Reduced Expression of CLEC4G in Neurons Is Associated with Alzheimer’s Disease
Abstract
:1. Introduction
2. Results
2.1. High Levels of CLEC4G Are Expressed in Both Mouse and Human Brain Tissues
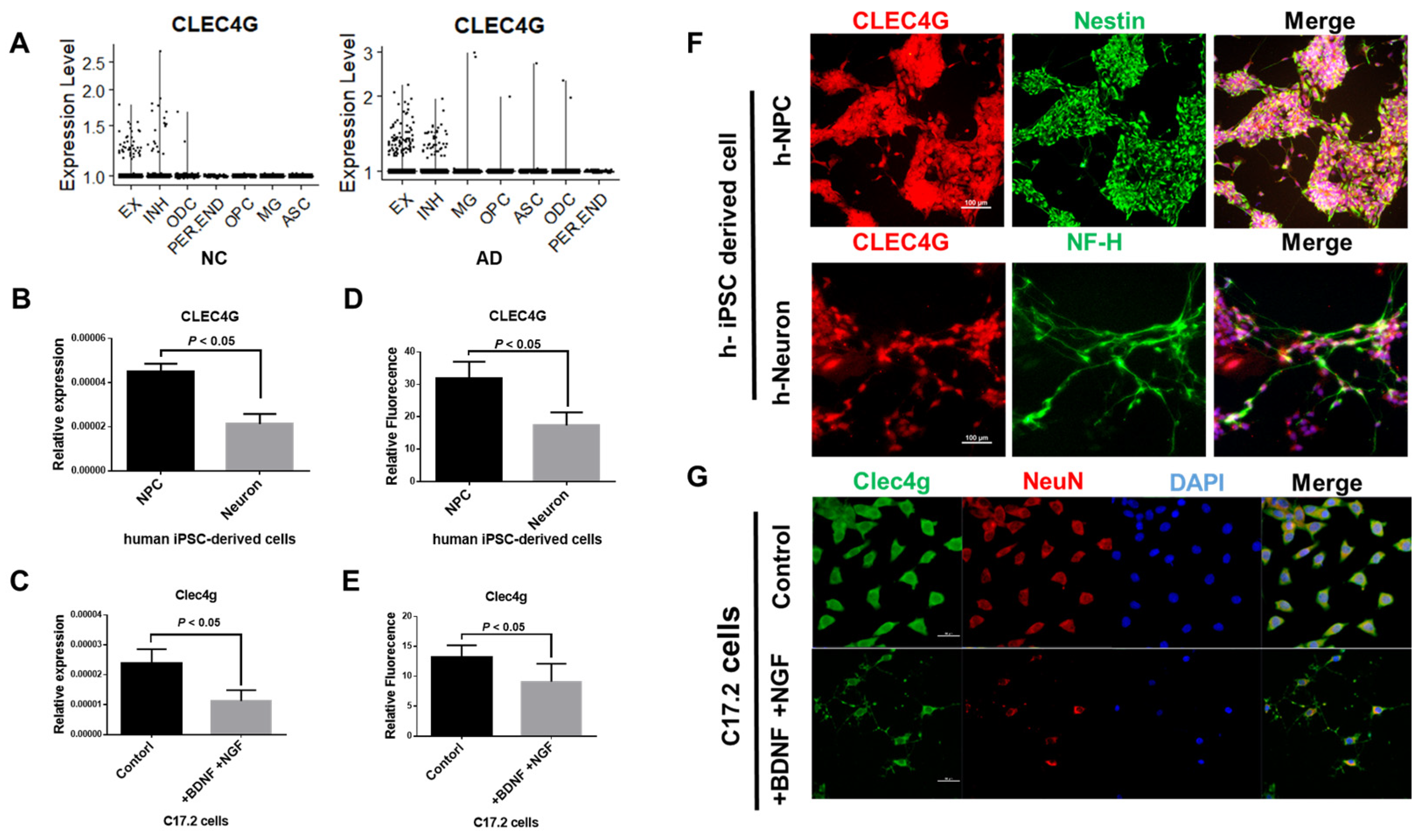
2.2. CLEC4G Is Mainly Expressed in Neurons
2.3. Reduced Expression of CLEC4G in Patients with AD and APP/PS1 Mice with Increasing Age
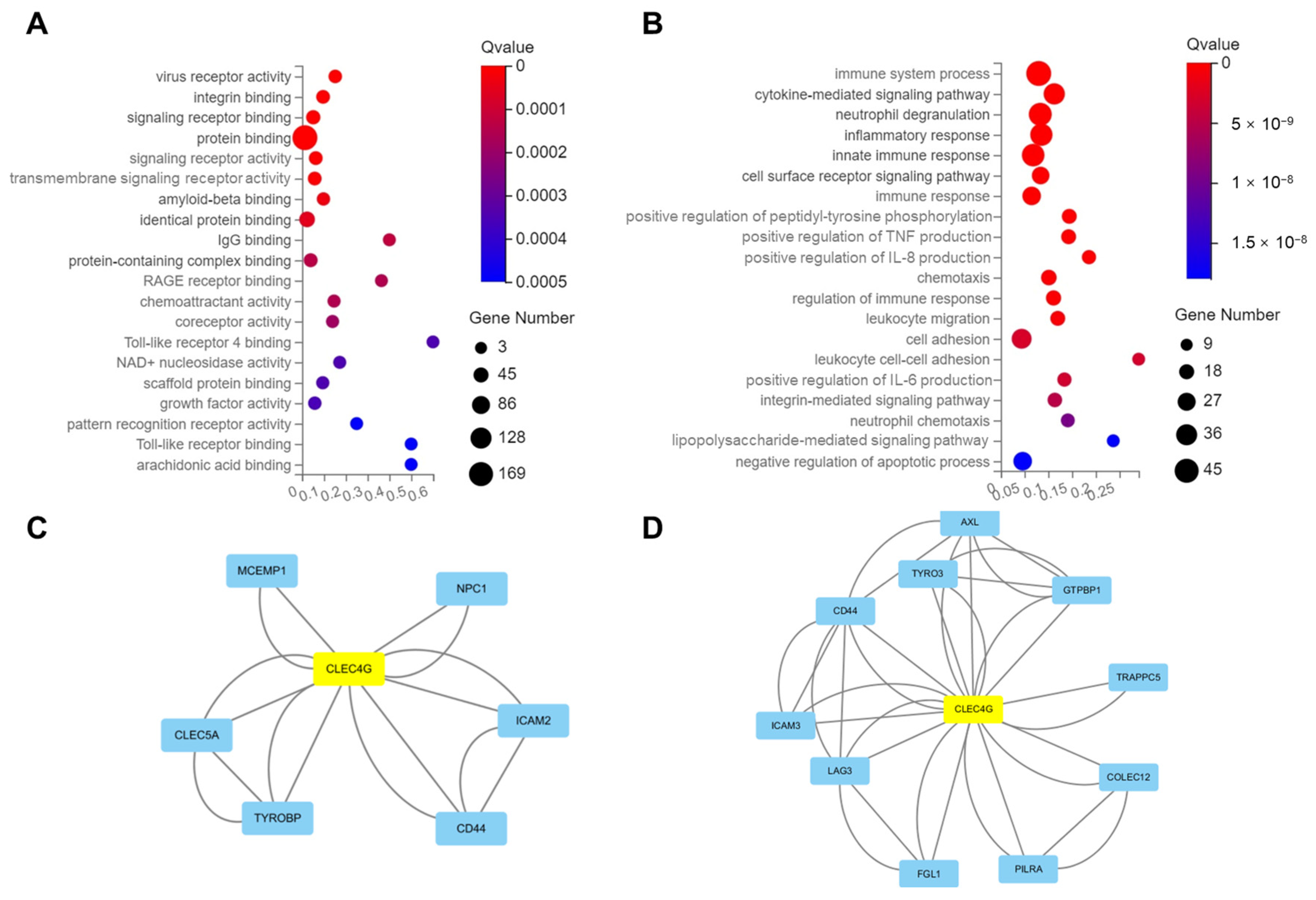
2.4. Functional Analysis of CLEC4G Protein in Neurons
2.5. CLEC4G Protein Plays a Positive Role in AD Progression
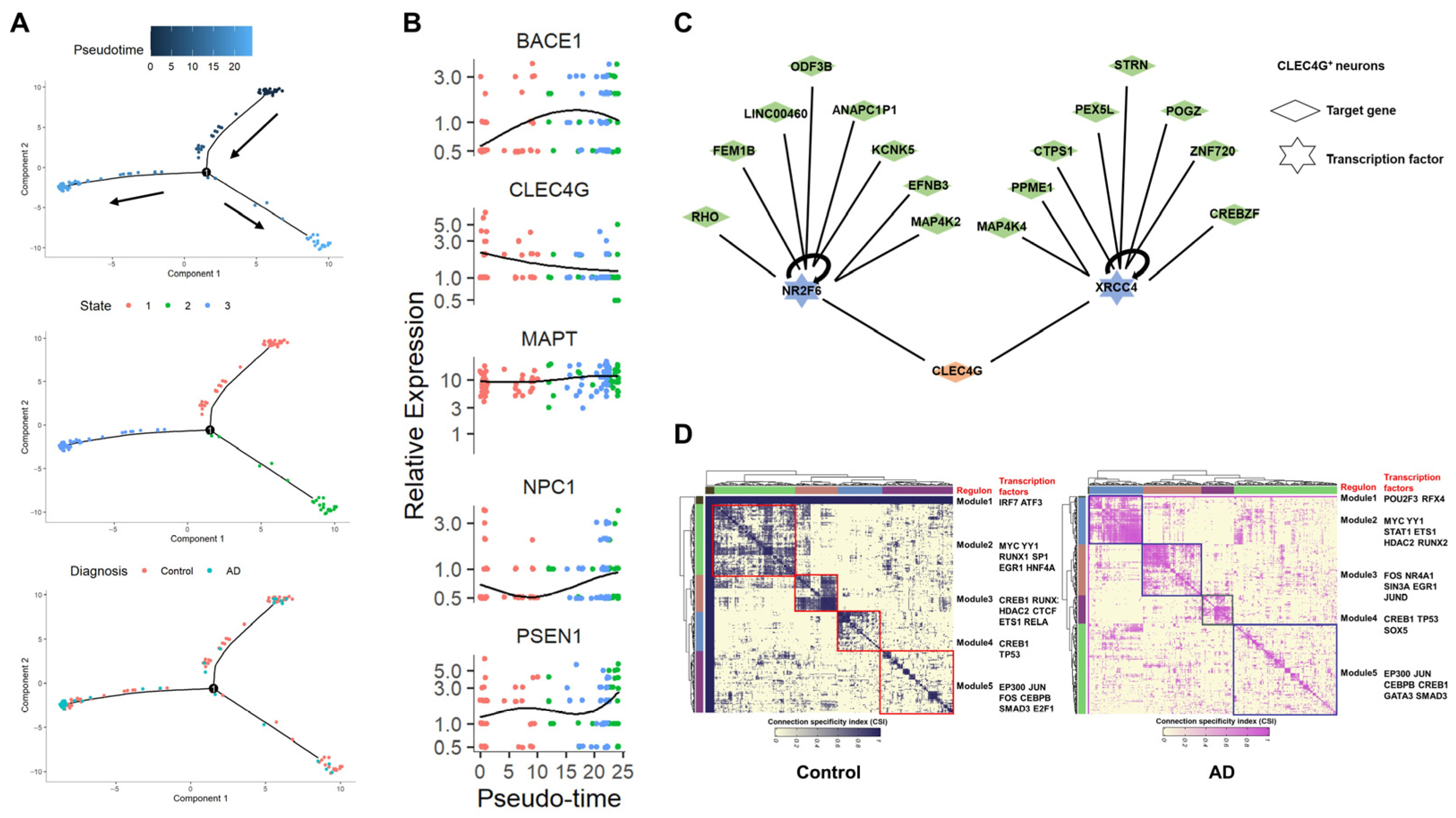
2.6. Regulatory Relationships of the CLEC4G Gene in Neurons
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. RT-PCR
4.3. Western Blot
4.4. Immunohistochemistry
4.5. Immunofluorescence
4.6. Animal Experiment
4.7. Transfection
4.8. Aβ40 ELISA
4.9. Data Resource and Description
4.10. Identification of CLEC4G-Related Genes
4.11. GO Biological Process Enrichment
4.12. Protein–Protein Interaction (PPI) Network Construction
4.13. Gene Regulatory Network Analysis of CLEC4G Gene
4.13.1. Trajectory Analysis
4.13.2. SCENIC Analysis
4.14. Pearson’s Correlation Analysis
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zhang, Y.; Ren, R.; Yang, L.; Zhang, H.; Shi, Y.; Okhravi, H.R.; Vitiello, M.V.; Sanford, L.D.; Tang, X. Sleep in Alzheimer’s Disease: A Systematic Review and Meta-Analysis of Polysomnographic Findings. Transl. Psychiatry 2022, 12, 136. [Google Scholar] [CrossRef] [PubMed]
- Volloch, V.; Rits-Volloch, S. Next Generation Therapeutic Strategy for Treatment and Prevention of Alzheimer’s Disease and Aging-Associated Cognitive Decline: Transient, Once-in-a-Lifetime-Only Depletion of Intraneuronal Aβ (iAβ) by Its Targeted Degradation via Augmentation of Intra-iAβ-Cleaving Activities of BACE1 and/or BACE2. Int. J. Mol. Sci. 2023, 24, 17586. [Google Scholar] [CrossRef] [PubMed]
- Volloch, V.; Rits-Volloch, S. The Amyloid Cascade Hypothesis 2.0 for Alzheimer’s Disease and Aging-Associated Cognitive Decline: From Molecular Basis to Effective Therapy. Int. J. Mol. Sci. 2023, 24, 12246. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yang, J.; Tang, X.; Ying, W.; Qian, X.; He, F. The DC-SIGN Family Member LSECtin Is a Novel Ligand of CD44 on Activated T Cells. Eur. J. Immunol. 2010, 40, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, Q.; Wang, X.; Jiang, X.; Zhao, D.; Lin, X.; He, F.; Tang, L. C-Type Lectin Receptor LSECtin-Mediated Apoptotic Cell Clearance by Macrophages Directs Intestinal Repair in Experimental Colitis. Proc. Natl. Acad. Sci. USA 2018, 115, 11054–11059. [Google Scholar] [CrossRef]
- Rahimi, N. C-Type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors. Biology 2020, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-Type Lectins in Immunity and Homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Wohlfeil, S.A.; Häfele, V.; Dietsch, B.; Schledzewski, K.; Winkler, M.; Zierow, J.; Leibing, T.; Mohammadi, M.M.; Heineke, J.; Sticht, C. Hepatic Endothelial Notch Activation Protects against Liver Metastasis by Regulating Endothelial-Tumor Cell Adhesion Independent of Angiocrine Signaling. Cancer Res. 2019, 79, 598–610. [Google Scholar] [CrossRef]
- Kizuka, Y.; Kitazume, S.; Sato, K.; Taniguchi, N. Clec4g (LSECtin) Interacts with BACE1 and Suppresses Aβ Generation. FEBS Lett. 2015, 589, 1418–1422. [Google Scholar] [CrossRef]
- Moreno-Jiménez, E.P.; Flor-García, M.; Terreros-Roncal, J.; Rábano, A.; Cafini, F.; Pallas-Bazarra, N.; Ávila, J.; Llorens-Martín, M. Adult Hippocampal Neurogenesis Is Abundant in Neurologically Healthy Subjects and Drops Sharply in Patients with Alzheimer’s Disease. Nat. Med. 2019, 25, 554–560. [Google Scholar] [CrossRef]
- Terreros-Roncal, J.; Flor-García, M.; Moreno-Jiménez, E.P.; Rodríguez-Moreno, C.B.; Márquez-Valadez, B.; Gallardo-Caballero, M.; Rábano, A.; Llorens-Martín, M. Methods to Study Adult Hippocampal Neurogenesis in Humans and across the Phylogeny. Hippocampus 2023, 33, 271–306. [Google Scholar] [CrossRef]
- Meng, Y.; Heybrock, S.; Neculai, D.; Saftig, P. Cholesterol Handling in Lysosomes and Beyond. Trends Cell Biol. 2020, 30, 452–466. [Google Scholar] [CrossRef]
- Rogers, M.A.; Chang, C.C.; Maue, R.A.; Melton, E.M.; Peden, A.A.; Garver, W.S.; Lee, J.; Schroen, P.; Huang, M.; Chang, T.-Y. Acat1/Soat1 Knockout Extends the Mutant Npc1 Mouse Lifespan and Ameliorates Functional Deficiencies in Multiple Organelles of Mutant Cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2201646119. [Google Scholar] [CrossRef]
- Kågedal, K.; Kim, W.S.; Appelqvist, H.; Chan, S.; Cheng, D.; Agholme, L.; Barnham, K.; McCann, H.; Halliday, G.; Garner, B. Increased Expression of the Lysosomal Cholesterol Transporter NPC1 in Alzheimer’s Disease. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2010, 1801, 831–838. [Google Scholar]
- Li, D.; Zhang, J.; Liu, Q. Brain Cell Type-Specific Cholesterol Metabolism and Implications for Learning and Memory. Trends Neurosci. 2022, 45, 401–414. [Google Scholar] [CrossRef]
- Haure-Mirande, J.-V.; Audrain, M.; Ehrlich, M.E.; Gandy, S. Microglial TYROBP/DAP12 in Alzheimer’s Disease: Transduction of Physiological and Pathological Signals across TREM2. Mol. Neurodegener. 2022, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Davey, K.; Tsartsalis, S.; Khozoie, C.; Fancy, N.; Tang, S.S.; Liaptsi, E.; Weinert, M.; McGarry, A.; Muirhead, R.C.J.; et al. Diverse Human Astrocyte and Microglial Transcriptional Responses to Alzheimer’s Pathology. Acta Neuropathol. 2022, 143, 75–91. [Google Scholar] [CrossRef]
- Audrain, M.; Haure-Mirande, J.-V.; Mleczko, J.; Wang, M.; Griffin, J.K.; St George-Hyslop, P.H.; Fraser, P.; Zhang, B.; Gandy, S.; Ehrlich, M.E. Reactive or Transgenic Increase in Microglial TYROBP Reveals a TREM2-Independent TYROBP-APOE Link in Wild-Type and Alzheimer’s-Related Mice. Alzheimers Dement. 2021, 17, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Tena, J.; Di Lucente, J.; Maezawa, I.; Harvey, D.J.; Jin, L.-W.; Lebrilla, C.B.; Zivkovic, A.M. Transcriptomic and Glycomic Analyses Highlight Pathway-Specific Glycosylation Alterations Unique to Alzheimer’s Disease. Sci. Rep. 2023, 13, 7816. [Google Scholar] [CrossRef]
- Kizuka, Y.; Kitazume, S.; Taniguchi, N. N-Glycan and Alzheimer’s Disease. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 2447–2454. [Google Scholar] [CrossRef]
- Biswas, B.; Batista, F.; Sundaram, S.; Stanley, P. MGAT1 and Complex N-Glycans Regulate ERK Signaling during Spermatogenesis. Sci. Rep. 2018, 8, 2022. [Google Scholar] [CrossRef] [PubMed]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.-C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-Cell Regulatory Network Inference and Clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ren, S.; Zuo, Y. DC-SIGN, DC-SIGNR and LSECtin: C-Type Lectins for Infection. Int. Rev. Immunol. 2014, 33, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.T.; Lepenies, B. Myeloid C-Type Lectin Receptors in Viral Recognition and Antiviral Immunity. Viruses 2017, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, A.L.; Chevalier, M.T.; Russo, L.; Pandit, A. Role and Therapeutic Implications of Protein Glycosylation in Neuroinflammation. Trends Mol. Med. 2022, 28, 270–289. [Google Scholar] [CrossRef]
- Kizuka, Y.; Taniguchi, N. Neural Functions of Bisecting GlcNAc. Glycoconj. J. 2018, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Kitazume, S.; Fujinawa, R.; Saito, T.; Iwata, N.; Saido, T.C.; Nakano, M.; Yamaguchi, Y.; Hashimoto, Y.; Staufenbiel, M. An Aberrant Sugar Modification of BACE 1 Blocks Its Lysosomal Targeting in A Lzheimer’s Disease. EMBO Mol. Med. 2015, 7, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Hodges, A.K.; Piers, T.M.; Collier, D.; Cousins, O.; Pocock, J.M. Pathways Linking Alzheimer’s Disease Risk Genes Expressed Highly in Microglia. Neuroimmunol. Neuroinflamm. 2021, 8, 245. [Google Scholar] [CrossRef]
- Andrade-Guerrero, J.; Santiago-Balmaseda, A.; Jeronimo-Aguilar, P.; Vargas-Rodríguez, I.; Cadena-Suárez, A.R.; Sánchez-Garibay, C.; Pozo-Molina, G.; Méndez-Catalá, C.F.; Cardenas-Aguayo, M.-C.; Diaz-Cintra, S.; et al. Alzheimer’s Disease: An Updated Overview of Its Genetics. Int J Mol Sci 2023, 24, 3754. [Google Scholar] [CrossRef]
- Mossad, O.; Erny, D. The Microbiota–Microglia Axis in Central Nervous System Disorders. Brain Pathol. 2020, 30, 1159–1177. [Google Scholar] [CrossRef]
- Wang, S.; Sudan, R.; Peng, V.; Zhou, Y.; Du, S.; Yuede, C.M.; Lei, T.; Hou, J.; Cai, Z.; Cella, M.; et al. TREM2 Drives Microglia Response to Amyloid-β via SYK-Dependent and -Independent Pathways. Cell 2022, 185, 4153–4169.e19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Han, X.; Zheng, X.; Wang, H.; Yang, Z.; Liu, D.; Han, K.; Liu, J.; Wang, X.; Yang, W. The Myeloid LSECtin Is a DAP12-Coupled Receptor That Is Crucial for Inflammatory Response Induced by Ebola Virus Glycoprotein. PLoS Pathog. 2016, 12, e1005487. [Google Scholar]
- Bettcher, B.M.; Tansey, M.G.; Dorothée, G.; Heneka, M.T. Peripheral and Central Immune System Crosstalk in Alzheimer Disease—A Research Prospectus. Nat. Rev. Neurol. 2021, 17, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Kummer, M.P.; Hammerschmidt, T.; Martinez, A.; Terwel, D.; Eichele, G.; Witten, A.; Figura, S.; Stoll, M.; Schwartz, S.; Pape, H.-C.; et al. Ear2 Deletion Causes Early Memory and Learning Deficits in APP/PS1 Mice. J. Neurosci. 2014, 34, 8845–8854. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Liu, Z.; Xu, J.; Xiong, Y.; Zhang, X.; Qi, J.; Lin, X.; Chu, C.; Shen, L.; Liu, G.; et al. Black Phosphorus Nanosheets Suppress Oxidative Damage of Stem Cells for Improved Neurological Recovery. Chem. Eng. J. 2023, 451, 138737. [Google Scholar] [CrossRef]
- Quinn, T.P.; Crowley, T.M.; Richardson, M.F. Benchmarking Differential Expression Analysis Tools for RNA-Seq: Normalization-Based vs. Log-Ratio Transformation-Based Methods. BMC Bioinform. 2018, 19, 274. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Young, R.L.; Xue, H.; Wagner, G.P. Transcriptomic Analysis of Avian Digits Reveals Conserved and Derived Digit Identities in Birds. Nature 2011, 477, 583–586. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Qi, F.; Huang, Y.; Zhang, G.; Deng, W. Reduced Expression of CLEC4G in Neurons Is Associated with Alzheimer’s Disease. Int. J. Mol. Sci. 2024, 25, 4621. https://doi.org/10.3390/ijms25094621
Feng X, Qi F, Huang Y, Zhang G, Deng W. Reduced Expression of CLEC4G in Neurons Is Associated with Alzheimer’s Disease. International Journal of Molecular Sciences. 2024; 25(9):4621. https://doi.org/10.3390/ijms25094621
Chicago/Turabian StyleFeng, Xinwei, Fangfang Qi, Yuying Huang, Ge Zhang, and Wenbin Deng. 2024. "Reduced Expression of CLEC4G in Neurons Is Associated with Alzheimer’s Disease" International Journal of Molecular Sciences 25, no. 9: 4621. https://doi.org/10.3390/ijms25094621




