scholar] [CrossRef]
- Fadil, H.; Borazanci, A.; Haddou, E.A.B.; Yahyaoui, M.; Korniychuk, E.; Jaffe, S.L.; Minagar, A. Early onset dementia. Int. Rev. Neurobiol. 2009, 84, 245–262. [Google Scholar] [PubMed]
- Tahami Monfared, A.A.; Byrnes, M.J.; White, L.A.; Zhang, Q. Alzheimer’s disease: Epidemiology and clinical progression. Neurol. Ther. 2022, 11, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S. Molecular and cellular basis of neurodegeneration in Alzheimer’s disease. Mol. Cells 2017, 40, 613. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2023, 23, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.R.; Praveen, M.; Narasimhan, R.; Khamar, P.; D’souza, S.; Sinha-Roy, A.; Sethu, S.; Shetty, R.; Ghosh, A. Tear biomarkers in dry eye disease: Progress in the last decade. Indian J. Ophthalmol. 2023, 71, 1190. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L. Host-pathogen interactions: The attributes of virulence. J. Infect. Dis. 2001, 184, 337–344. [Google Scholar] [CrossRef]

 = cell debris;
= cell debris;  = EV.
= EV.
 = cell debris;
= cell debris;  = EV.
= EV.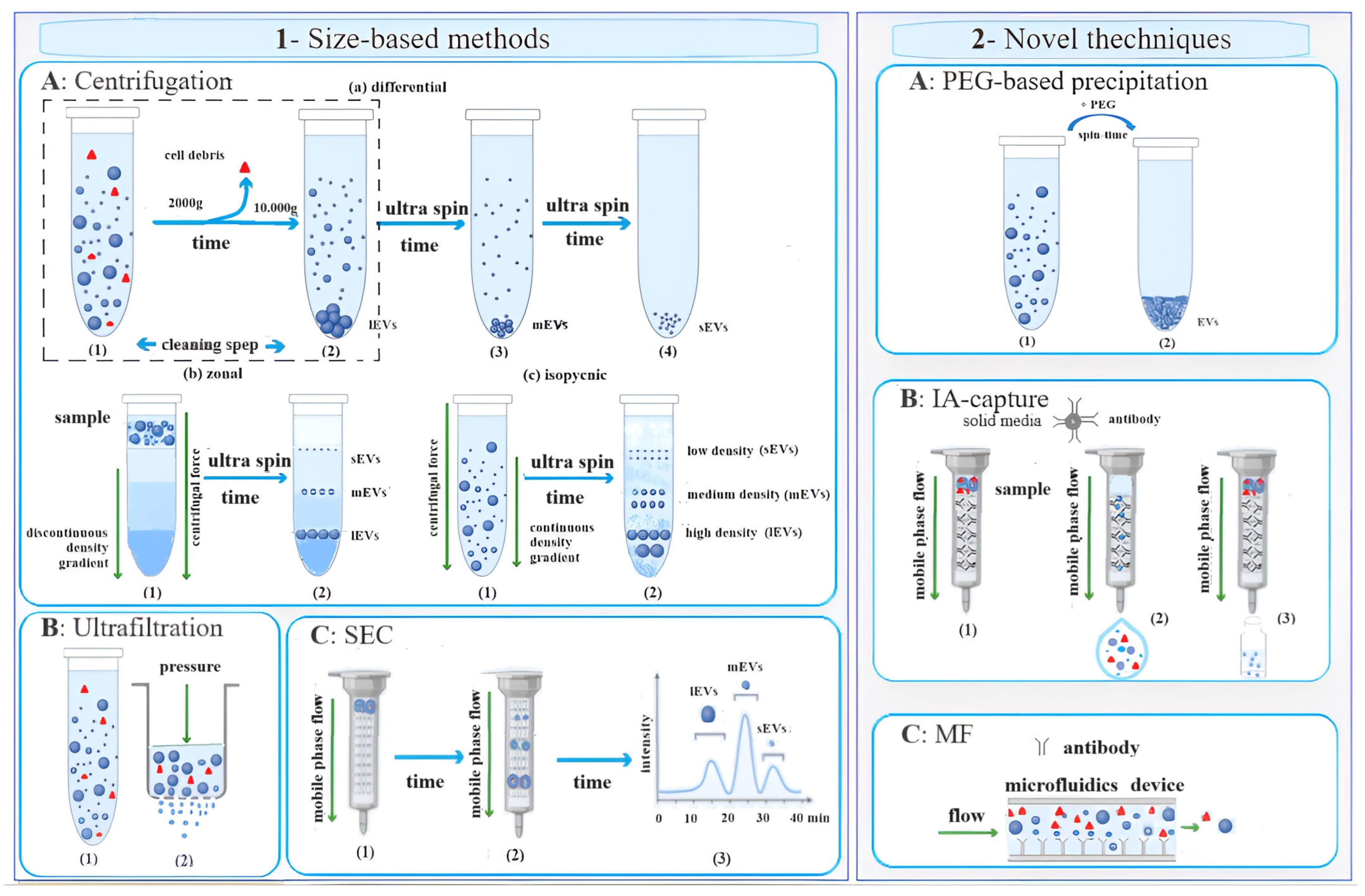


| Authors | Objective | Techniques | Ref |
|---|---|---|---|
| Peterka et al. | Lipidomic characterization of exosomes isolated from human plasma. | Ultra-high-performance supercritical fluid chromatography–mass spectrometry (UHPSFC/MS), ultra-high-performance liquid chromatography–mass spectrometry (UHPLC/MS), and MALDI/MS. | [43] |
| Banliat et al. | Evaluation of the changes induced by oviduct fluid extracellular vesicles on embryo phospholipids. | Intact cell MALDI-TOF/MS. | [44] |
| Madonna et al. | Understanding the mechanisms of the therapeutic effects of stem/progenitor cells, including adipose-tissue-derived mesenchymal stromal cells (AT-MSCs). | Two-dimensional gel electrophoresis (2-DE) with MALDI-TOF/TOF. | [45] |
| Nguyen et al. | Proteomic characterization of exosomes in biological fluids. | MALDI combined with Fourier transform ion cyclotron resonance mass spectrometry (FTICR/MS). | [46] |
| Burkova, et al. | Search for protein biomarkers in human placenta exosomes. | MALDI/MS and MALDI/MS/MS of protein tryptic hydrolysates from SDS-PAGE and 2-DE. | [47,48] |
| Saraswat et al. | Investigation of the role of N-glycoproteome of urinary exosomes. | Collision-induced dissociation–MS/MS (CID–tandem MS) and MALDI/MS. | [49] |
| Zhao et al. | Biomarker screening for viral myocarditis through proteomic analysis. | MALDI-TOF/TOF mass spectrometry, validation using ELISA analysis. | [50] |
| Xie et al. | Comprehensive proteomic profile of serum exosomes from children with coronary artery aneurysms caused by Kawasaki disease. | 2-DE with MALDI-TOF/TOF/MS. | [51] |
| Song et al. | N-glycan and sulfated N-glycan compositions in urine EVs for noninvasive investigations into the pathophysiological states of the urinary system. | Capillary electrophoresis–mass spectrometry (CE-MS), MALDI/MS, and capillary liquid chromatography–tandem mass spectrometry (LC-MS/MS). | [52] |
| Korenevsky et al. | Investigation on distant communications of cells and their regulatory mechanisms through the proteomic study of microvesicles derived from THP-1 monocyte cells. | MALDI/MS. | [53] |
| Sedykh et al. | Morphology and the protein content of major horse milk exosomes. | MALDI/MS and MS/MS spectrometry. | [54] |
| Sedykh et al. | Preparation of crude vesicles from horse milk for analysis of peptides and small proteins. | Standard methods of centrifugation, ultracentrifugation and gel filtration. Extra-purification with affinity chromatography on anti-CD81-Sepharose. Detection using MALDI-TOF/MS. | [55] |
| Kaminska et al. | Check the relationship between the density of urinary EVs, their size distribution, and the progress of early renal damage in type 2 diabetic patients (DMt2). | MALDI-TOF-MS/MS. | [56] |
| Gu et al. | Search for potential urine biomarkers for the diagnosis of prediabetes and early diabetic nephropathy. | Characterization using transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and Western blotting of the tumor susceptibility gene product TSG101. Two-dimensional DIGE (2D-DIGE) with MS analysis. | [57] |
| Wu et al. | CaTiO3/Al3+/Pr3+/Sm3+ nanocomposite was synthesized and applied for highly selective and efficient separation of exosomes. | 2-DE with MALDI TOF/TOF/MS. | [58] |
| Zhang et al. | To analyze peptidome/proteome profiles of tears and EVs for rapid dry eye diagnosis. | MALDI TOF/MS. | [59] |
| Paingankar et al. | To identify the host cellular factors that interact with Hepatitis E virus’ 5′ and 3′ untranslated regions (UTRs). | RNA pull-down and matrix-assisted laser desorption/ionization (MALDI) TOF. | [60] |
| Medvedeva et al. | To compare the cellular and vesicular proteomes of A. laidlawii strains with differing susceptibility to melittin (an antimicrobial peptide from bee venom). | 2-DE with MALDI-TOF/TOF MS. | [61] |
| Mouzykantov et al. | To compare the genome profiles of ciprofloxacin-resistant A. laidlawii strains PG8r1 and PG8r3 selected under different in vitro conditions when a ciprofloxacin-sensitive A. laidlawii PG8B strain was cultured at increasing concentrations of ciprofloxacin in a broth medium alone and with vesicles derived from the ciprofloxacin-resistant A. laidlawii PG8R10c-2 strain, respectively. | 2-DE with MALDI-TOF/TOF/MS. | [62] |
| Wang et al. | To explore the mechanism underlying the molecular immune response of macrophages stimulated with exosome [(+)exosome] from macrophages after Mycobacterium avium (M. avium) infection and analyze the differential protein component of the exosome. | 2-DE with MALDI TOF/TOF MS. | [63] |
| Wang et al. | To study the role of exosomes shed from Mycobacterium avium sp. paratuberculosis-infected macrophages in intercellular communication processes. | MALDI-TOF/TOF. | [64] |
| Sviben et al. | To identify which virus-coded proteins are present in measles and mumps virus virions and to try to detect which host cell proteins, if any, are incorporated into the virions or adsorbed on their outer surface and which are more likely to be contamination from co-purified ECVs. | MALDI-TOF/TOF-MS. | [65] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aresta, A.M.; De Vietro, N.; Zambonin, C. Analysis and Characterization of the Extracellular Vesicles Released in Non-Cancer Diseases Using Matrix-Assisted Laser Desorption Ionization/Mass Spectrometry. Int. J. Mol. Sci. 2024, 25, 4490. https://doi.org/10.3390/ijms25084490
Aresta AM, De Vietro N, Zambonin C. Analysis and Characterization of the Extracellular Vesicles Released in Non-Cancer Diseases Using Matrix-Assisted Laser Desorption Ionization/Mass Spectrometry. International Journal of Molecular Sciences. 2024; 25(8):4490. https://doi.org/10.3390/ijms25084490
Chicago/Turabian StyleAresta, Antonella Maria, Nicoletta De Vietro, and Carlo Zambonin. 2024. "Analysis and Characterization of the Extracellular Vesicles Released in Non-Cancer Diseases Using Matrix-Assisted Laser Desorption Ionization/Mass Spectrometry" International Journal of Molecular Sciences 25, no. 8: 4490. https://doi.org/10.3390/ijms25084490






