Isolation, Characterization, Genome Annotation, and Evaluation of Hyaluronidase Inhibitory Activity in Secondary Metabolites of Brevibacillus sp. JNUCC 41: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation
Abstract
:1. Introduction
2. Results and Discussion
2.1. General Characteristics of the Genome of Brevibacillus sp. JNUCC 41
2.1.1. COGs Database Annotations
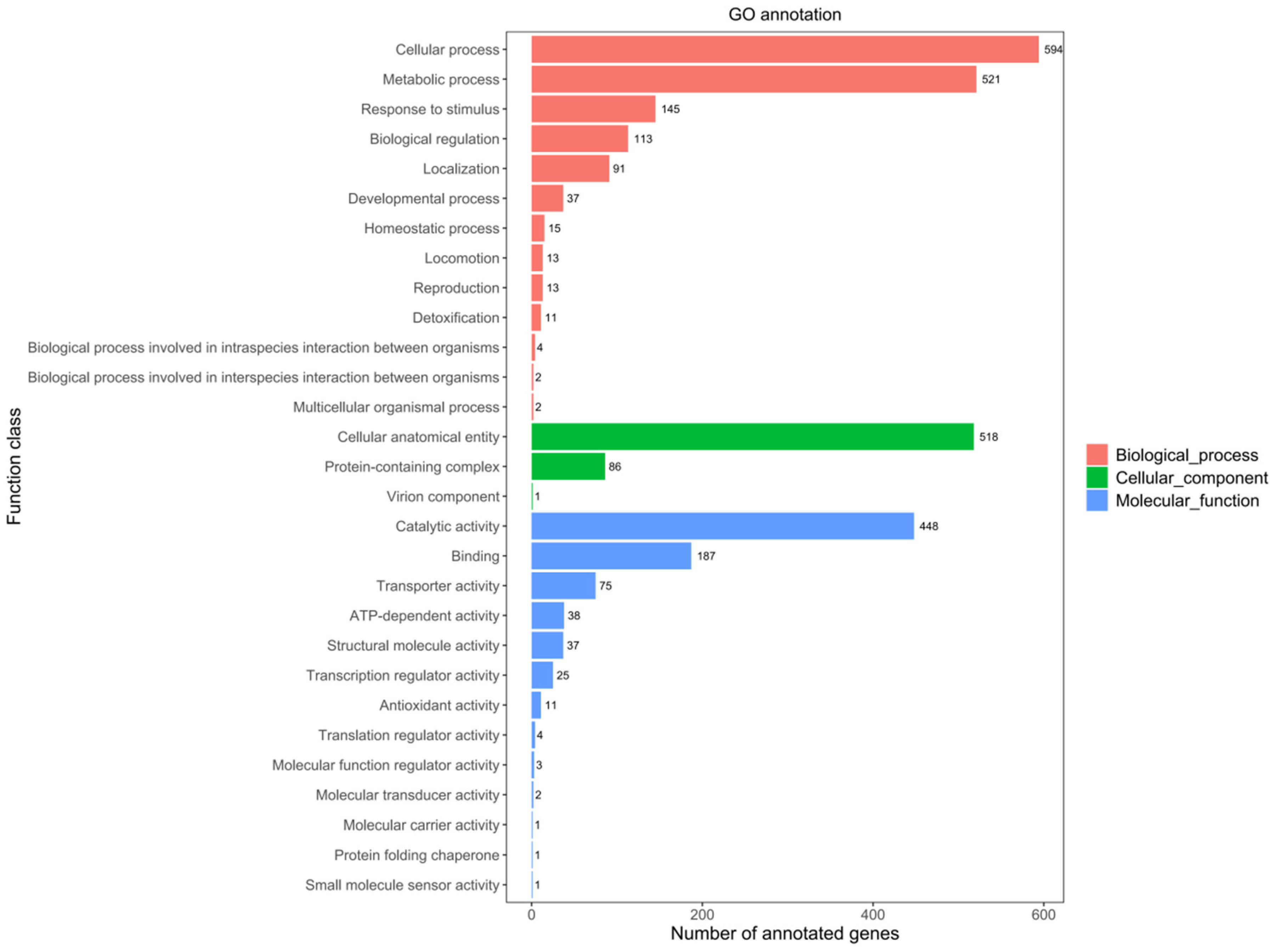
2.1.2. GO Database Annotations
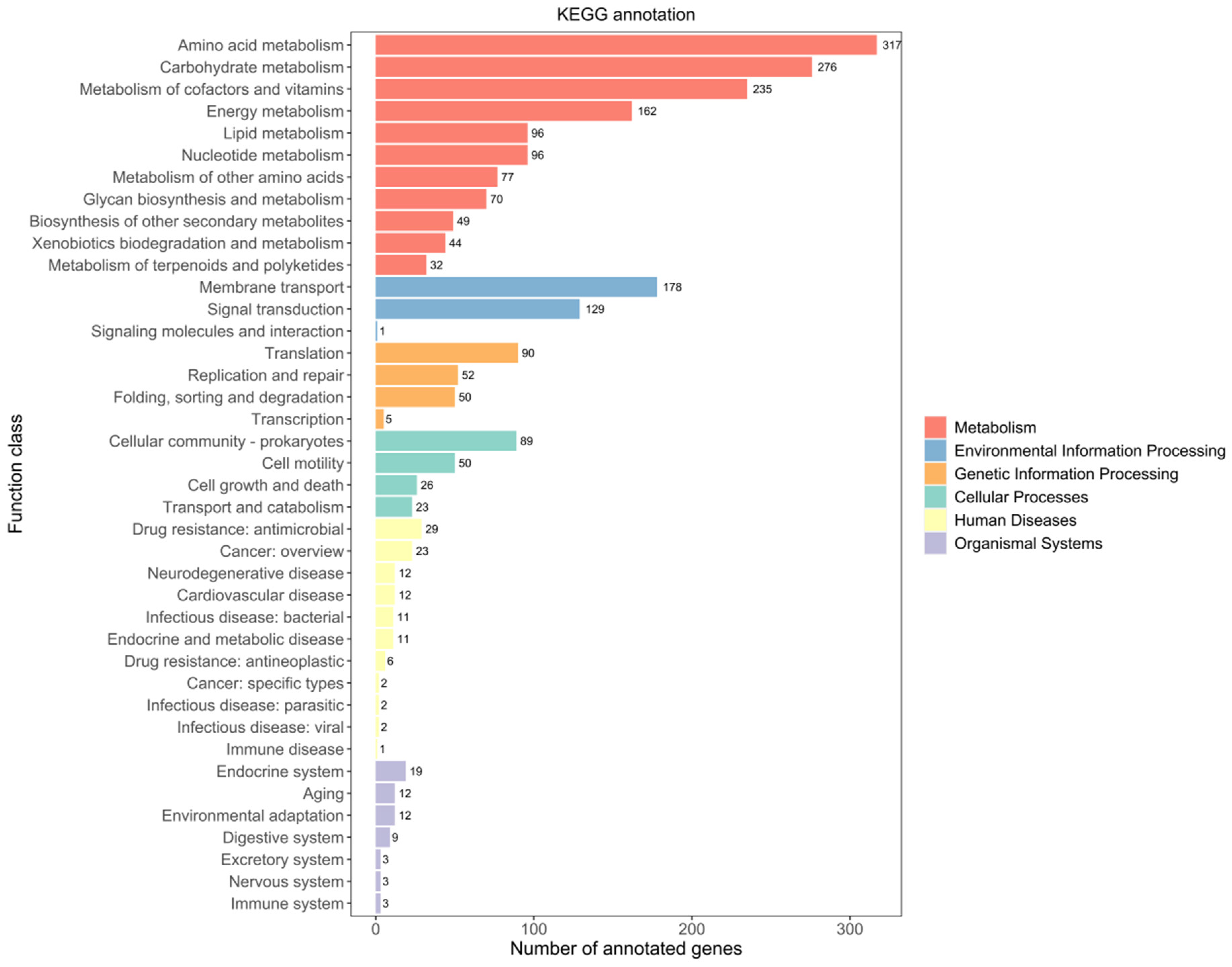
2.1.3. KEGG Database Annotations
2.1.4. CAZy Database Annotations
2.1.5. Virulence Factor Gene Mining
2.2. Secondary Metabolites Isolated from Brevibacillus sp. JNUCC 41
2.3. Hyaluronidase Inhibitory Activity of the Isolated Secondary Metabolites
2.4. Molecular Properties and Drug Likeness
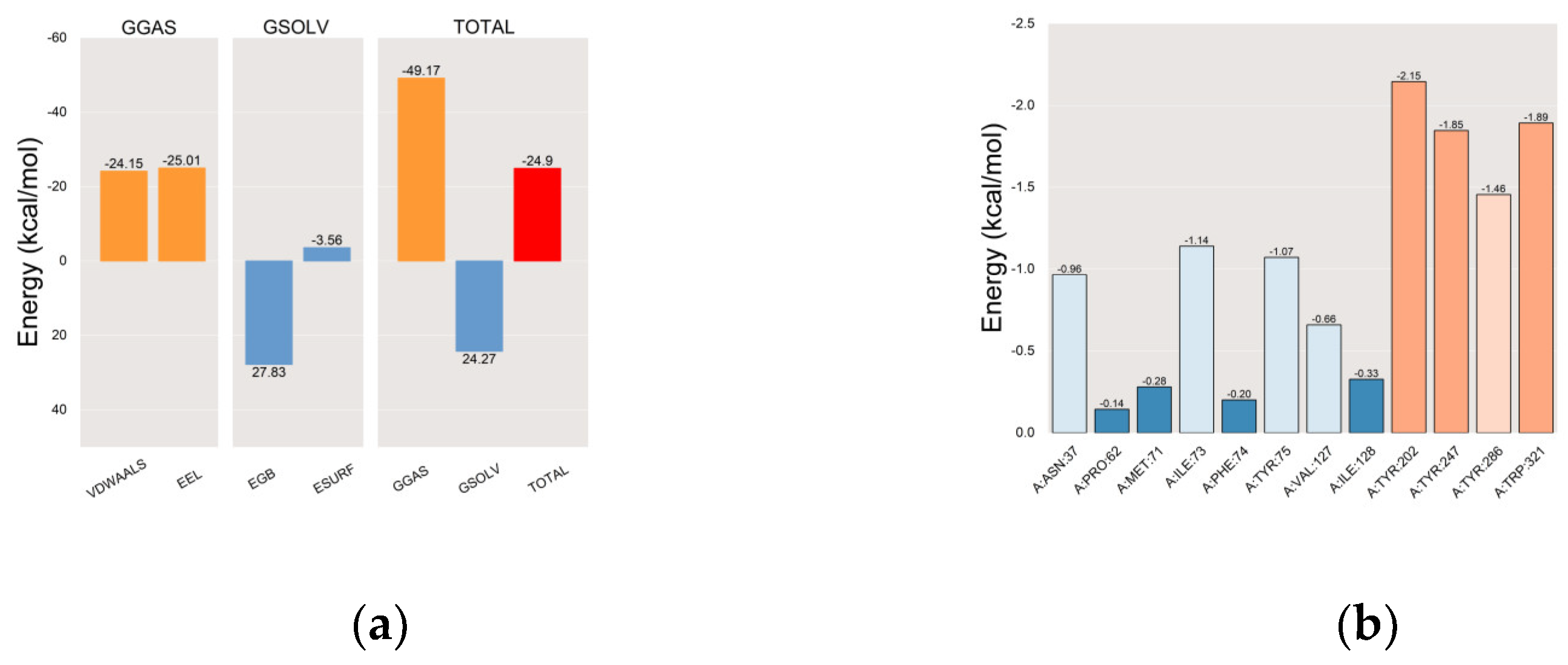
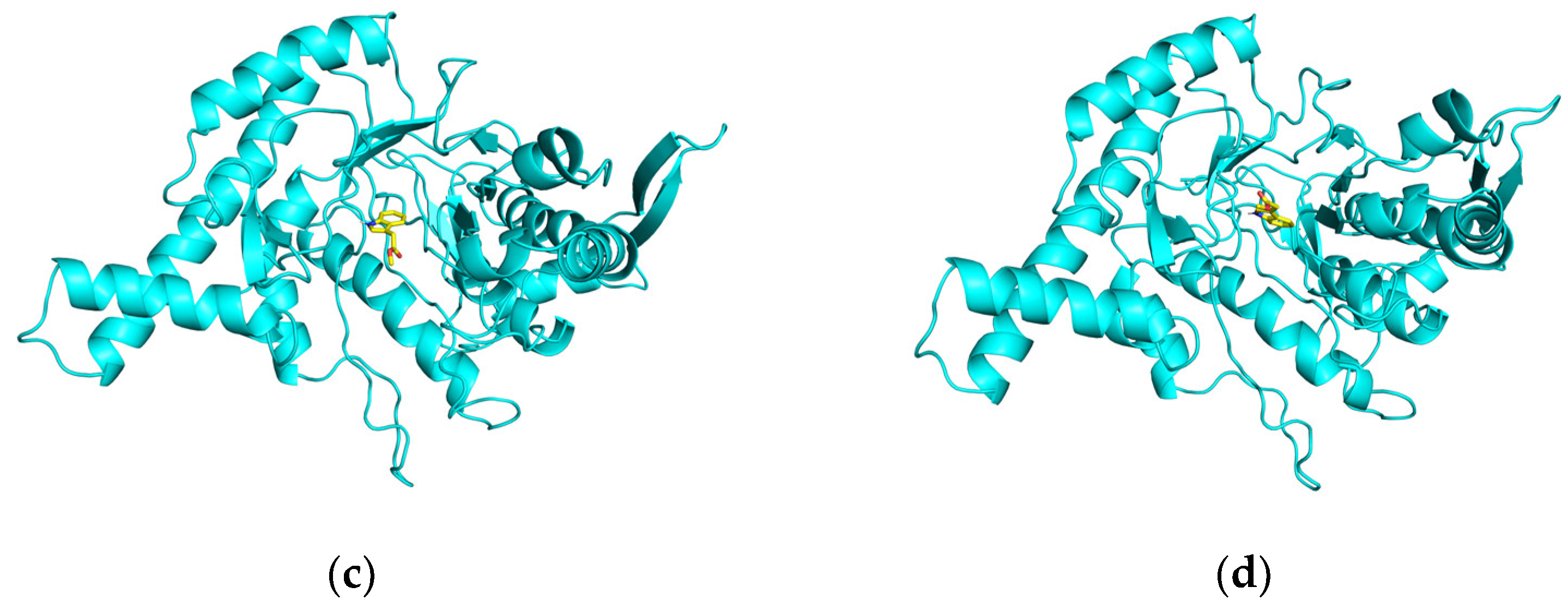
2.5. Docking and Molecular Dynamics (MD) Simulations
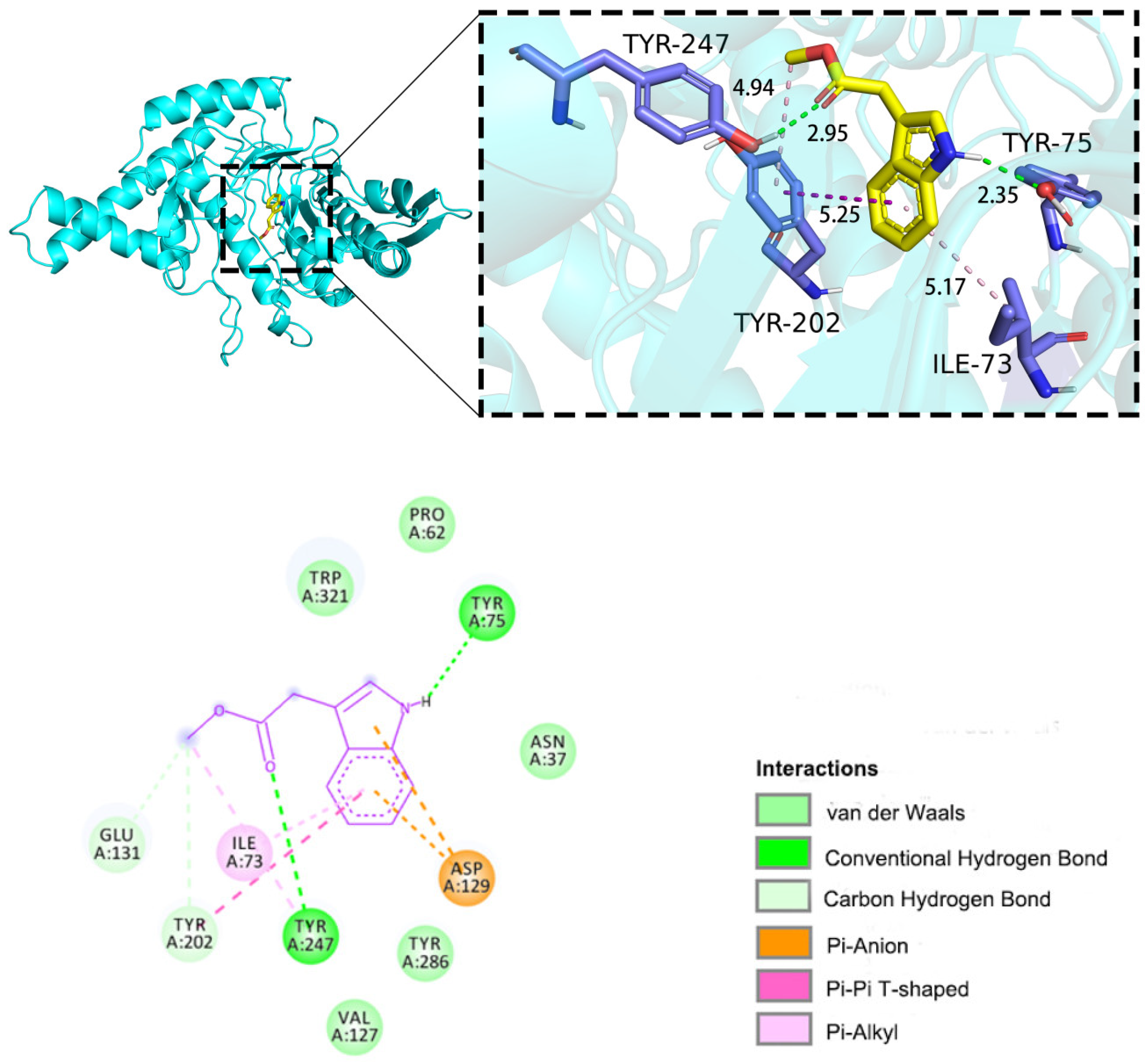
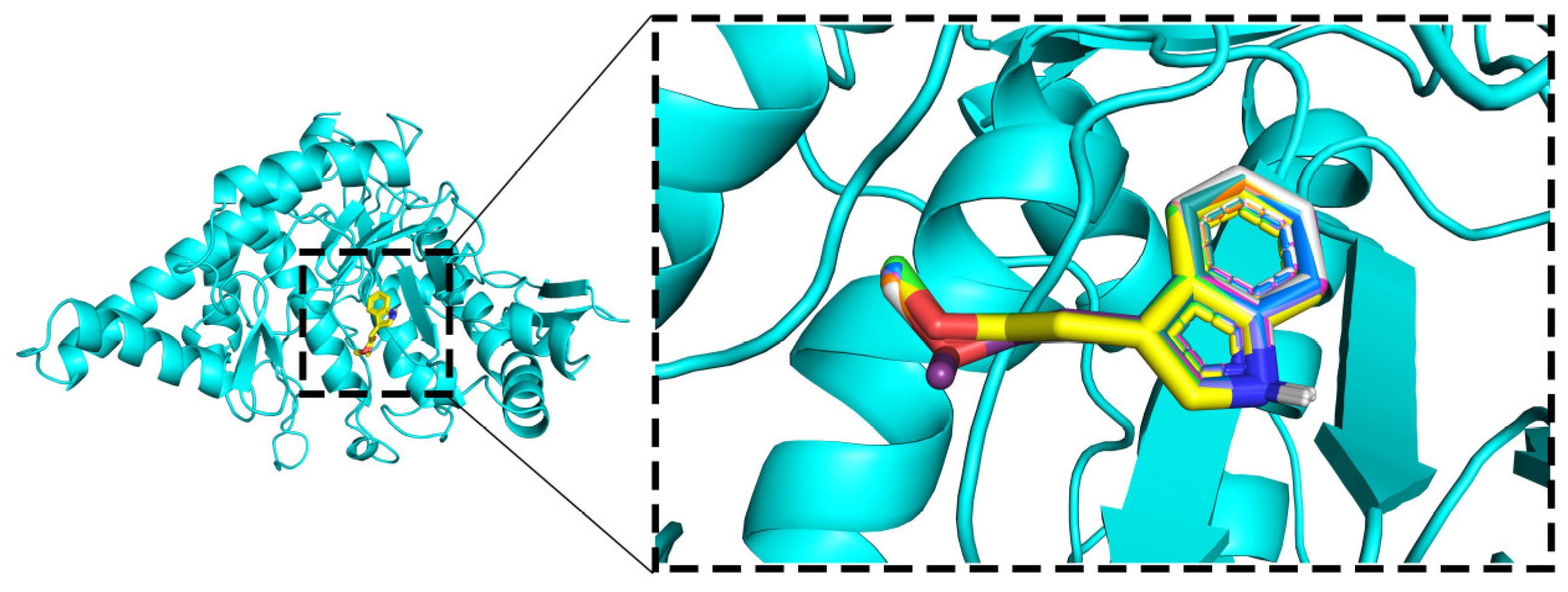
2.5.1. Molecular Docking
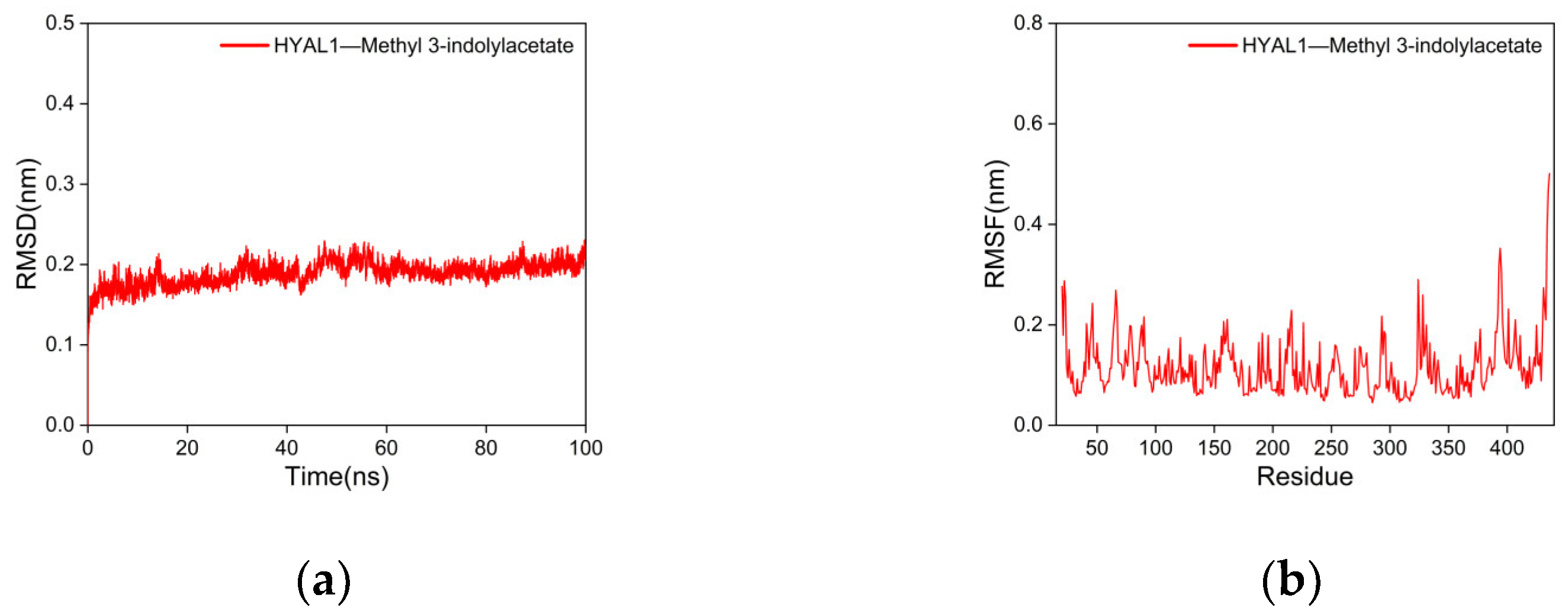
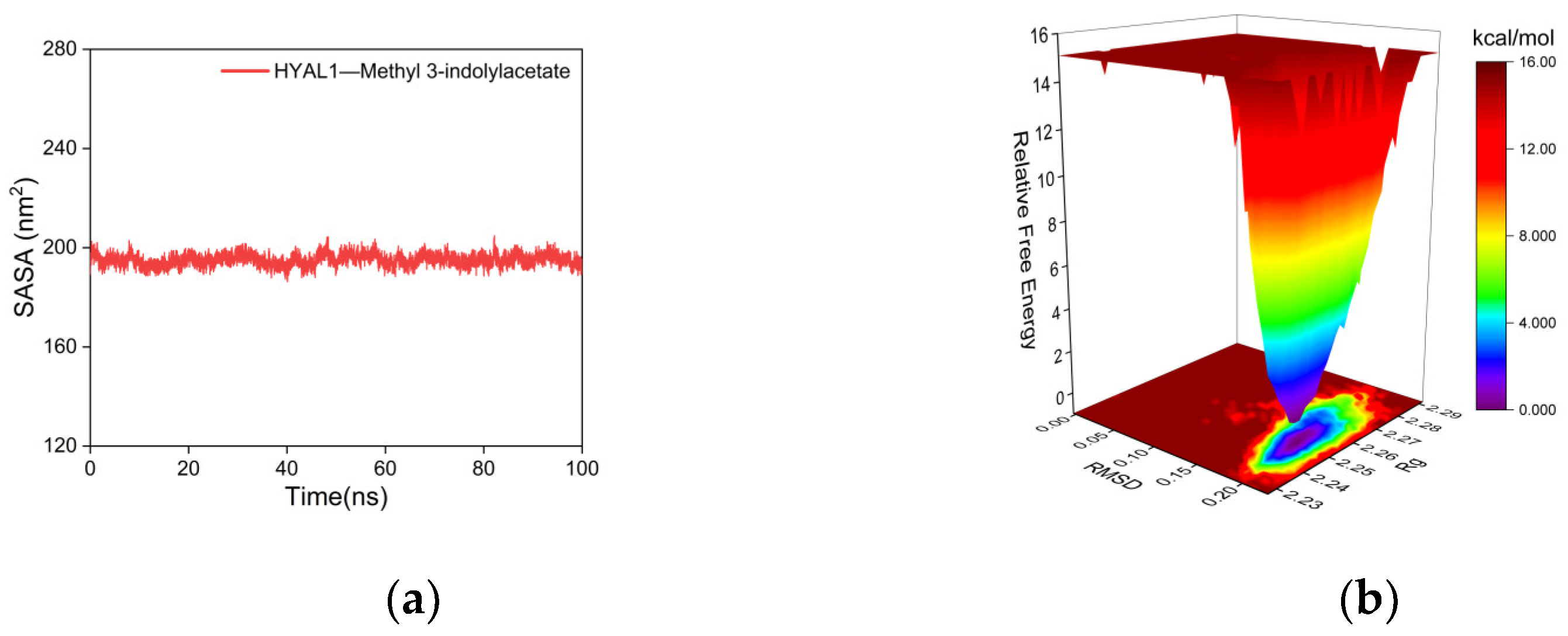
2.5.2. Molecular Dynamics (MD) Simulations
3. Materials and Methods
3.1. Isolation of the Bacterial Brevibacillus sp. JNUCC 41
3.2. Genome Extraction and Sequencing
3.3. Genome Annotation
3.4. General Experimental Procedures
3.5. Fermentation, Extraction, and Isolation
3.5.1. Methyl Indole-3-Acetate
3.5.2. Dibutyl Phthalate
3.5.3. Daidzein
3.5.4. Maculosin
3.5.5. N-Acetyl-L-Tryptophan
3.6. Hyaluronidase Inhibitory Activity of Secondary Metabolites
3.7. Molecular Properties and Drug Likeness
3.8. Molecular Docking Simulation
3.9. Molecular Dynamics (MD) Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shida, O.; Takagi, H.; Kadowaki, K. Proposal for two new genera, Brevibacillus gen. nov. and Aneurinibacillus gen. nov. Int. J. Syst. Evol. Microbiol. 1996, 46, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Kinkle, B.K.; Sadowsky, M.J.; Johnstone, K. Tellurium and selenium resistance in rhizobia and its potential use for direct isolation of Rhizobium meliloti from soil. Appl. Environ. Microbiol. 1994, 60, 1674–1677. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.I.S. Biological control of potato brown leaf spot disease caused by Alternaria alternata using Brevibacillus formosus strain DSM 9885 and Brevibacillus brevis strain NBRC 15304. J. Plant Pathol. Microbiol. 2017, 8, 6. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, M.; Wang, J. Biocontrol effects of Brevibacillus laterosporus AMCC100017 on potato common scab and its impact on rhizosphere bacterial communities. Biol. Control 2017, 106, 89–98. [Google Scholar] [CrossRef]
- Li, C.; Shi, W.; Wu, D. Biocontrol of potato common scab by Brevibacillus laterosporus BL12 is related to the reduction of pathogen and changes in soil bacterial community. Biol. Control 2021, 153, 104496. [Google Scholar] [CrossRef]
- Girish, N.; Umesha, S. Effect of plant growth promoting rhizobacteria on bacterial canker of tomato. Arch. Phytopathol. Plant Prot. 2005, 38, 235–243. [Google Scholar] [CrossRef]
- Chandel, S.; Allan, E.J.; Woodward, S. Biological control of Fusarium oxysporum f. sp. lycopersici on tomato by Brevibacillus brevis. J. Phytopathol. 2010, 158, 470–478. [Google Scholar]
- Wani, P.A.; Rafi, N.; Wani, U. Simultaneous bioremediation of heavy metals and biodegradation of hydrocarbons by metal resistant Brevibacillus parabrevis OZF5 improves plant growth promotion. Bioremediat. J. 2023, 27, 20–31. [Google Scholar] [CrossRef]
- Lim, J.G.; Park, D.H. Degradation of polyvinyl alcohol by Brevibacillus laterosporus: Metabolic pathway of polyvinyl alcohol to acetate. J. Microbiol. Biotechnol. 2001, 11, 928–933. [Google Scholar]
- Jeyaseelan, A.; Sivashanmugam, K.; Jayaraman, K. Comparative applications of bioreactor and shake flask system for the biodegradation of tannin and biotreatment of composite tannery effluents. Pollut. Res. 2008, 27, 371–375. [Google Scholar]
- Reda, A.B.; Ashra, T.A.H. Optimization of bacterial biodegradation of toluene and phenol under different nutritional and environmental conditions. J. Appl. Sci. Res. 2010, 8, 1086–1095. [Google Scholar]
- Scheloske, S.; Maetz, M.; Schneider, T. Element distribution in mycorrhizal and nonmycorrhizal roots of the halophyte Aster tripolium determined by proton induced X-ray emission. Protoplasma 2004, 223, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Vivas, A.; Barea, J.M.; Biró, B. Effectiveness of autochthonous bacterium and mycorrhizal fungus on Trifolium growth, symbiotic development and soil enzymatic activities in Zn contaminated soil. J. Appl. Microbiol. 2006, 100, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Vivas, A.; Biró, B.; Németh, T. Nickel-tolerant Brevibacillus brevis and arbuscular mycorrhizal fungus can reduce metal acquisition and nickel toxicity effects in plant growing in nickel supplemented soil. Soil Boil. Biochem. 2006, 38, 2694–2704. [Google Scholar] [CrossRef]
- Yang, X.; Yousef, A.E. Antimicrobial peptides produced by Brevibacillus spp.: Structure, classification and bioactivity: A mini review. World J. Microbiol. Biotechnol. 2018, 34, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Liu, Q.; Guo, H. Tostadin, a novel antibacterial peptide from an antagonistic microorganism Brevibacillus brevis XDH. Bioresour. Technol. 2012, 111, 504–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qin, L.; Pace, C.J. Solubilized gramicidin A as potential systemic antibiotics. ChemBioChem 2012, 13, 51–55. [Google Scholar] [CrossRef]
- Hill, S.R.; Bonjouklian, R.; Powis, G. A multisample assay for inhibitors of phosphatidylinositol phospholipase C: Identification of naturally occurring peptide inhibitors with antiproliferative activity. Anti-Cancer Drug Des. 1994, 9, 353–361. [Google Scholar]
- Takeuchi, T.; IINUMA, H.; KUNIMOTO, S. A new antitumor antibiotic, spergualin: Isolation and antitumor activity. J Antibiot. 1981, 34, 1619–1621. [Google Scholar] [CrossRef]
- Desjardine, K.; Pereira, A.; Wright, H. Tauramamide, a lipopeptide antibiotic produced in culture by Brevibacillus laterosporus isolated from a marine habitat: Structure elucidation and synthesis. J. Nat. Prod. 2007, 70, 1850–1853. [Google Scholar] [CrossRef]
- Yang, X.; Huang, E.; Yesil, M. Draft genome sequence of Brevibacillus laterosporus OSY-I1, a strain that produces brevibacillin, which combats drug-resistant Gram-positive bacteria. Genome Announc. 2017, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Shoji, J.; Kato, T. The structure of brevistin studies on antibiotics from the genus bacillus. J. Antibiot. 1976, 29, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Huang, E.; Yuan, C. Isolation and structural elucidation of brevibacillin, an antimicrobial lipopeptide from Brevibacillus laterosporus that combats drug-resistant Gram-positive bacteria. Appl. Environ Microbiol. 2016, 82, 2763–2772. [Google Scholar] [CrossRef] [PubMed]
- Ghadbane, M.; Harzallah, D.; Laribi, A.I. Purification and biochemical characterization of a highly thermostable bacteriocin isolated from Brevibacillus brevis strain GM100. Biosci. Biotechnol. Biochem. 2013, 77, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Chittpurna, A. Identification, purification and characterization of laterosporulin, a novel bacteriocin produced by Brevibacillus sp. strain GI-9. PLoS ONE 2012, 7, 31498. [Google Scholar] [CrossRef] [PubMed]
- Baindara, P.; Singh, N.; Ranjan, M. Laterosporulin10: A novel defensin like class IId bacteriocin from Brevibacillus sp. strain SKDU10 with inhibitory activity against microbial pathogens. Microbiology 2016, 162, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.S.; Sharma, D.; Singh, C. Brevicillin, a novel lanthipeptide from the genus Brevibacillus with antimicrobial, antifungal, and antiviral activity. J. Appl. Microbiol. 2023, 134, 54. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Asano, D.; Sawamura, M. Ulbactins F and G, polycyclic thiazoline derivatives with tumor cell migration inhibitory activity from Brevibacillus sp. Org. Lett. 2016, 18, 1658–1661. [Google Scholar] [CrossRef] [PubMed]
- Jianmei, C.; Bo, L.; Zheng, C. Identification of ethylparaben as the antimicrobial substance produced by Brevibacillus brevis FJAT-0809-GLX. Microbiol. Res. 2015, 172, 48–56. [Google Scholar] [CrossRef]
- Falaleeva, M.; Zurek, O.W.; Watkins, R.L. Transcription of the Streptococcus pyogenes hyaluronic acid capsule biosynthesis operon is regulated by previously unknown upstream elements. Infect. Immun. 2014, 82, 5293–5307. [Google Scholar] [CrossRef]
- Weigel, P.H. Functional characteristics and catalytic mechanisms of the bacterial hyaluronan synthases. IUBMB Life 2002, 54, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Spicer, A.P.; McDonald, J.A. Characterization and molecular evolution of a vertebrate hyaluronan synthase gene family. J. Biol. Chem. 1998, 273, 1923–1932. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Spicer, A.P. Hyaluronan: A multifunctional, megaDalton, stealth molecule. Curr. Opin. Cell Biol. 2000, 12, 581–586. [Google Scholar] [CrossRef]
- Toole, B.P. Hyaluronan: From extracellular glue to pericellular cue. Nat. Rev. Cancer 2004, 4, 528–539. [Google Scholar] [CrossRef]
- Krupkova, O.; Greutert, H.; Boos, N. Expression and activity of hyaluronidases HYAL-1, HYAL-2 and HYAL-3 in the human intervertebral disc. Eur. Spine J. 2020, 29, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Stern, R. Hyaluronidases in cancer biology. In Hyaluronan in Cancer Biology; Academic Press: San Diego, CA, USA, 2008; pp. 207–220. [Google Scholar]
- Gireesh, T.; Kamble, R.R.; Kattimani, P.P. Synthesis of sydnone substituted Biginelli derivatives as hyaluronidase inhibitors. Arch. Pharm. 2013, 346, 645–653. [Google Scholar] [CrossRef]
- Girish, K.S.; Kemparaju, K.; Nagaraju, S. Hyaluronidase inhibitors: A biological and therapeutic perspective. Curr. Med. Chem. 2009, 16, 2261–2288. [Google Scholar] [CrossRef]
- Maksimenko, A.V.; Beabealashvili, R.S. Effect of the hyaluronidase microe nvironment on the enzyme structure–function relationship and computational study of the in silico molecular docking of chondroitin sulfate and heparin short fragments to hyaluronidase. Russ. Chem. Bull. 2018, 67, 636–646. [Google Scholar] [CrossRef]
- Li, X.; Xu, R.; Cheng, Z. Comparative study on the interaction between flavonoids with different core structures and hyaluronidase. Spectrochim. Acta A 2021, 262, 120079. [Google Scholar] [CrossRef]
- Zeng, H.; Yang, R.; You, J. Spectroscopic and Docking Studies on the Binding of Liquiritigenin with Hyaluronidase for Antiallergic Mechanism. Scientifica 2016, 2016, 9178097. [Google Scholar] [CrossRef]
- Sunitha, K.; Suresh, P.; Santhosh, M.S. Inhibition of hyaluronidase by N-acetyl cysteine and glutathione: Role of thiol group in hyaluronan protection. Int. J. Biol. Macromol. 2013, 55, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Fatoki, T.H.; Ajiboye, B.O.; Aremu, A.O. In Silico Evaluation of the Antioxidant, Anti-Inflammatory, and Dermatocosmetic Activities of Phytoconstituents in Licorice (Glycyrrhiza glabra L.). Cosmetics 2023, 10, 69. [Google Scholar] [CrossRef]
- Zeng, H.; Hu, G.; You, J. Spectroscopic and molecular modeling investigation on the interactions between hyaluronidase and baicalein and chrysin. Process. Biochem. 2015, 50, 738–745. [Google Scholar] [CrossRef]
- Zeng, H.; You, J.; Yang, R. Molecular interaction of silybin with hyaluronidase: A spectroscopic and molecular docking study. Spectr. Lett. 2017, 50, 515–521. [Google Scholar] [CrossRef]
- Elattar, E.M.; Galala, A.A.; Saad, H.E.A. Hyaluronidase Inhibitory Activity and In Silico Docking Study of New Eugenol 1, 2, 3-triazole Derivatives. Chem. Select. 2022, 7, e202202194. [Google Scholar] [CrossRef]
- Luo, Z.; He, H.; Tang, T. Synthesis and Biological Evaluations of Betulinic Acid Derivatives with Inhibitory Activity on Hyaluronidase and Anti-Inflammatory Effects Against Hyaluronic Acid Fragment Induced Inflammation. Front. Chem. 2022, 10, 892554. [Google Scholar] [CrossRef] [PubMed]
- Arming, S.; Strobl, B.; Wechselberger, C. In vitro mutagenesis of PH-20 hyaluronidase from human sperm. Eur. J. Biochem. 1997, 247, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Bharadwaj, A.G.; Casper, A. Hyaluronidase activity of human Hyal1 requires active site acidic and tyrosine residues. J. Biol. Chem. 2009, 284, 9433–9442. [Google Scholar] [CrossRef]
- Jason, R.G.; Paul, S. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K. Microbial genome analysis: The COG approach. Brief. Bioinform. 2019, 20, 1063–1070. [Google Scholar] [CrossRef]
- Wei, X.; Jiang, S.; Chen, Y. Cirrhosis related functionality characteristic of the fecal microbiota as revealed by a metaproteomic approach. BMC Gastroenterol. 2016, 16, 121. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.O.; van der Horst, D.J. Lipid transport biochemistry and its role in energy production. Annu. Rev. Entomol. 2000, 45, 233–260. [Google Scholar] [CrossRef] [PubMed]
- Glatz, J.F.C.; Luiken, J.J.F.P.; Bonen, A. Membrane fatty acid transporters as regulators of lipid metabolism: Implications for metabolic disease. Physiol. Rev. 2010, 90, 367–417. [Google Scholar] [CrossRef] [PubMed]
- Horne, J.E.; Brockwell, D.J.; Radford, S.E. Role of the lipid bilayer in outer membrane protein folding in Gram-negative bacteria. J. Biol. Chem. 2020, 295, 10340–10367. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ge, Q.; Yan, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, 115–121. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Krüger, K.; Chafee, M.; Ben Francis, T. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J. 2019, 13, 2800–2816. [Google Scholar] [CrossRef]
- Sichert, A.; Cordero, O.X. Polysaccharide-bacteria interactions from the lens of evolutionary ecology. Front. Microbiol. 2021, 12, 705082. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, 95–101. [Google Scholar] [CrossRef]
- Razv, I.E.; Whitfield, G.B.; Reichhardt, C. Glycoside hydrolase processing of the Pel polysaccharide alters biofilm biomechanics and Pseudomonas aeruginosa virulence. NPJ Biofilms Microbiomes 2023, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Mizan, S.; Henk, A.; Stallings, A. Cloning and characterization of sialidases with 2–6′ and 2–3′ sialyl lactose specificity from Pasteurella multocida. J. Bacteriol. 2000, 182, 6874–6883. [Google Scholar] [CrossRef]
- Friedlander, A.M. The anthrax capsule: Role in pathogenesis and target for vaccines and therapeutics. In The Challenge of Highly Pathogenic Microorganisms: Mechanisms of Virulence and Novel Medical Countermeasures; Springer: Dordrecht, The Netherlands, 2010; pp. 1–9. [Google Scholar]
- Roberts, I.S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 1996, 50, 285–315. [Google Scholar] [CrossRef]
- Wang, L.; Si, W.; Xue, H. A fibronectin-binding protein (FbpA) of Weissella cibaria inhibits colonization and infection of Staphylococcus aureus in mammary glands. Cell Microbiol. 2017, 19, 12731. [Google Scholar] [CrossRef]
- Souza, R.F.S.; Jardin, J.; Cauty, C. Contribution of sortase SrtA2 to Lactobacillus casei BL23 inhibition of Staphylococcus aureus internalization into bovine mammary epithelial cells. PLoS ONE 2017, 12, e0174060. [Google Scholar] [CrossRef]
- Chen, P.N.; Hao, M.J.; Li, H.J. Biotransformations of anthranilic acid and phthalimide to potent antihyperlipidemic alkaloids by the marine-derived fungus Scedosporium apiospermum F41–1. Bioorg. Chem. 2021, 116, 105375. [Google Scholar] [CrossRef] [PubMed]
- Ruikar, A.D.; Gadkari, T.V.; Phalgune, U.D. Dibutyl phthalate, a secondary metabolite from Mimusops elengi. Chem. Nat. Compd. 2011, 46, 955–956. [Google Scholar] [CrossRef]
- Shaaban, K.A.; Shepherd, M.D.; Ahmed, T.A. Pyramidamycins AD and 3-hydroxyquinoline-2-carboxamide; cytotoxic benzamides from Streptomyces sp. DGC1. J. Antibiot. 2012, 65, 615–622. [Google Scholar] [CrossRef]
- Driche, E.H.; Badji, B.; Bijani, C. A new saharan strain of Streptomyces sp. GSB-11 produces maculosin and N-acetyltyramine active against multidrug-resistant pathogenic bacteria. Curr. Microbiol. 2022, 79, 298. [Google Scholar] [CrossRef]
- Liu, D.; Shi, X.; Wang, Z. Anticancer constituents of ethyl acetate extract from Euphorbia helioscopia. Chem. Nat. Compd. 2015, 51, 969–971. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef]
- Polkam, N.; Ramaswamy, V.R.; Rayam, P. Synthesis, molecular properties prediction and anticancer, antioxidant evaluation of new edaravone derivatives. Bioorg. Med. Chem. Lett. 2016, 26, 2562–2568. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Chao, K.L.; Muthukumar, L.; Herzberg, O. Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme involved in tumor growth and angiogenesis. Biochemistry 2007, 46, 6911–6920. [Google Scholar] [CrossRef]
- Daniel, S.; Bert, L.D.G. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar]
- Mariana, P.B.; Cleydson, B.R.S.; Leonardo, B.F. Pharmacophore and structure-based drug design, molecular dynamics and admet/tox studies to design novel potential pad4 inhibitors. J. Biomol. Struct. Dyn. 2018, 37, 966–981. [Google Scholar]
- Li, X.; Liu, H.; Yang, Z. Study on the interaction of hyaluronidase with certain flavonoids. J. Mol. Struct. 2021, 1241, 130686. [Google Scholar] [CrossRef]
- Sargsyan, K.; Grauffel, C.; Lim, C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef]
- Benson, N.C.; Daggett, V. A comparison of multiscale methods for the analysis of molecular dynamics simulations. J. Phys. Chem. B 2012, 116, 8722–8731. [Google Scholar] [CrossRef]
- Yamamoto, E.; Akimoto, T.; Mitsutake, A. Universal relation between instantaneous diffusivity and radius of gyration of proteins in aqueous solution. Phys. Rev. Lett. 2021, 126, 128101. [Google Scholar] [CrossRef]
- Martı, J. Analysis of the hydrogen bonding and vibrational spectra of supercritical model water by molecular dynamics simulations. J. Chem. Phys. 1999, 110, 6876–6886. [Google Scholar] [CrossRef]
- Ausaf, A.S.; Hassan, I.; Islam, A. A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr. Protein Pept. Sci. 2014, 15, 456–476. [Google Scholar]
- Fu, H.; Chen, H.; Blazhynska, M. Accurate determination of protein: Ligand standard binding free energies from molecular dynamics simulations. Nat. Protoc. 2022, 17, 1114–1141. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Dis. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, Z.; Zhang, J. Molecular dynamics simulation of the interaction of food proteins with small molecules. Food Chem. 2023, 405, 134824. [Google Scholar] [CrossRef]
- Noel, M.O.B.; Michael, B.; Craig, A.J.; Chris, M.; Tim, V.; Geoffrey, R.H. Open Babel: An open chemical toolbox. J. Chem. 2011, 3, 33. [Google Scholar]
- Garrett, M.M.; Ruth, H.; Arthur, J.O. Using AutoDock for Ligand-Receptor Docking. Curr. Protoc. Bioinform. 2008, 24, 8–14. [Google Scholar]

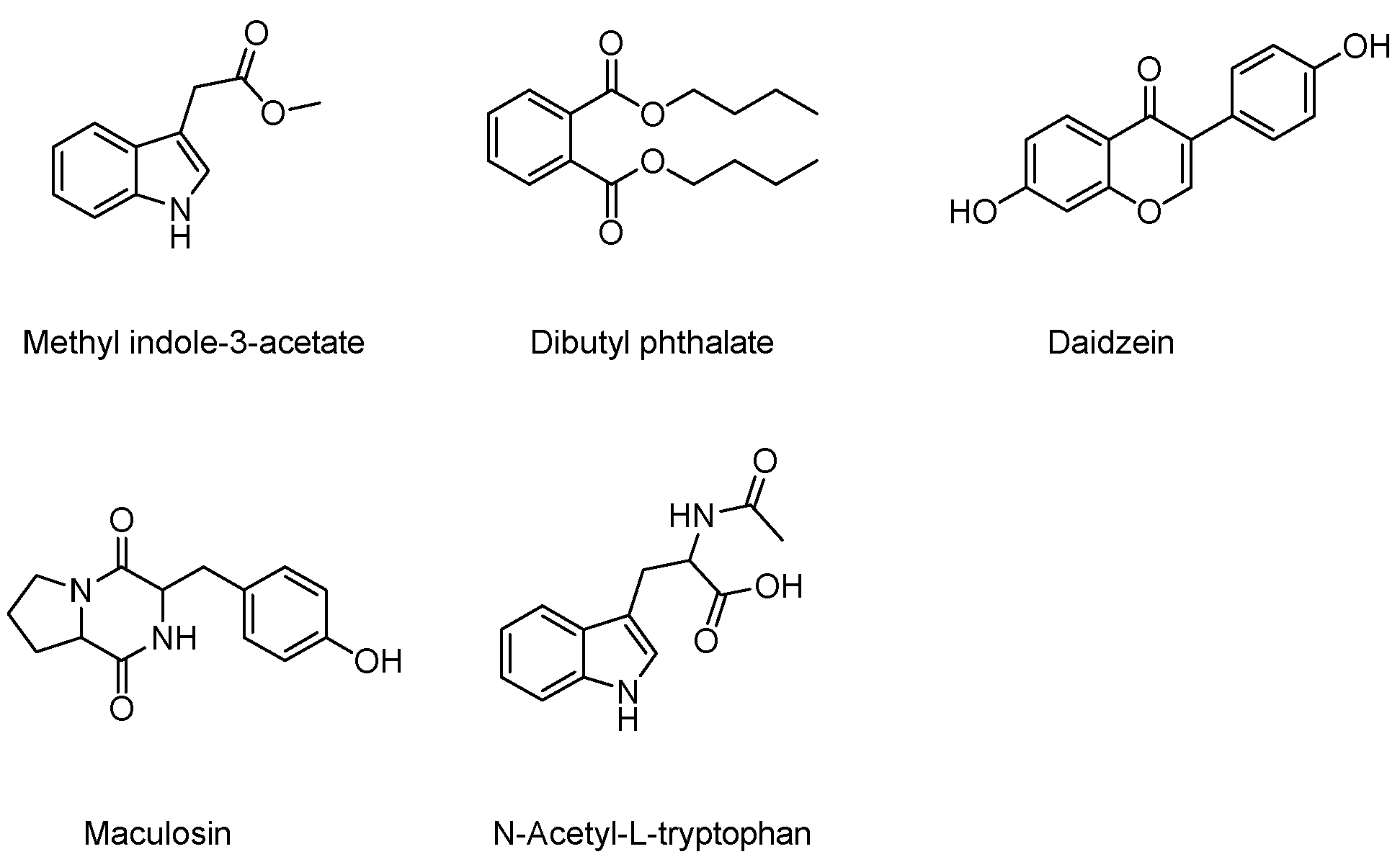
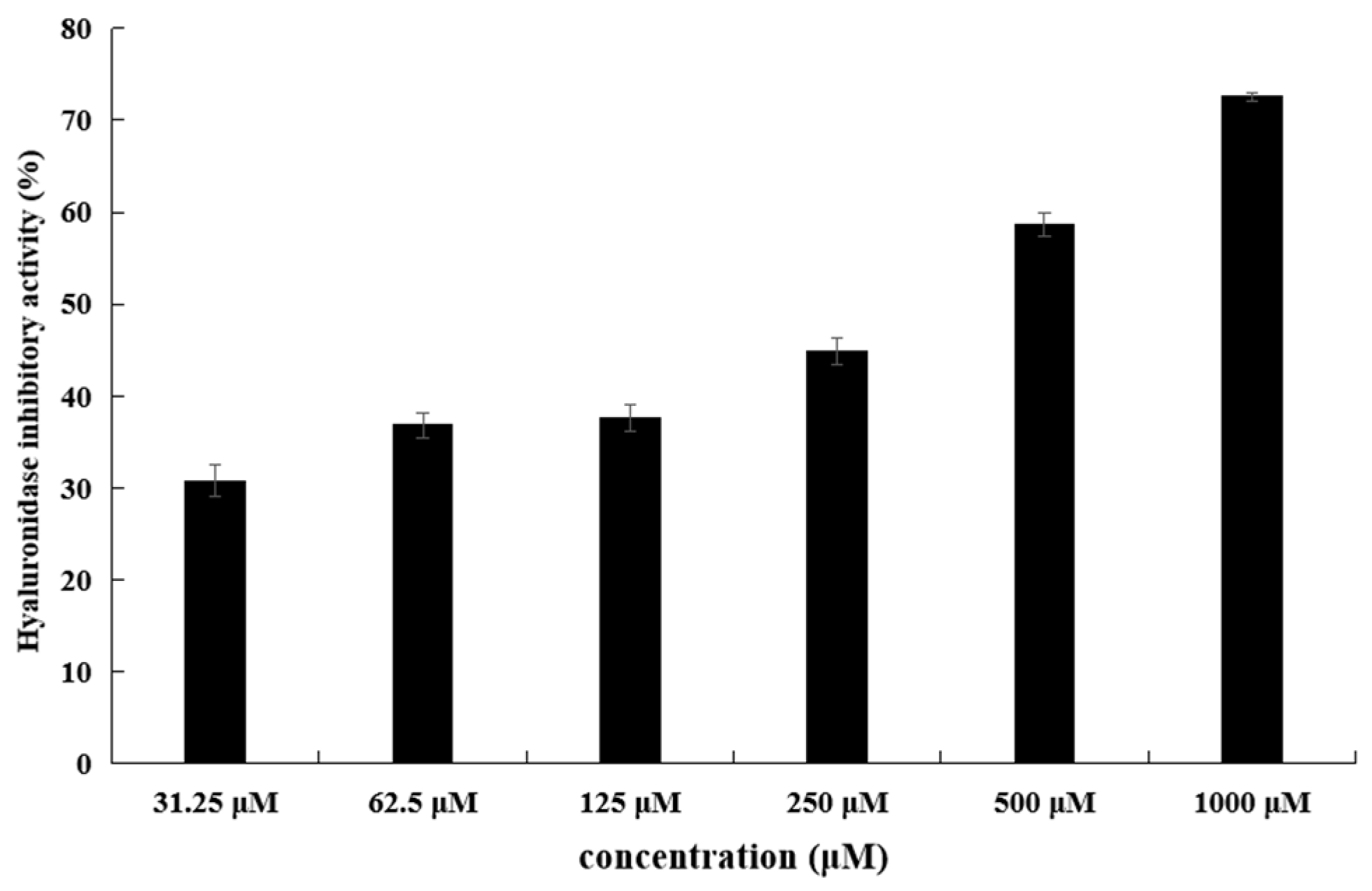


| Compounds | Hyaluronidase-Inhibitory Activity IC50 (μM) |
| Methyl indole-3-acetate | 343.9 |
| Dibutyl phthalate | - |
| Maculosin | - |
| N-Acetyl-L-tryptophan | - |
| EGCG | 172.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Liang, X.; Hyun, C.-G. Isolation, Characterization, Genome Annotation, and Evaluation of Hyaluronidase Inhibitory Activity in Secondary Metabolites of Brevibacillus sp. JNUCC 41: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation. Int. J. Mol. Sci. 2024, 25, 4611. https://doi.org/10.3390/ijms25094611
Xu Y, Liang X, Hyun C-G. Isolation, Characterization, Genome Annotation, and Evaluation of Hyaluronidase Inhibitory Activity in Secondary Metabolites of Brevibacillus sp. JNUCC 41: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation. International Journal of Molecular Sciences. 2024; 25(9):4611. https://doi.org/10.3390/ijms25094611
Chicago/Turabian StyleXu, Yang, Xuhui Liang, and Chang-Gu Hyun. 2024. "Isolation, Characterization, Genome Annotation, and Evaluation of Hyaluronidase Inhibitory Activity in Secondary Metabolites of Brevibacillus sp. JNUCC 41: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation" International Journal of Molecular Sciences 25, no. 9: 4611. https://doi.org/10.3390/ijms25094611








