Harnessing Nature’s Defence: The Antimicrobial Efficacy of Pasteurised Cattle Milk-Derived Extracellular Vesicles on Staphylococcus aureus ATCC 25923
Abstract
:1. Introduction
2. Results
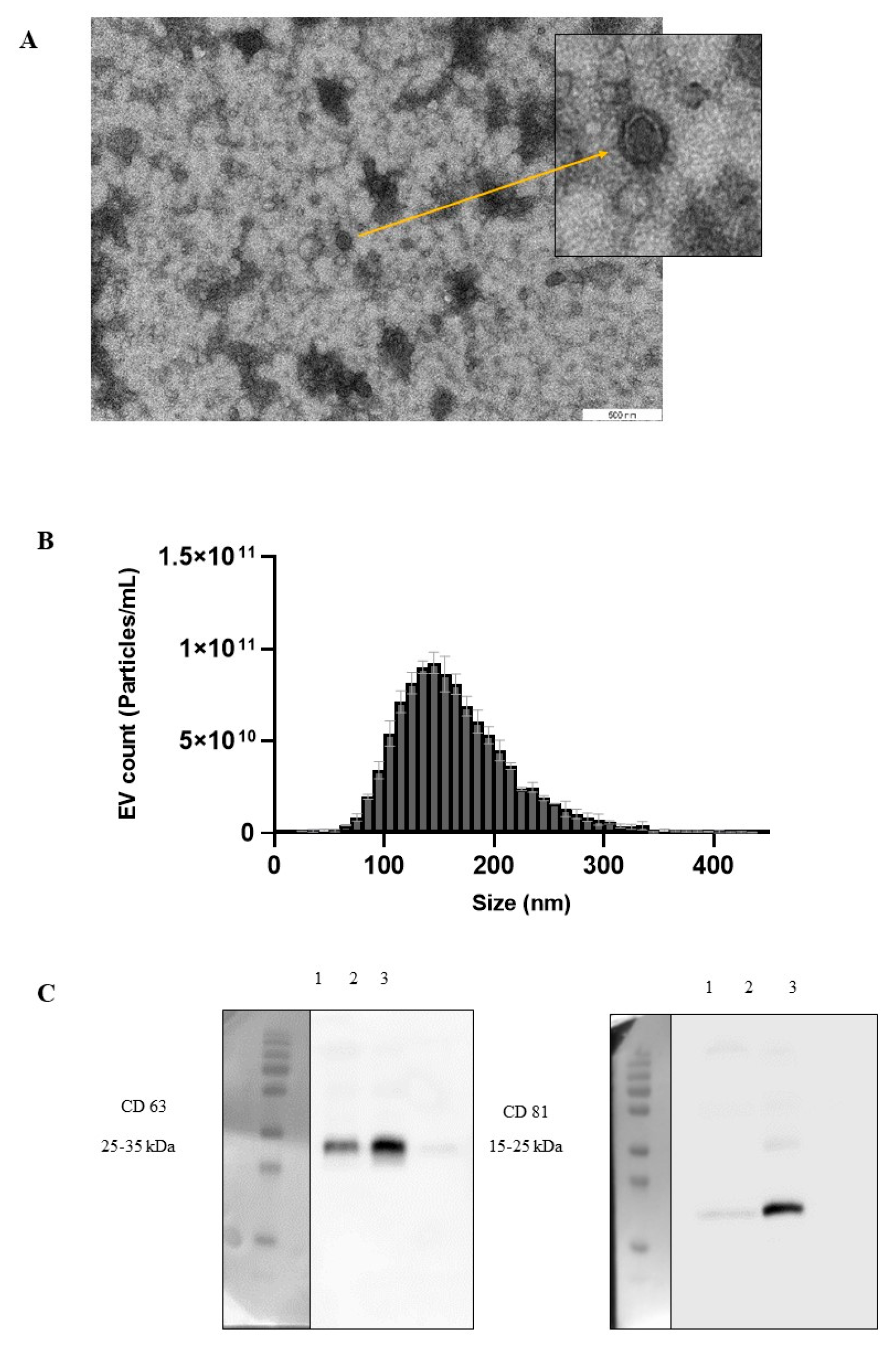
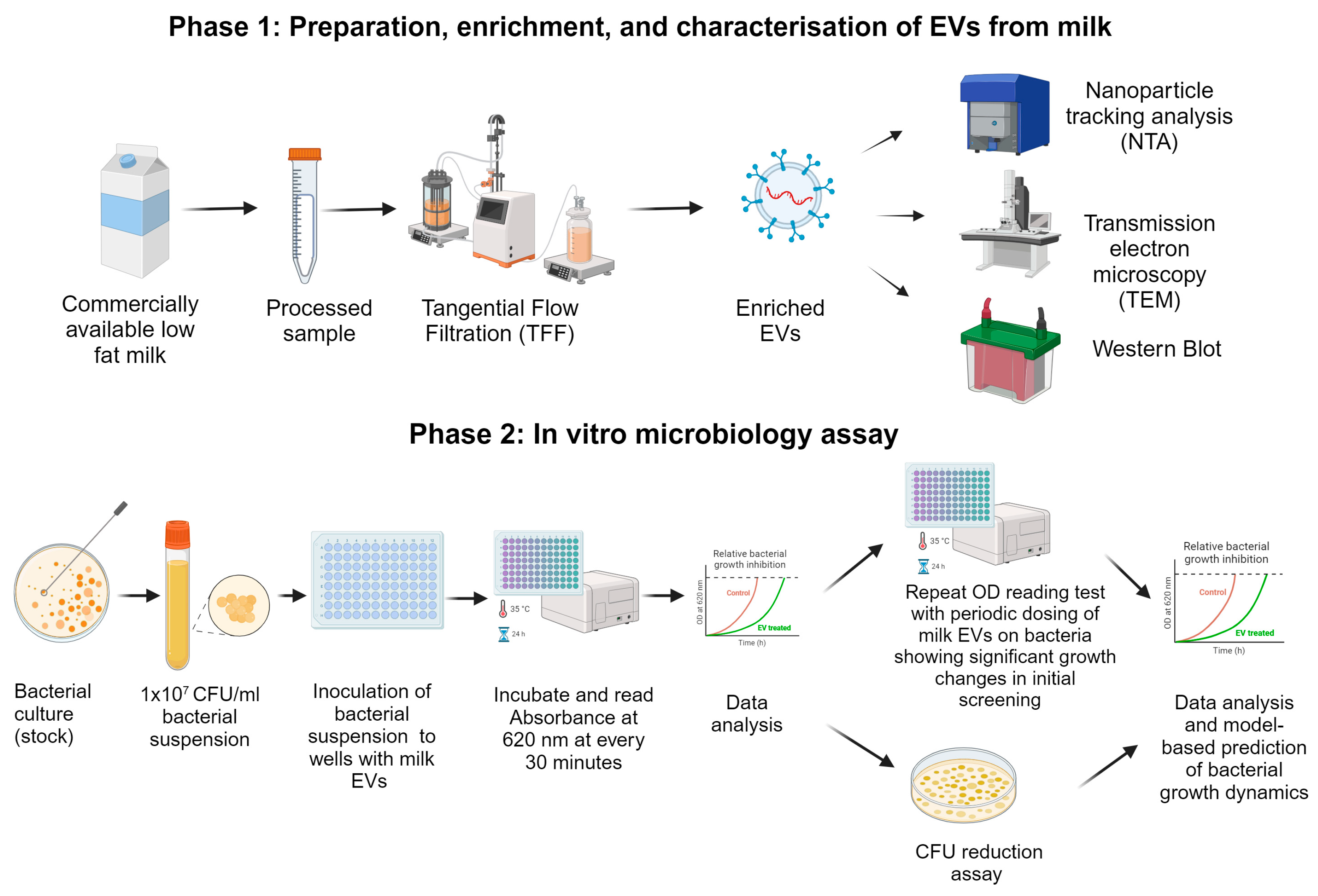
2.1. Isolation and Characterisation of Pasteurised Milk EVs
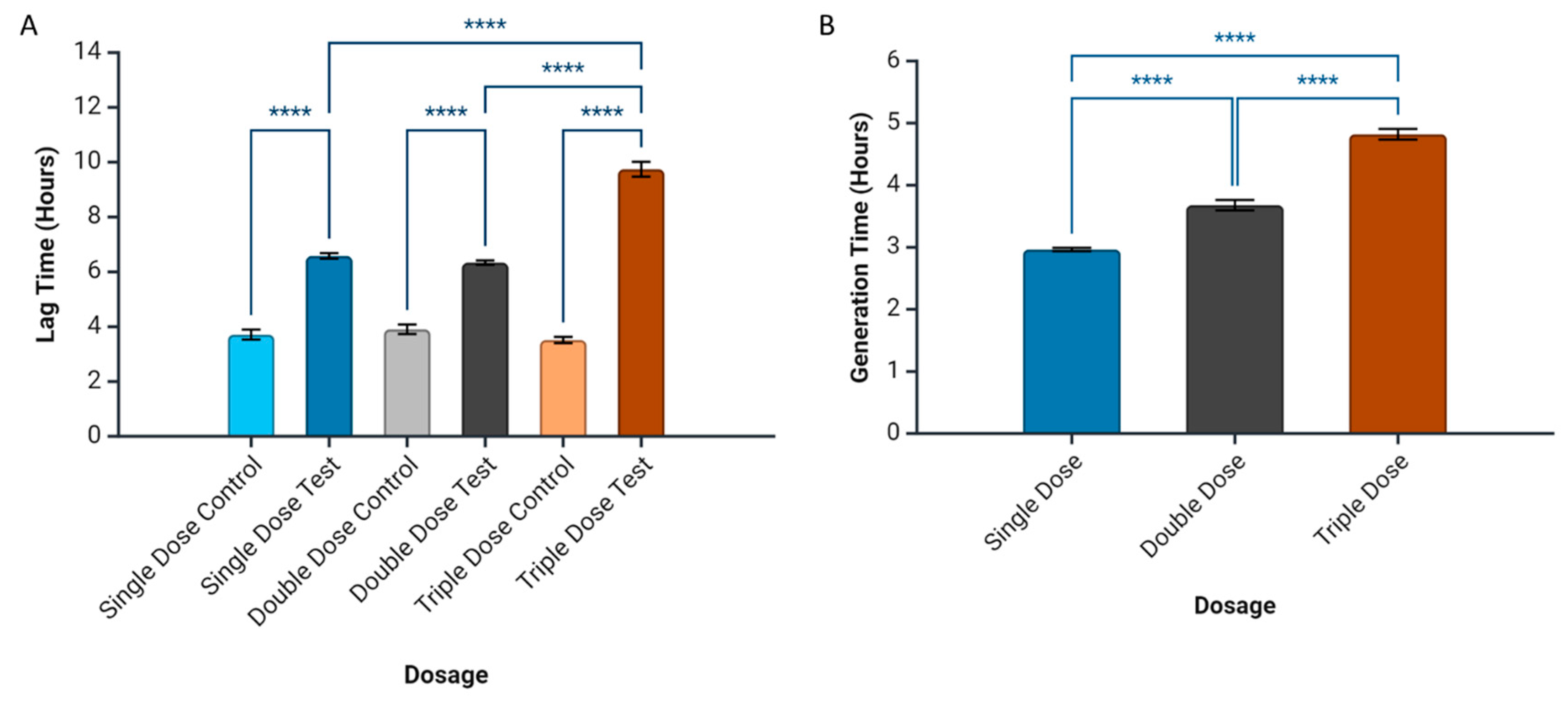
2.2. Milk EV Activity on the Bacterial Growth Curve
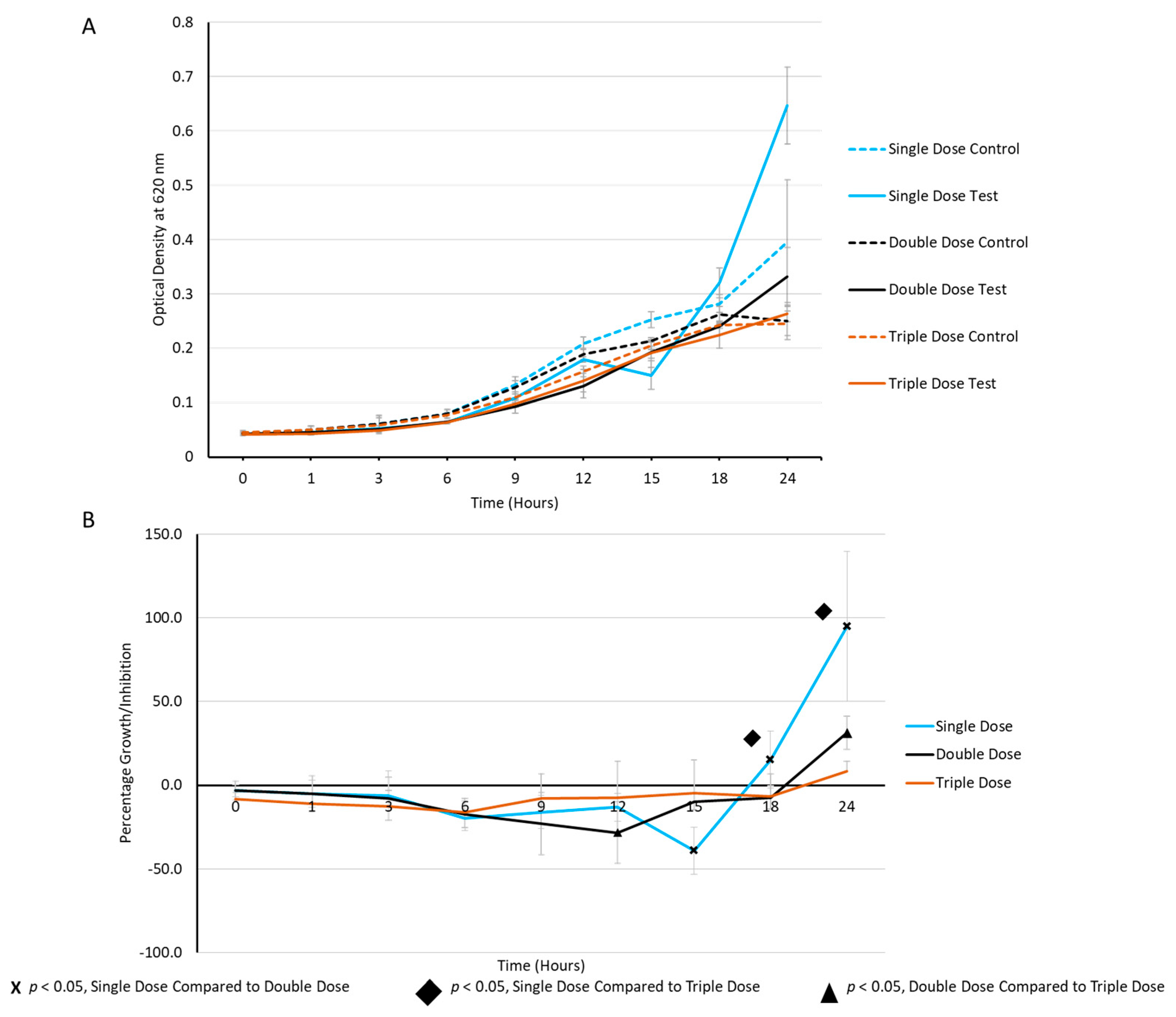
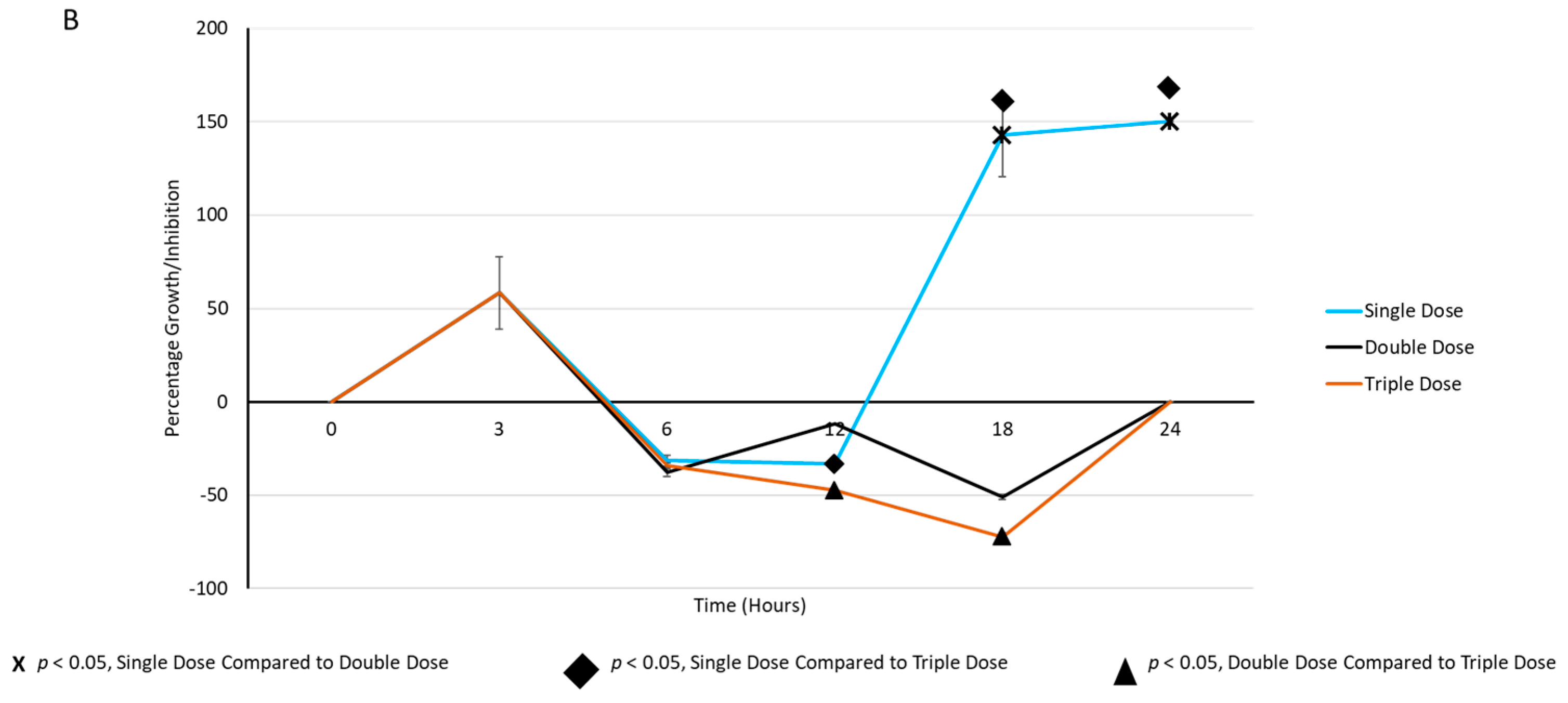
2.3. Periodic Dosing of Milk EVs on S. aureus Growth and CFU Reduction Assay
2.4. Prediction of Bacterial Growth-Related Parameters
3. Discussion
4. Materials and Methods
4.1. Preparation, Enrichment, and Characterisation of Milk EVs
4.1.1. Extracellular Vesicle Enrichment from Pasteurised Milk
4.1.2. Nanoparticle Tracking Analysis (NTA)
4.1.3. Transmission Electron Microscopy (TEM)
4.1.4. SDS–PAGE and Western Blotting
4.2. Microbiological Experiments and Model-Based Prediction of Bacterial Growth Dynamics
4.2.1. Bacterial Strains and Culture Conditions
4.2.2. Bacterial Growth Curve Measurements with a Microplate Reader and Colony-Forming Unit Assay
4.2.3. Model-Based Prediction of Time Derivatives of Bacterial Growth and Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- de Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance#:~:text=It%20is%20estimated%20that%20bacterial,development%20of%20drug%2Dresistant%20pathogens. (accessed on 18 April 2024).
- World Health Organization. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 4 April 2024).
- Rex, J.H.; Lynch, H.F.; Cohen, I.G.; Darrow, J.J.; Outterson, K. Designing development programs for non-traditional antibacterial agents. Nat. Commun. 2019, 10, 3416. [Google Scholar] [CrossRef]
- Hadizadeh, N.; Bagheri, D.; Shamsara, M.; Hamblin, M.R.; Farmany, A.; Xu, M.; Liang, Z.; Razi, F.; Hashemi, E. Extracellular vesicles biogenesis, isolation, manipulation and genetic engineering for potential in vitro and in vivo therapeutics: An overview. Front. Bioeng. Biotechnol. 2022, 10, 1019821. [Google Scholar] [CrossRef]
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.; Vader, P. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 2020, 159, 332–343. [Google Scholar] [CrossRef]
- Feng, X.; Chen, X.; Zheng, X.; Zhu, H.; Qi, Q.; Liu, S.; Zhang, H.; Che, J. Latest trend of milk derived exosomes: Cargos, functions, and applications. Front. Nutr. 2021, 8, 747294. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; You, L.; Zhang, Z.; Cui, X.; Zhong, H.; Sun, X.; Ji, C.; Chi, X. Biological properties of milk-derived extracellular vesicles and their physiological functions in infant. Front. Cell Dev. Biol. 2021, 9, 693534. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Zimmermann, A.-K.; Rivieccio, F.; Visser, C.; Blango, M.G. Host-derived extracellular vesicles for antimicrobial defense. microLife 2021, 2, uqab003. [Google Scholar] [CrossRef]
- Buzas, E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2022, 23, 236–250. [Google Scholar] [CrossRef]
- Kuipers, M.E.; Hokke, C.H.; Smits, H.H.; Nolte-‘t Hoen, E.N. Pathogen-derived extracellular vesicle-associated molecules that affect the host immune system: An overview. Front. Microbiol. 2018, 9, 2182. [Google Scholar] [CrossRef]
- Saad, M.G.; Beyenal, H.; Dong, W.-J. Dual roles of the conditional extracellular vesicles derived from pseudomonas aeruginosa biofilms: Promoting and inhibiting bacterial biofilm growth. Biofilm 2024, 7, 100183. [Google Scholar] [CrossRef]
- Bose, S.; Aggarwal, S.; Singh, D.V.; Acharya, N. Extracellular vesicles: An emerging platform in gram-positive bacteria. Microb. Cell 2020, 7, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Sanwlani, R.; Fonseka, P.; Chitti, S.V.; Mathivanan, S. Milk-derived extracellular vesicles in inter-organism, cross-species communication and Drug Delivery. Proteomes 2020, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Mun, D.; Oh, S.; Kim, Y. Perspectives on bovine milk-derived extracellular vesicles for therapeutic applications in Gut Health. Food Sci. Anim. Resour. 2022, 42, 197–209. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.F.; Fitzgerald, J.R.; Penadés, J.R. Staphylococcus aureus in animals. Microbiol. Spectr. 2019, 7, 731–746. [Google Scholar] [CrossRef]
- Johnson, A.P. Methicillin-resistant Staphylococcus aureus: The European Landscape. J. Antimicrob. Chemother. 2011, 66, iv43–iv48. [Google Scholar] [CrossRef]
- Tang, K.W.; Millar, B.C.; Moore, J.E. Antimicrobial resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Gales, A.C.; Laxminarayan, R.; Dodd, P.C. Antimicrobial resistance: Addressing a global threat to humanity. PLoS Med. 2023, 20, e1004264. [Google Scholar] [CrossRef]
- Adebisi, Y.A.; Ogunkola, I.O. The global antimicrobial resistance response effort must not exclude marginalised populations. Trop. Med. Health 2023, 51, 33. [Google Scholar] [CrossRef]
- Kodam, S.P.; Ullah, M. Diagnostic and therapeutic potential of extracellular vesicles. Technol. Cancer Res. Treat. 2021, 20, 153303382110412. [Google Scholar] [CrossRef]
- Tetta, C.; Ghigo, E.; Silengo, L.; Deregibus, M.C.; Camussi, G. Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine 2012, 44, 11–19. [Google Scholar] [CrossRef]
- He, Z.; Greven, J.; Shi, Y.; Qin, K.; Zhao, Q.; Zhang, X.; Buhl, E.M.; Eschweiler, J.; Hildebrand, F.; Balmayor, E.R. Extracellular vesicles derived from endothelial cells modulate macrophage phenotype in vitro. Eur. J. Med. Res. 2023, 28, 506. [Google Scholar] [CrossRef]
- Yu, S.; Zhao, Z.; Xu, X.; Li, M.; Li, P. Characterization of three different types of extracellular vesicles and their impact on bacterial growth. Food Chem. 2019, 272, 372–378. [Google Scholar] [CrossRef]
- Aarts, J.; Boleij, A.; Pieters, B.C.; Feitsma, A.L.; van Neerven, R.J.; ten Klooster, J.P.; M’Rabet, L.; Arntz, O.J.; Koenders, M.I.; van de Loo, F.A. Flood control: How milk-derived extracellular vesicles can help to improve the intestinal barrier function and break the gut–joint axis in rheumatoid arthritis. Front. Immunol. 2021, 12, 703277. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Hao, H.; Zhang, X.; Zhang, Z.; Lv, Y.; Zhang, L.; Yi, H. Oral administration of bovine milk-derived extracellular vesicles alters the gut microbiota and enhances intestinal immunity in mice. Mol. Nutr. Food Res. 2020, 64, 1901251. [Google Scholar] [CrossRef] [PubMed]
- Sprouffske, K.; Wagner, A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016, 17, 172. [Google Scholar] [CrossRef]
- Blazanin, M. Gcplyr: An R package for microbial growth curve data analysis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Singh, A.; Duche, R.T.; Wandhare, A.G.; Sian, J.K.; Singh, B.P.; Sihag, M.K.; Singh, K.S.; Sangwan, V.; Talan, S.; Panwar, H. Milk-derived antimicrobial peptides: Overview, applications, and future perspectives. Probiotics Antimicrob. Proteins 2022, 15, 44–62. [Google Scholar] [CrossRef]
- Piibor, J.; Waldmann, A.; Dissanayake, K.; Andronowska, A.; Ivask, M.; Prasadani, M.; Kavak, A.; Kodithuwakku, S.; Fazeli, A. Uterine fluid extracellular vesicles proteome is altered during the estrous cycle. Mol. Cell. Proteom. 2023, 22, 100642. [Google Scholar] [CrossRef]
- Stevenson, K.; McVey, A.F.; Clark, I.B.; Swain, P.S.; Pilizota, T. General calibration of microbial growth in microplate readers. Sci. Rep. 2016, 6, 38828. [Google Scholar] [CrossRef] [PubMed]
- Leiva-Sabadini, C.; Alvarez, S.; Barrera, N.P.; Schuh, C.M.; Aguayo, S. Antibacterial effect of honey-derived exosomes containing antimicrobial peptides against oral streptococci. Int. J. Nanomed. 2021, 16, 4891–4900. [Google Scholar] [CrossRef] [PubMed]
- Kettering, J.D.; Stephens, J.A.; Muñoz-Viveros, C.A.; Naylor, W.P. Reducing bacterial counts in dental unit waterlines: Tap water vs. distilled water. J. Contemp. Dent. Pract. 2002, 3, 1–11. [Google Scholar] [CrossRef]
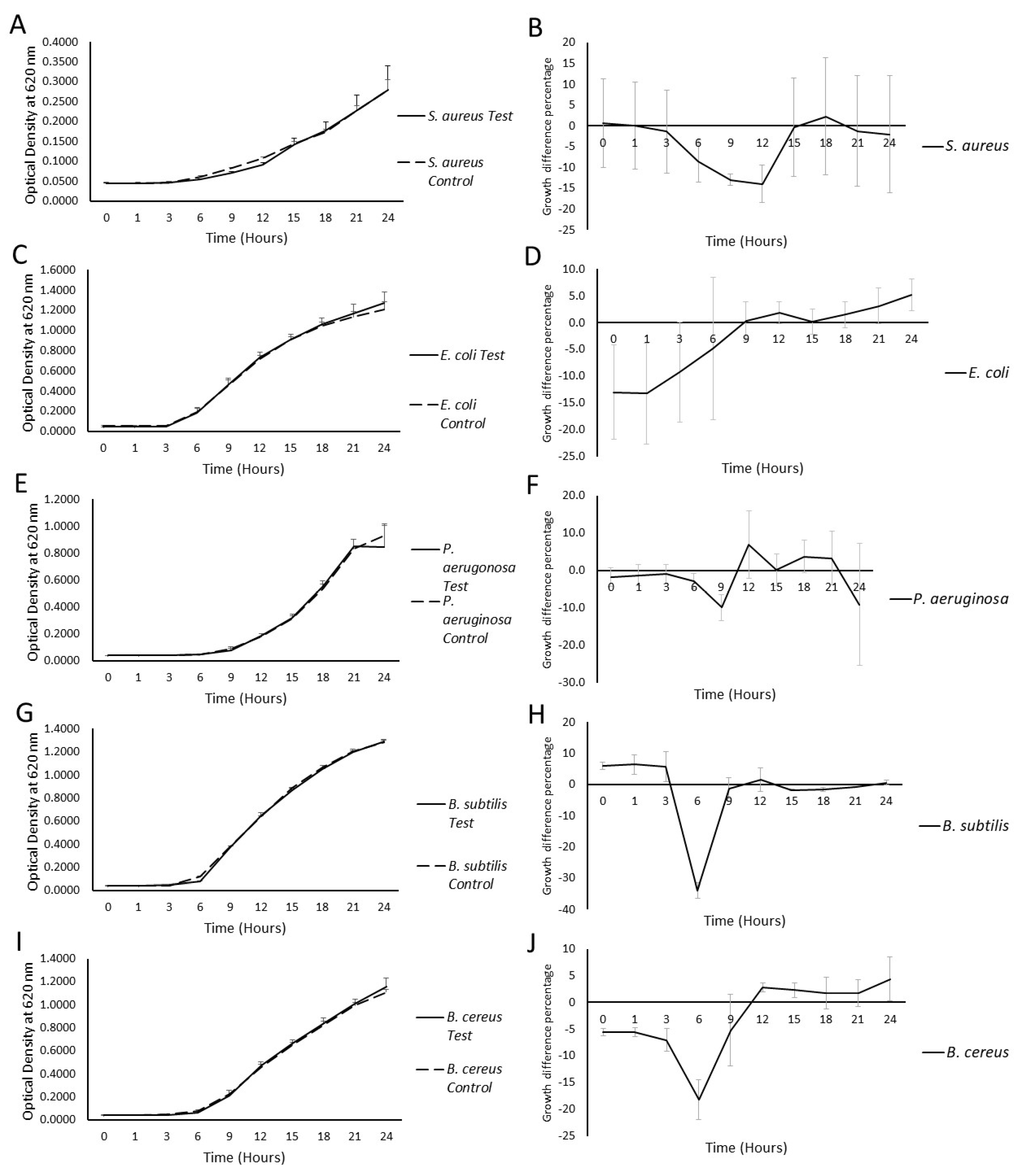

| Bacterial Strain | Maximum Percentage Growth Difference (%, p-Value) | |||||
|---|---|---|---|---|---|---|
| 1 h | 3 h | 6 h | 9 h | 12 h | 15 h | |
| Staphylococcus aureus ATCC 25923 | +0.10 ± 10.49, (0.8856) | −1.39 ± 9.96, (0.7397) | −8.44 ± 5.11, (0.2445) | −12.9 ± 1.39, (0.0222) | −13.96 ± 4.48, (0.04506) | −0.31 ± 11.9, (0.9454) |
| Bacillus subtilis ATCC 6633 | +6.46 ± 3.10, (0.1484) | +5.69 ± 4.84, (0.2738) | −33.95 ± 2.42, (0.0106) | −1.23 ± 3.37, (0.6450) | +1.59 ± 3.81, (0.6739) | −1.80 ± 0.29, (0.3768) |
| Bacillus cereus ATCC 11778 | −5.51 ± 0.82, (0.2983) | −7.05 ± 2.17, (0.2168) | −18.17 ± 3.72, (0.4195) | −5.21 ± 6.72, (0.4195) | +2.81 ± 0.79, (0.7679) | +2.29 ± 1.34, (0.7535) |
| Escherichia coli ATCC 53868 | −13.3 ± 9.37, (0.2811) | −9.2 ± 9.39, (0.3730) | −4.8 ± 13.26, (0.9499) | +0.3 ± 3.68, (0.9585) | +1.9 ± 2.03, (0.8131) | +0.2 ± 2.41, (0.9611) |
| Pseudomonas aeruginosa ATCC 27853 | −1.29 ± 2.81, (0.7469) | −1.01 ± 2.67, (0.8640) | −2.88 ± 13.26, (0.8283) | +9.90 ± 3.53, (0.7168) | +6.94 ± 9.09, (0.7452) | +0.16 ± 4.27, (0.9587) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapugahawatte, D.N.; Godakumara, K.; Mäesaar, M.; Ekanayake, G.; Midekessa, G.B.; Prasadani, M.; Kodithuwakku, S.; Roasto, M.; Andronowska, A.; Fazeli, A. Harnessing Nature’s Defence: The Antimicrobial Efficacy of Pasteurised Cattle Milk-Derived Extracellular Vesicles on Staphylococcus aureus ATCC 25923. Int. J. Mol. Sci. 2024, 25, 4759. https://doi.org/10.3390/ijms25094759
Sapugahawatte DN, Godakumara K, Mäesaar M, Ekanayake G, Midekessa GB, Prasadani M, Kodithuwakku S, Roasto M, Andronowska A, Fazeli A. Harnessing Nature’s Defence: The Antimicrobial Efficacy of Pasteurised Cattle Milk-Derived Extracellular Vesicles on Staphylococcus aureus ATCC 25923. International Journal of Molecular Sciences. 2024; 25(9):4759. https://doi.org/10.3390/ijms25094759
Chicago/Turabian StyleSapugahawatte, Dulmini Nanayakkara, Kasun Godakumara, Mihkel Mäesaar, Gayandi Ekanayake, Getnet Balcha Midekessa, Madhusha Prasadani, Suranga Kodithuwakku, Mati Roasto, Aneta Andronowska, and Alireza Fazeli. 2024. "Harnessing Nature’s Defence: The Antimicrobial Efficacy of Pasteurised Cattle Milk-Derived Extracellular Vesicles on Staphylococcus aureus ATCC 25923" International Journal of Molecular Sciences 25, no. 9: 4759. https://doi.org/10.3390/ijms25094759






