Change in Tissue Microbiome and Related Human Beta Defensin Levels Induced by Antibiotic Use in Bladder Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
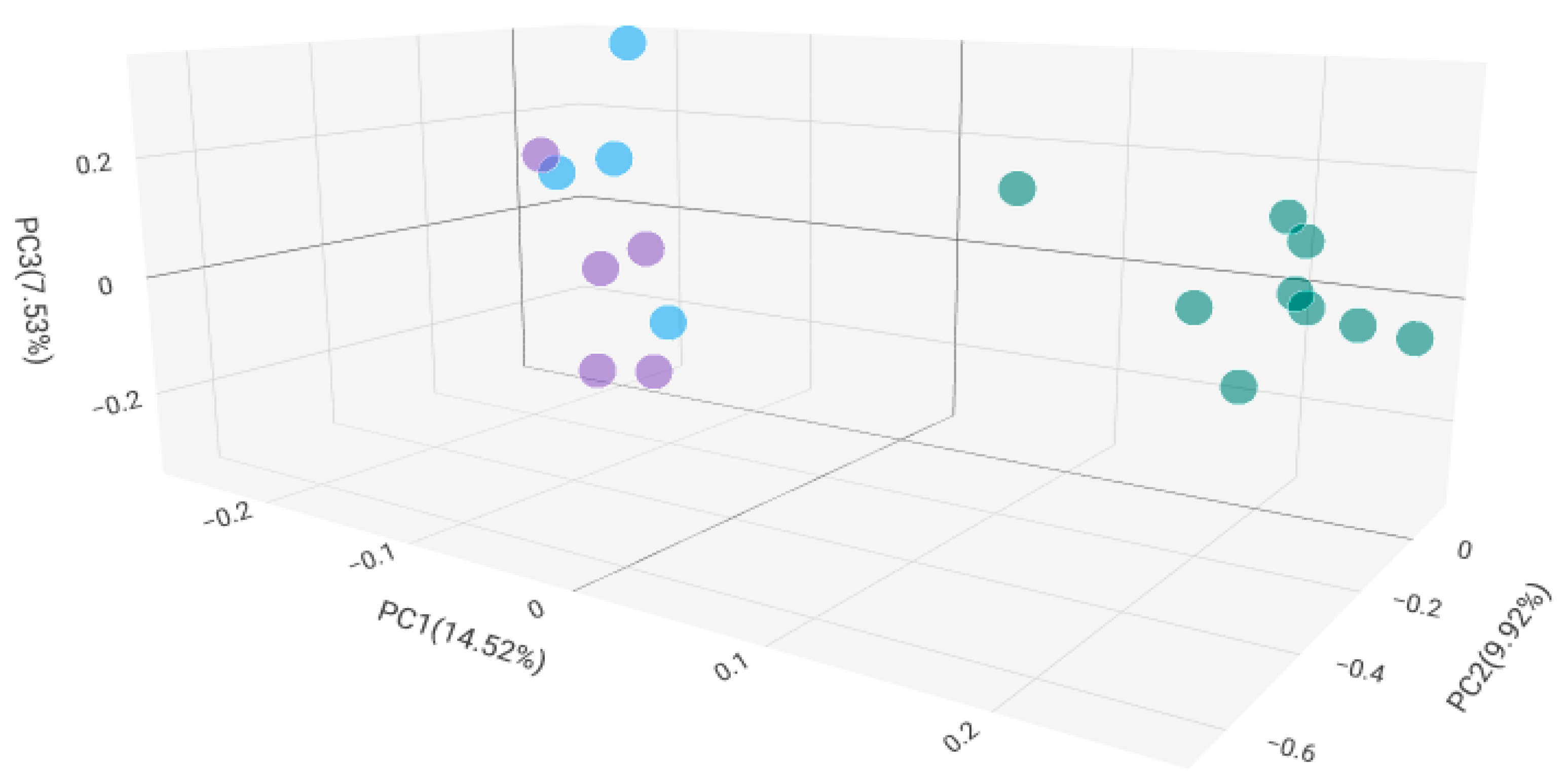
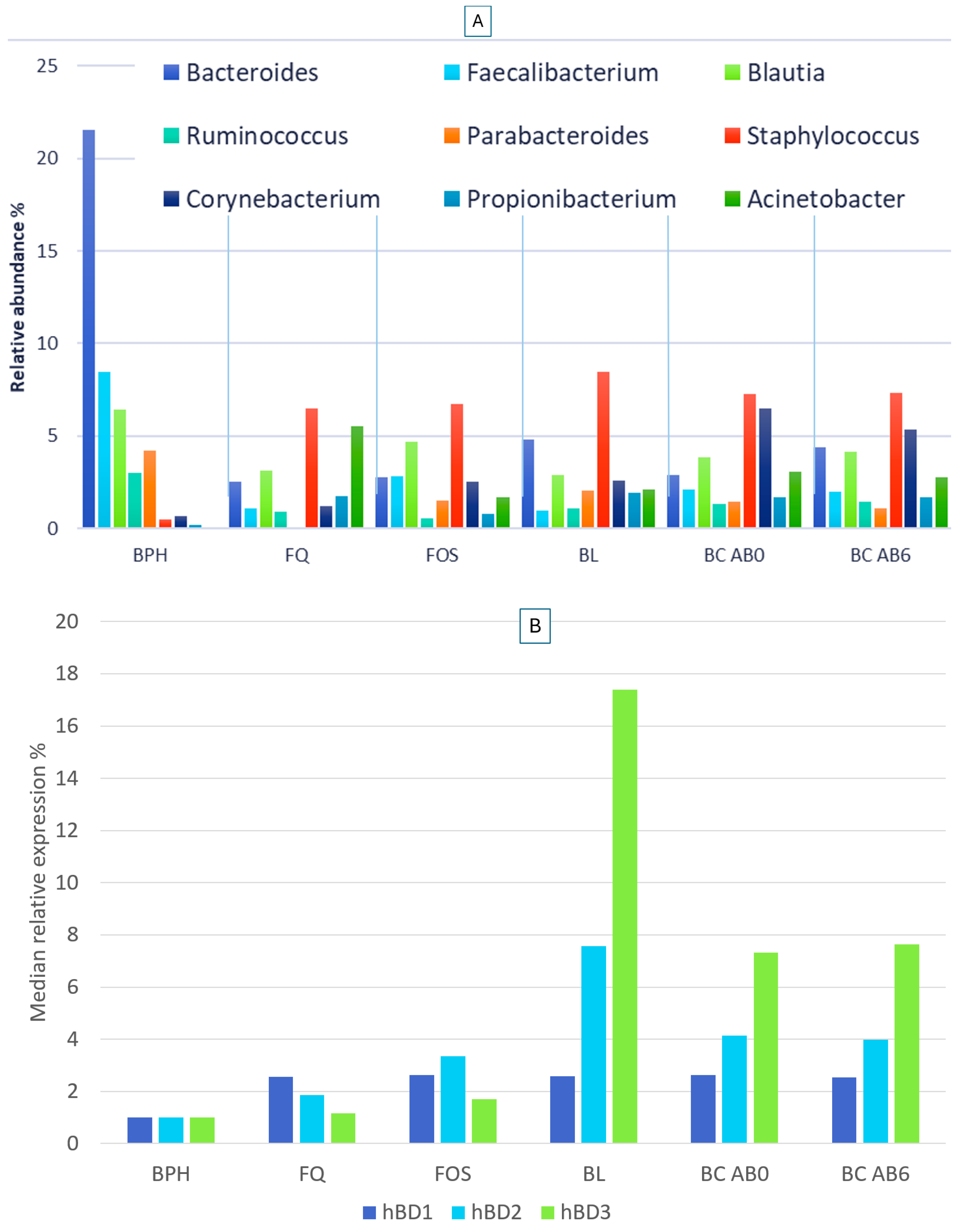
2.2. Microbiome Composition in Healthy, Antibiotic-Untreated Bladder Cancer and Antibiotic-Treated Bladder Cancer Tissues
2.3. Human Beta Defensin Expression and the Relationship with Microbiome Composition
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. DNA Isolation, 16S rRNA Gene Libary Preparation, and MiSeq Sequencing
4.3. Defensin Expression Assays
4.4. Statistical Analysis
4.5. Ethical Considerations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| AB0 | AB6 | AB3 with BL | AB3 with FQ | AB3 with FOS | |
|---|---|---|---|---|---|
| Staphylococcus | p < 0.0001 | p < 0.0001 | p < 0.001 | p = 0.004 | p = 0.002 |
| Corynebacterium | p < 0.0001 | p = 0.006 | X | X | p = 0.003 |
| Acinetobacter | p < 0.0001 | p = 0.001 | p < 0.0001 | p = 0.011 | p = 0.002 |
| Propionibacterium | p < 0.0001 | p = 0.001 | p < 0.0001 | p = 0.004 | p = 0.015 |
| Bacteroides | p < 0.0001 | p = 0.001 | p < 0.001 | p = 0.004 | p = 0.004 |
| Faecalibacterium | p < 0.0001 | p = 0.007 | p < 0.001 | p = 0.011 | p = 0.009 |
| Blautia | p = 0.007 | p = 0.025 | X | X | X |
| Ruminococcus | p = 0.009 | p = 0.001 | p = 0.003 | X | X |
| Parabacteroides | p = 0.008 | p < 0.0001 | X | X | p = 0.03 |
References
- Bucevic Popovic, V.; Situm, M.; Chow, C.T.; Chan, L.S.; Roje, B.; Terzic, J. The urinary microbiome associated with bladder cancer. Sci. Rep. 2018, 8, 12157. [Google Scholar] [CrossRef] [PubMed]
- Hussein, A.A.; Smith, G.; Guru, K.A. The Association between the Urinary Microbiome and Bladder Cancer. Urol. Clin. North. Am. 2023, 50, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Tian, Y.; Song, C.; Li, J.; Liu, T.; Chen, Z.; Chen, C.; Huang, Y.; Zhang, Y. Urinary microbiota—A potential biomarker and therapeutic target for bladder cancer. J. Med. Microbiol. 2019, 68, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Mansour, B.; Monyok, A.; Makra, N.; Gajdacs, M.; Vadnay, I.; Ligeti, B.; Juhasz, J.; Szabo, D.; Ostorhazi, E. Bladder cancer-related microbiota: Examining differences in urine and tissue samples. Sci. Rep. 2020, 10, 11042. [Google Scholar] [CrossRef] [PubMed]
- Mansour, B.; Monyok, A.; Gajdacs, M.; Stercz, B.; Makra, N.; Penzes, K.; Vadnay, I.; Szabo, D.; Ostorhazi, E. Bladder Tissue Microbiome Composition in Patients of Bladder Cancer or Benign Prostatic Hyperplasia and Related Human Beta Defensin Levels. Biomedicines 2022, 10, 1758. [Google Scholar] [CrossRef] [PubMed]
- Li, W.T.; Iyangar, A.S.; Reddy, R.; Chakladar, J.; Bhargava, V.; Sakamoto, K.; Ongkeko, W.M.; Rajasekaran, M. The Bladder Microbiome Is Associated with Epithelial-Mesenchymal Transition in Muscle Invasive Urothelial Bladder Carcinoma. Cancers 2021, 13, 3649. [Google Scholar] [CrossRef]
- Liu, F.; Liu, A.; Lu, X.; Zhang, Z.; Xue, Y.; Xu, J.; Zeng, S.; Xiong, Q.; Tan, H.; He, X.; et al. Dysbiosis signatures of the microbial profile in tissue from bladder cancer. Cancer Med. 2019, 8, 6904–6914. [Google Scholar] [CrossRef] [PubMed]
- Pederzoli, F.; Ferrarese, R.; Amato, V.; Locatelli, I.; Alchera, E.; Luciano, R.; Nebuloni, M.; Briganti, A.; Gallina, A.; Colombo, R.; et al. Sex-specific Alterations in the Urinary and Tissue Microbiome in Therapy-naive Urothelial Bladder Cancer Patients. Eur. Urol. Oncol. 2020, 3, 784–788. [Google Scholar] [CrossRef]
- Bisacchi, G.S.; Hale, M.R. A “Double-Edged” Scaffold: Antitumor Power within the Antibacterial Quinolone. Curr. Med. Chem. 2016, 23, 520–577. [Google Scholar] [CrossRef]
- Chrysostomou, S.; Roy, R.; Prischi, F.; Thamlikitkul, L.; Chapman, K.L.; Mufti, U.; Peach, R.; Ding, L.; Hancock, D.; Moore, C.; et al. Repurposed floxacins targeting RSK4 prevent chemoresistance and metastasis in lung and bladder cancer. Sci. Transl. Med. 2021, 13, eaba4627. [Google Scholar] [CrossRef]
- Karp, I.; Lyakhovich, A. Targeting cancer stem cells with antibiotics inducing mitochondrial dysfunction as an alternative anticancer therapy. Biochem. Pharmacol. 2022, 198, 114966. [Google Scholar] [CrossRef]
- Jing, Y.; Chen, X.; Li, K.; Liu, Y.; Zhang, Z.; Chen, Y.; Liu, Y.; Wang, Y.; Lin, S.H.; Diao, L.; et al. Association of antibiotic treatment with immune-related adverse events in patients with cancer receiving immunotherapy. J. Immunother. Cancer 2022, 10, e003779. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, S.; Lin, M.; Hou, C.; Li, C.; Li, G. Analysis of interactions of immune checkpoint inhibitors with antibiotics in cancer therapy. Front. Med. 2022, 16, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Fidelle, M.; Rauber, C.; Alves Costa Silva, C.; Tian, A.L.; Lahmar, I.; de La Varende, A.M.; Zhao, L.; Thelemaque, C.; Lebhar, I.; Messaoudene, M.; et al. A microbiota-modulated checkpoint directs immunosuppressive intestinal T cells into cancers. Science 2023, 380, eabo2296. [Google Scholar] [CrossRef] [PubMed]
- Pak, S.; Kim, S.Y.; Kim, S.H.; Joung, J.Y.; Park, W.S.; Chung, J.; Lee, K.H.; Seo, H.K. Association Between Antibiotic Treatment and the Efficacy of Intravesical BCG Therapy in Patients With High-Risk Non-Muscle Invasive Bladder Cancer. Front. Oncol. 2021, 11, 570077. [Google Scholar] [CrossRef]
- Beerepoot, M.A.; ter Riet, G.; Nys, S.; van der Wal, W.M.; de Borgie, C.A.; de Reijke, T.M.; Prins, J.M.; Koeijers, J.; Verbon, A.; Stobberingh, E.; et al. Lactobacilli vs antibiotics to prevent urinary tract infections: A randomized, double-blind, noninferiority trial in postmenopausal women. Arch. Intern. Med. 2012, 172, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Mulder, M.; Radjabzadeh, D.; Hassing, R.J.; Heeringa, J.; Uitterlinden, A.G.; Kraaij, R.; Stricker, B.H.; Verbon, A. The effect of antimicrobial drug use on the composition of the genitourinary microbiota in an elderly population. BMC Microbiol. 2019, 19, 9. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Wu, X.; Wang, Z.; Wu, X.; Wang, S.; Jing, G.; Yan, T. Fecal Microbiota Transplantation as an Effective Treatment for Carbapenem-Resistant Klebsiella pneumoniae Infection in a Renal Transplant Patient. Infect. Drug Resist. 2021, 14, 1805–1811. [Google Scholar] [CrossRef]
- Xu, D.; Lu, W. Defensins: A Double-Edged Sword in Host Immunity. Front. Immunol. 2020, 11, 764. [Google Scholar] [CrossRef]
- Wiesner, J.; Vilcinskas, A. Antimicrobial peptides: The ancient arm of the human immune system. Virulence 2010, 1, 440–464. [Google Scholar] [CrossRef]
- Ghosh, S.K.; McCormick, T.S.; Weinberg, A. Human Beta Defensins and Cancer: Contradictions and Common Ground. Front. Oncol. 2019, 9, 341. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Wang, R.; Sun, C.; Jin, X.; Liu, D.; Zhao, X.; Wang, L.; Ji, N.; Li, J.; Zhou, Y.; et al. Human beta-defensin-1 suppresses tumor migration and invasion and is an independent predictor for survival of oral squamous cell carcinoma patients. PLoS ONE 2014, 9, e91867. [Google Scholar] [CrossRef] [PubMed]
- Adyns, L.; Proost, P.; Struyf, S. Role of Defensins in Tumor Biology. Int. J. Mol. Sci. 2023, 24, 5268. [Google Scholar] [CrossRef]
- Sun, C.Q.; Arnold, R.S.; Hsieh, C.L.; Dorin, J.R.; Lian, F.; Li, Z.; Petros, J.A. Discovery and mechanisms of host defense to oncogenesis: Targeting the beta-defensin-1 peptide as a natural tumor inhibitor. Cancer Biol. Ther. 2019, 20, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.H.; Martinez Velazquez, M.; Prado Montes de Oca, E. Human beta-defensin 1 update: Potential clinical applications of the restless warrior. Int. J. Biochem. Cell Biol. 2018, 104, 133–137. [Google Scholar] [CrossRef]
- Whang, Y.M.; Jin, S.B.; Park, S.I.; Chang, I.H. MEK inhibition enhances efficacy of bacillus Calmette-Guerin on bladder cancer cells by reducing release of Toll-like receptor 2-activated antimicrobial peptides. Oncotarget 2017, 8, 53168–53179. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, S.J.; Lee, K.M.; Chang, I.H. Human beta-defensin 2 may inhibit internalisation of bacillus Calmette-Guerin (BCG) in bladder cancer cells. BJU Int. 2013, 112, 781–790. [Google Scholar] [CrossRef]
- Meade, K.G.; O‘Farrelly, C. beta-Defensins: Farming the Microbiome for Homeostasis and Health. Front. Immunol. 2018, 9, 3072. [Google Scholar] [CrossRef]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef]
- Parra-Grande, M.; Ore-Arce, M.; Martinez-Priego, L.; D‘Auria, G.; Rossello-Mora, R.; Lillo, M.; Sempere, A.; Lumbreras, B.; Sanchez-Hellin, V. Profiling the Bladder Microbiota in Patients With Bladder Cancer. Front. Microbiol. 2021, 12, 718776. [Google Scholar] [CrossRef]
- Mizuhashi, S.; Kajihara, I.; Sawamura, S.; Kanemaru, H.; Makino, K.; Aoi, J.; Makino, T.; Masuguchi, S.; Fukushima, S.; Ihn, H. Skin microbiome in acral melanoma: Corynebacterium is associated with advanced melanoma. J. Dermatol. 2021, 48, e15–e16. [Google Scholar] [CrossRef]
- Hasan, R.; Bose, S.; Roy, R.; Paul, D.; Rawat, S.; Nilwe, P.; Chauhan, N.K.; Choudhury, S. Tumor tissue-specific bacterial biomarker panel for colorectal cancer: Bacteroides massiliensis, Alistipes species, Alistipes onderdonkii, Bifidobacterium pseudocatenulatum, Corynebacterium appendicis. Arch. Microbiol. 2022, 204, 348. [Google Scholar] [CrossRef]
- Sheweita, S.A.; Alsamghan, A.S. Molecular Mechanisms Contributing Bacterial Infections to the Incidence of Various Types of Cancer. Mediators Inflamm. 2020, 2020, 4070419. [Google Scholar] [CrossRef]
- Oresta, B.; Braga, D.; Lazzeri, M.; Frego, N.; Saita, A.; Faccani, C.; Fasulo, V.; Colombo, P.; Guazzoni, G.; Hurle, R.; et al. The Microbiome of Catheter Collected Urine in Males with Bladder Cancer According to Disease Stage. J. Urol. 2021, 205, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Shannon, B.A.; Garrett, K.L.; Cohen, R.J. Links between Propionibacterium acnes and prostate cancer. Future Oncol. 2006, 2, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, W.; Gong, D.; Shang, R.; Wang, J.; Yu, H. Propionibacterium acnes overabundance in gastric cancer promote M2 polarization of macrophages via a TLR4/PI3K/Akt signaling. Gastric Cancer 2021, 24, 1242–1253. [Google Scholar] [CrossRef]
- Kavoussi, L.R.; Brown, E.J.; Ritchey, J.K.; Ratliff, T.L. Fibronectin-mediated Calmette-Guerin bacillus attachment to murine bladder mucosa. Requirement for the expression of an antitumor response. J. Clin. Investig. 1990, 85, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Ratliff, T.L.; Kavoussi, L.R.; Catalona, W.J. Role of fibronectin in intravesical BCG therapy for superficial bladder cancer. J. Urol. 1988, 139, 410–414. [Google Scholar] [CrossRef]
- Kloskowski, T.; Frackowiak, S.; Adamowicz, J.; Szeliski, K.; Rasmus, M.; Drewa, T.; Pokrywczynska, M. Quinolones as a Potential Drug in Genitourinary Cancer Treatment-A Literature Review. Front. Oncol. 2022, 12, 890337. [Google Scholar] [CrossRef]
- Chen, C.; Huang, Z.; Huang, P.; Li, K.; Zeng, J.; Wen, Y.; Li, B.; Zhao, J.; Wu, P. Urogenital Microbiota:Potentially Important Determinant of PD-L1 Expression in Male Patients with Non-muscle Invasive Bladder Cancer. BMC Microbiol. 2022, 22, 7. [Google Scholar] [CrossRef]
- Du, Y.; Yang, Y.; Zhang, W.; Yang, C.; Xu, P. Human beta-defensin-3 and nuclear factor-kappa B p65 synergistically promote the cell proliferation and invasion of oral squamous cell carcinoma. Transl. Oncol. 2023, 27, 101582. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, B.; Liao, C.; Zhang, W.; Wang, W.; Chang, Y.; Shao, Y. Human beta-defensin 3 contributes to the carcinogenesis of cervical cancer via activation of NF-kappaB signaling. Oncotarget 2016, 7, 75902–75913. [Google Scholar] [CrossRef] [PubMed]
- Uraki, S.; Sugimoto, K.; Shiraki, K.; Tameda, M.; Inagaki, Y.; Ogura, S.; Kasai, C.; Nojiri, K.; Yoneda, M.; Yamamoto, N.; et al. Human beta-defensin-3 inhibits migration of colon cancer cells via downregulation of metastasis-associated 1 family, member 2 expression. Int. J. Oncol. 2014, 45, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wi, Y.M.; Thoendel, M.J.; Raval, Y.S.; Greenwood-Quaintance, K.E.; Abdel, M.P.; Jeraldo, P.R.; Chia, N.; Patel, R. Evaluation of the CosmosID Bioinformatics Platform for Prosthetic Joint-Associated Sonicate Fluid Shotgun Metagenomic Data Analysis. J. Clin. Microbiol. 2019, 57, e01182-18. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Antibiotic Treatment Date | Number of Patients | Age Median (ICR) | Male/Female | Stage Ta/T1/T2/T3/T4 | Grade G1/G2/G3 | |
|---|---|---|---|---|---|---|
| BPH | untreated | 10 | 72 (17) | 10/0 | n.a. | n.a. |
| BC AB0 | untreated | 26 | 73 (15) | 20/6 | 7/8/4/7/0 | 11/5/10 |
| BC AB6 | 3 months<, 6 months> | 8 | 74 (16) | 6/2 | 2/3/1/2/0 | 3/3/2 |
| BC AB3 | 3 months> | 26 | 71 (19) | 18/8 | 8/8/3/7/0 | 12/5/9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monyók, Á.; Mansour, B.; Vadnay, I.; Makra, N.; Dunai, Z.A.; Nemes-Nikodém, É.; Stercz, B.; Szabó, D.; Ostorházi, E. Change in Tissue Microbiome and Related Human Beta Defensin Levels Induced by Antibiotic Use in Bladder Carcinoma. Int. J. Mol. Sci. 2024, 25, 4562. https://doi.org/10.3390/ijms25084562
Monyók Á, Mansour B, Vadnay I, Makra N, Dunai ZA, Nemes-Nikodém É, Stercz B, Szabó D, Ostorházi E. Change in Tissue Microbiome and Related Human Beta Defensin Levels Induced by Antibiotic Use in Bladder Carcinoma. International Journal of Molecular Sciences. 2024; 25(8):4562. https://doi.org/10.3390/ijms25084562
Chicago/Turabian StyleMonyók, Ádám, Bassel Mansour, István Vadnay, Nóra Makra, Zsuzsanna A. Dunai, Éva Nemes-Nikodém, Balázs Stercz, Dóra Szabó, and Eszter Ostorházi. 2024. "Change in Tissue Microbiome and Related Human Beta Defensin Levels Induced by Antibiotic Use in Bladder Carcinoma" International Journal of Molecular Sciences 25, no. 8: 4562. https://doi.org/10.3390/ijms25084562






