Assessing Genetic Diversity and Population Differentiation in Wild Hop (Humulus lupulus) from the Region of Central Greece via SNP-NGS Genotyping
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Sample Collection
2.2. DNA Extraction
2.3. PCR Amplification
2.4. Next-Generation Sequencing (NGS) of Amplicon
2.5. Data Analysis
2.5.1. Genotyping Protocol
2.5.2. Genetic and Statistical Analyses
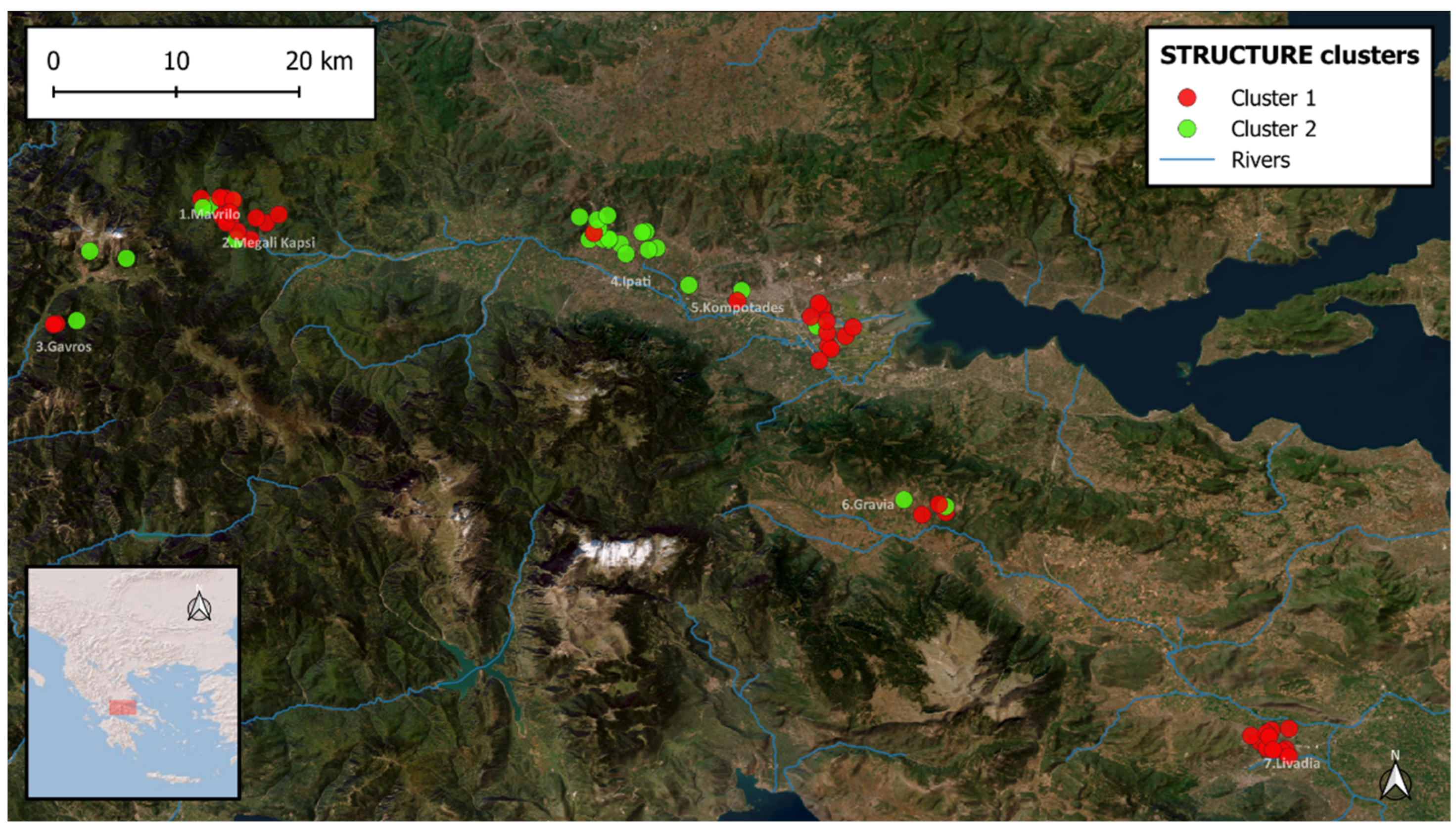
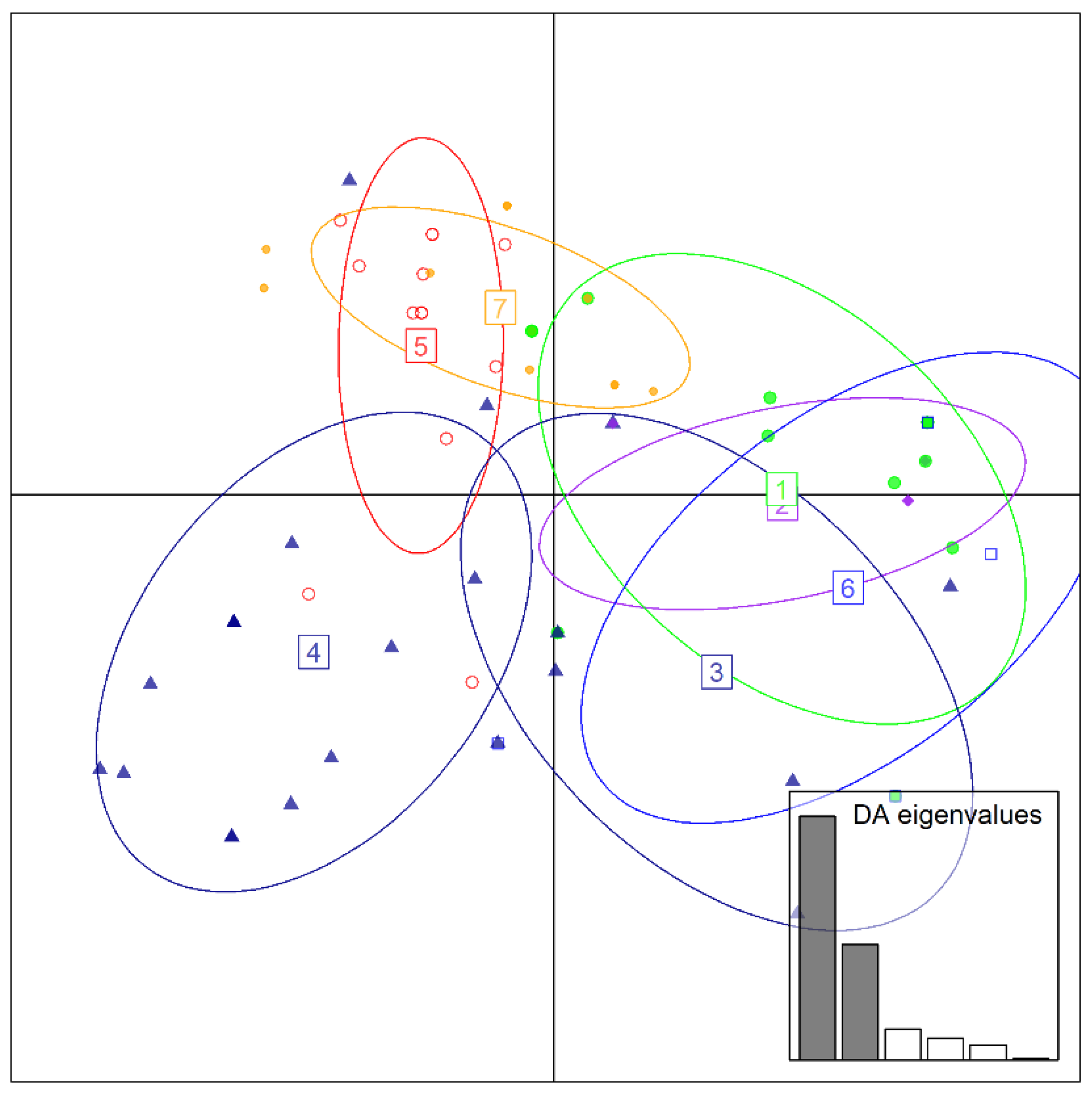
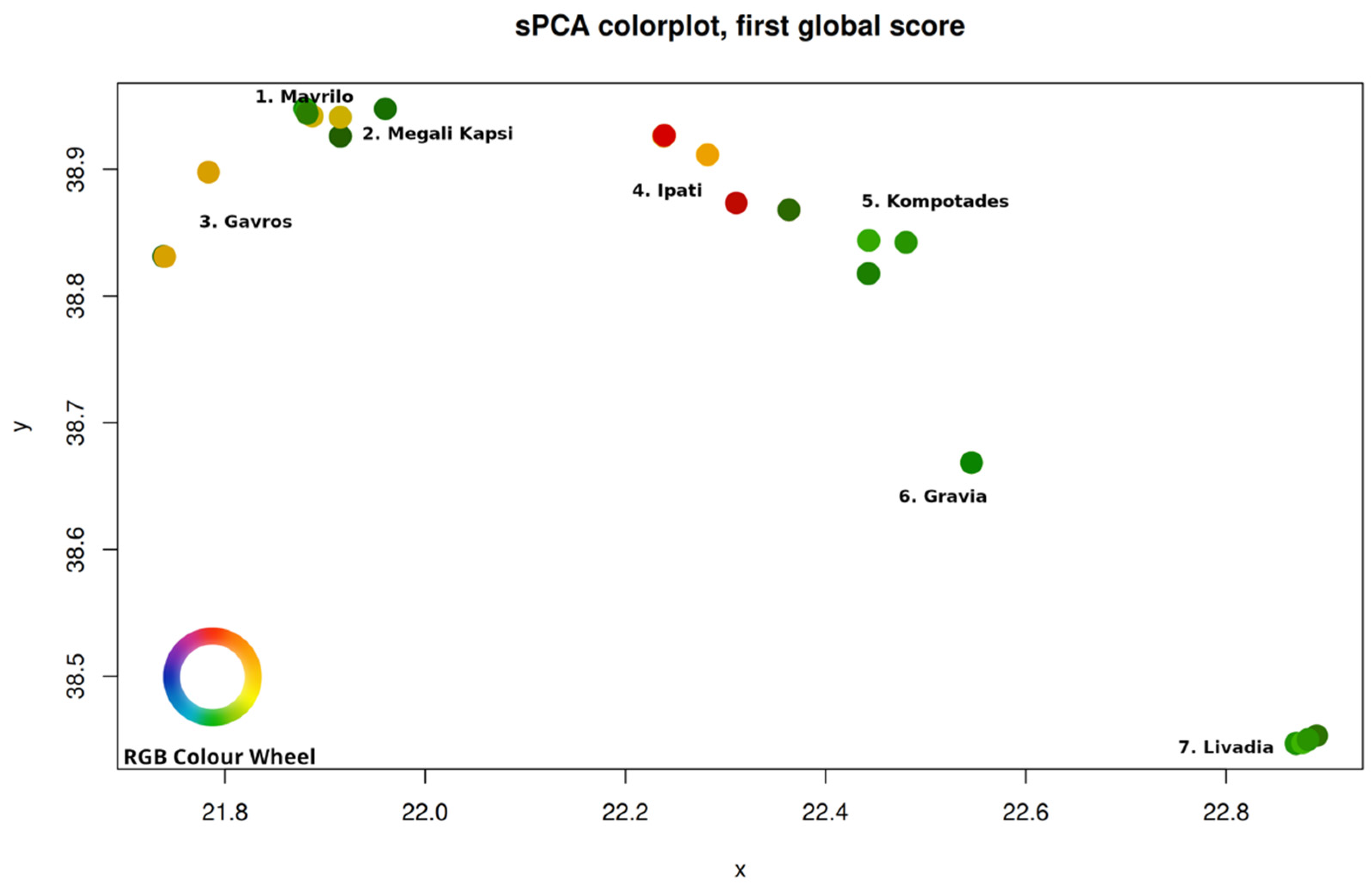
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, Z.K.; Bartholomew, B. Cannabaceae. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2003. [Google Scholar]
- Neve, R.A. Botany. In Hops, 1st ed.; Springer: Dordrecht, The Netherlands, 1991. [Google Scholar]
- Murakami, A.; Darby, P.; Javornik, B.; Pais, M.S.S.; Seigner, E.; Lutz, A.; Svoboda, P. Molecular phylogeny of wild Hops, Humulus lupulus L. Heredity 2006, 97, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Karabín, M.M.; Hudcová, T.; Jelínek, L.; Dostálek, P. Biologically Active Compounds from Hops and Prospects for Their Use. Compr. Rev. Food Sci. Food Saf. 2016, 15, 542–567. [Google Scholar] [CrossRef] [PubMed]
- Small, E. A Numerical and Nomenclatural Analysis of Morpho-Geographic Taxa of Humulus. Syst. Bot. 1978, 3, 37–76. Available online: http://www.jstor.org/stable/2418532 (accessed on 15 May 2016). [CrossRef]
- Fu, J.Q. Humulus lupulus var. fengxianensis. Acta Bot. Boreal Occid. Sin. 1992, 12, 70. [Google Scholar]
- Briggs, D.E.; Boulton, C.A.; Brookes, P.A.; Stevens, R. Hops. In Brewing: Science and Practice; Woodhead Publishing Limited: Sawston, UK; CRC Press LLC: Cambridge, UK, 2004. [Google Scholar]
- Stevens, J.F.; Page, J.E. Xanthohumol and related prenylflavonoids from hops and beer: To your good health! Phytochemistry 2004, 65, 1317–1330. [Google Scholar] [CrossRef]
- Stevens, J.F.; Miranda, C.L.; Buhler, D.R.; Deinzer, M.L. Chemistry and Biology of Hop Flavonoids. J. Am. Soc. Brew. Chem. 1998, 56, 136–145. [Google Scholar] [CrossRef]
- Van Cleemput, M.; Cattoor, K.; De Bosscher, K.; Haegeman, G.; De Keukeleire, D.; Heyerick, A. Hop (Humulus lupulus)-Derived Bitter Acids as Multipotent Bioactive Compounds Marjan. J. Nat. Prod. 2009, 72, 1220–1230. [Google Scholar] [CrossRef]
- De Keukeleire, J.; Ooms, G.; Heyerick, A.; Roldan-Ruiz, I.; Van Bockstaele, E.; De Keukeleire, D. Formation and accumulation of α-acids, β-acids, desmethylxanthohumol, and xanthohumol during flowering of hops (Humulus lupulus L.). J. Agric. Food Chem. 2003, 51, 4436–4441. [Google Scholar] [CrossRef] [PubMed]
- Kavalier, A.R.; Litt, A.; Ma, C.; Pitra, N.J.; Coles, M.C.; Kennelly, E.J.; Matthews, P.D. Phytochemical and morphological characterization of hop (Humulus lupulus L.) cones over five developmental stages using high performance liquid chromatography coupled to time-of-flight mass spectrometry, ultrahigh performance liquid chromatography photodiode array detection, and light microscopy techniques. J. Agric. Food Chem. 2011, 59, 4783–4793. [Google Scholar]
- Bolton, J.L.; Dunlap, T.L.; Hajirahimkhan, A.; Mbachu, O.; Chen, S.-N.; Chadwick, L.; Nikolic, D.; van Breemen, R.B.; Pauli, G.F.; Dietz, B.M. The Multiple Biological Targets of Hops and Bioactive Compounds. Chem. Res. Toxicol. 2019, 32, 222–233. [Google Scholar] [CrossRef]
- Kyrou, I.; Christou, A.; Panagiotakos, D.; Stefanaki, C.; Skenderi, K.; Katsana, K.; Tsigos, C. Effects of a hops (Humulus lupulus L.) dry extract supplement on self-reported depression, anxiety and stress levels in apparently healthy young adults: A randomized, placebo-controlled, double-blind, crossover pilot study. Hormones 2017, 16, 171–180. [Google Scholar]
- Šuštar-Vozlič, J.; Javornik, B. Genetic relationships in cultivars of hop, Humulus lupulus L., determined by RAPD analysis. Plant Breed. 1999, 118, 175–181. [Google Scholar] [CrossRef]
- Pillay, M.; Kenny, S.T. Structure and inheritance of ribosomal DNA variants in cultivated and wild hop, Humulus lupulus L. Theor. Appl. Genet. 1996, 93, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Koelling, J.; Coles, M.C.; Matthews, P.D.; Schwekendiek, A. Development of new microsatellite markers (SSRs) for Humulus lupulus. Mol. Breed. 2012, 30, 479–484. [Google Scholar] [CrossRef]
- Patzak, J. Comparison of RAPD, STS, ISSR and AFLP molecular methods used for assessment of genetic diversity in hop (Humulus lupulus L.). Euphytica 2001, 121, 9–18. [Google Scholar] [CrossRef]
- Abbott, M.S.; Fedele, M.J. A DNA-based varietal identification procedure for hop leaf tissue. J. Inst. Brew. 1994, 100, 283–285. [Google Scholar] [CrossRef]
- Padgitt-Cobb, L.K.; Kingan, S.B.; Wells, J.; Elser, J.; Kronmiller, B.; Moore, D.; Concepcion, G.; Peluso, P.; Rank, D.; Jaiswal, P.; et al. A draft phased assembly of the diploid Cascade hop (Humulus lupulus) genome. Plant Genome 2021, 14, e20072. [Google Scholar] [CrossRef] [PubMed]
- Matthews, P.D.; Coles, M.C.; Pitra, N.J. Next Generation Sequencing for a Plant of Great Tradition: Application of NGS to SNP Detection and Validation in Hops (Humulus lupulus L.). Brew. Sci. 2013, 66, 185–191. [Google Scholar]
- Deschamps, S.; Llaca, V.; May, G.D. Genotyping-by-sequencing in plants. Biology 2012, 1, 460–483. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Song, T.; Jiang, W.; Wang, D.; Pu, B.; Luan, C. SNP-Based Kompetitive Allele Specific PCR (KASPTM) Method for the Qualification and Quantification of Hop Varieties. J. Am. Soc. Brew. Chem. 2018, 76, 185–189. [Google Scholar] [CrossRef]
- Henning, J.A.; Coggins, J.; Peterson, M. Simple SNP-based minimal marker genotyping for Humulus lupulus L. identification and variety validation. BMC Res. Notes 2015, 8, 542. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.C.; Faria, M.A.; Barata, A.M.; da Silva, I.G.; Cerenak, A.; Ferreira, I.M. Portuguese wild hop diversity assessment by fast SNP genotyping using high-resolution melting. J. Appl. Genet. 2022, 63, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Mozny, M.; Tolasz, R.; Nekovar, J.; Sparks, T.; Trnka, M.; Zalud, Z. The impact of climate change on the yield and quality of Saaz hops in the Czech Republic. Agric. For. Meteorol. 2009, 149, 913–919. [Google Scholar] [CrossRef]
- Mozny, M.; Trnka, M.; Vlach, V.; Zalud, Z.; Cejka, T.; Hajkova, L.; Potopova, V.; Semenov, M.A.; Semeradova, D. Climate-induced decline in the quality and quantity of European hops calls for immediate adaptation measures. Nat. Commun. 2023, 14, 6028. [Google Scholar] [CrossRef] [PubMed]
- Horreo, J.L.; Peredo, E.L.; Olmedo, J.L.; Valladares, J.E.; García, E.; Revilla, M.A. Genetic diversity inferred from microsatellites of wild hops in Galicia (Spain). Brew. Sci. 2014, 67, 132–136. [Google Scholar]
- Karlsson Strese, E.M.; Lundström, M.; Hagenblad, J.; Leino, M.W. Genetic Diversity in Remnant Swedish Hop (Humulus lupulus L.) Yards from the 15th to 18th Century. Econ. Bot. 2014, 68, 231–245. [Google Scholar] [CrossRef]
- Riccioni, C.; Belfiori, B.; Sileoni, V.; Marconi, O.; Perretti, G.; Bellucci, M.; Rubini, A. High genetic and chemical diversity of wild hop populations from Central Italy with signals of a genetic structure influenced by both sexual and asexual reproduction. Plant Sci. 2021, 304, 110794. [Google Scholar] [CrossRef]
- Calvi, A.; Aci, M.M.; Lupini, A.; Preiti, G. Morphological and Genetic Analysis of Wild Hop (Humulus lupulus L.) Germplasm from Calabria Region in South Italy. Agronomy 2023, 13, 252. [Google Scholar] [CrossRef]
- Patzak, J.; Nesvadba, V.; Henychová, A.; Krofta, K. Assessment of the genetic diversity of wild hops (Humulus lupulus L.) in Europe using chemical and molecular analyses. Biochem. Syst. Ecol. 2010, 38, 136–145. [Google Scholar] [CrossRef]
- Patzak, J.; Nesvadba, V.; Krofta, K.; Henychova, A.; Marzoev, A.I.; Richards, K. Evaluation of genetic variability of wild hops (Humulus lupulus L.) in Canada and the Caucasus region by chemical and molecular methods. Genome 2010, 53, 545–557. [Google Scholar] [CrossRef]
- Edwardson, J.R. Hops: Their Botany, History, Production and Utilization. Econ. Bot. 1952, 6, 804–811. [Google Scholar] [CrossRef]
- Castro, C.B.; Whittock, L.D.; Whittock, S.P.; Leggett, G.; Koutoulis, A. DNA sequence and expression variation of hop (Humulus lupulus) valerophenone synthase (VPS), a key gene in bitter acid biosynthesis. Ann. Bot. 2008, 102, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Patrick, D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Seefelder, S.; Ehrmaier, H.; Schweizer, G.; Seigner, E. Genetic diversity and phylogenetic relationships among accessions of hop, Humulus lupulus, as determined by amplified fragment length polymorphism fingerprinting compared with pedigree data. Plant Breed. 2000, 119, 257–263. [Google Scholar] [CrossRef]
- Henning, J.A.; Steiner, J.J.; Hummer, K.E. Genetic diversity among world hop accessions grown in the USA. Crop Sci. 2004, 44, 411–417. [Google Scholar] [CrossRef]
- Grdiša, M.; Šatović, Z.; Liber, Z.; Jakše, J.; Varga, F.; Erhatić, R.; Srečec, S. High Genetic Diversity and Low Population Differentiation in Wild Hop (Humulus lupulus L.) from Croatia. Appl. Sci. 2021, 11, 6484. [Google Scholar] [CrossRef]
- Mafakheri, M.; Kordrostami, M.; Rahimi, M.; Matthews, P.D. Evaluating genetic diversity and structure of a wild hop (Humulus lupulus L.) germplasm using morphological and molecular characteristics. Euphytica 2020, 216, 58. [Google Scholar] [CrossRef]
- Rodolfi, M.; Silvanini, A.; Chiancone, B.; Marieschi, M.; Fabbri, A.; Bruni, R.; Ganino, T. Identification and genetic structure of wild Italian Humulus lupulus L. and comparison with European and American hop cultivars using nuclear microsatellite markers. Genet. Resour. Crop Evol. 2018, 65, 1405–1422. [Google Scholar] [CrossRef]



| Multiplex PCR Reaction | Forward (5′-3′) | Reverse (5′-3′) | Product Length |
|---|---|---|---|
| 1 (Amplicons 1 and 6) | GCATGATCAGCAGCCTTACTCG | ATGACGATGATGGGAATTGTGGCAT | 98 bp |
| GGGCCCTCGGAGAAACATT | TTTGGGAGATCAACCCTGGC | 238 bp | |
| 2 (Amplicons 2 and 3) | TGCAGCTTAAATGGATAAGGGGAAG | TCCTCATCTGTTGCATTTTCCTTCC | 115 bp |
| CAATGAGTGCCTCCTTTGACCGT | TCAGTGGGGTTCCTTCCTTTCCA | 80 bp | |
| 3 (Amplicons 4 and 5) | GACCCCCAATGTTGTTGCTATGGT | GATGGTGCATGGGTGCCTAAAGT | 106 bp |
| TTCTTCTCTGGTCCATGCGCCTT | AACGGAACAAACTGAGGGCGGT | 70 bp |
| SNP | WGS Accession Number | Amplicon Coordinates | SNP Position | Alleles |
|---|---|---|---|---|
| 1 | JALDWI010000197.1 | 365,042 to 365,139 | 365,109 | T/C |
| 2 | JALDWI010000023.1 | 2,509,758 to 2,509,872 | 2,509,809 | A/C |
| 3 | JALDWI010003715.1 | 46,926 to 47,011 | 46,965 | T/C |
| 4 | JALDWI010000347.1 | 413,416 to 413,522 | 413,465 | A/G |
| 5 | JALDWI010002821.1 | 93,676 to 93,745 | 93,714 | G/C |
| 6 | JALDWI010001295.1 | 695,872 to 696,109 | 695,964 | G/C |
| 7 | JALDWI010001295.1 | 695,872 to 696,109 | 695,965 | G/T |
| 8 | JALDWI010001295.1 | 695,872 to 696,109 | 696,029 | A/C |
| 9 | JALDWI010001295.1 | 695,872 to 696,109 | 696,051 | T/C |
| SNP | Na | Ne | Ho | He | F |
|---|---|---|---|---|---|
| 1 | 2.000 | 1.903 | 0.571 | 0.473 | −0.213 |
| 2 | 2.000 | 1.548 | 0.389 | 0.341 | −0.135 |
| 3 | 2.000 | 1.944 | 0.526 | 0.485 | −0.089 |
| 4 | 2.000 | 1.431 | 0.277 | 0.290 | −0.005 |
| 5 | 1.429 | 1.066 | 0.040 | 0.057 | 0.283 |
| 6 | 1.857 | 1.414 | 0.308 | 0.260 | −0.102 |
| 7 | 1.857 | 1.414 | 0.308 | 0.260 | −0.102 |
| 8 | 1.857 | 1.414 | 0.308 | 0.260 | −0.102 |
| 9 | 1.857 | 1.414 | 0.308 | 0.260 | −0.102 |
| Mean | 1.873 | 1.506 | 0.337 | 0.299 | −0.085 |
| SE | 0.042 | 0.044 | 0.030 | 0.021 | 0.047 |
| Population | Location | N | Na | Ne | Ho | He |
|---|---|---|---|---|---|---|
| 1 | Mavrilo | 13 | 2.000 | 1.401 | 0.299 | 0.259 |
| 2 | Megali Kapsi | 5 | 1.889 | 1.416 | 0.289 | 0.260 |
| 3 | Gavros | 5 | 2.000 | 1.690 | 0.444 | 0.400 |
| 4 | Ipati | 17 | 1.889 | 1.758 | 0.477 | 0.403 |
| 5 | Kompotades | 13 | 2.000 | 1.447 | 0.239 | 0.283 |
| 6 | Gravia | 5 | 1.889 | 1.472 | 0.378 | 0.291 |
| 7 | Livadia | 11 | 1.444 | 1.356 | 0.232 | 0.193 |
| Mean | - | 9.857 | 1.873 | 1.506 | 0.337 | 0.299 |
| SE | - | 0.574 | 0.042 | 0.044 | 0.030 | 0.021 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tegopoulos, K.; Fountas, D.V.; Andronidou, E.-M.; Bagos, P.G.; Kolovos, P.; Skavdis, G.; Pergantas, P.; Braliou, G.G.; Papageorgiou, A.C.; Grigoriou, M.E. Assessing Genetic Diversity and Population Differentiation in Wild Hop (Humulus lupulus) from the Region of Central Greece via SNP-NGS Genotyping. Diversity 2023, 15, 1171. https://doi.org/10.3390/d15121171
Tegopoulos K, Fountas DV, Andronidou E-M, Bagos PG, Kolovos P, Skavdis G, Pergantas P, Braliou GG, Papageorgiou AC, Grigoriou ME. Assessing Genetic Diversity and Population Differentiation in Wild Hop (Humulus lupulus) from the Region of Central Greece via SNP-NGS Genotyping. Diversity. 2023; 15(12):1171. https://doi.org/10.3390/d15121171
Chicago/Turabian StyleTegopoulos, Konstantinos, Dimitrios V. Fountas, Elisavet-Maria Andronidou, Pantelis G. Bagos, Petros Kolovos, George Skavdis, Panagiotis Pergantas, Georgia G. Braliou, Aristotelis C. Papageorgiou, and Maria E. Grigoriou. 2023. "Assessing Genetic Diversity and Population Differentiation in Wild Hop (Humulus lupulus) from the Region of Central Greece via SNP-NGS Genotyping" Diversity 15, no. 12: 1171. https://doi.org/10.3390/d15121171






