A Digital Microfluidics Platform for Loop-Mediated Isothermal Amplification Detection
Abstract
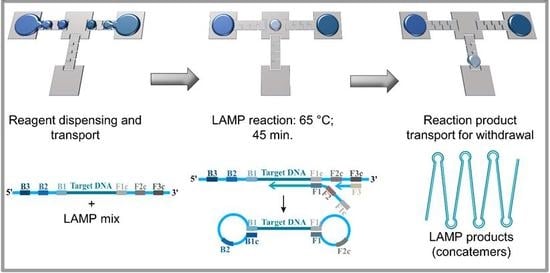
:1. Introduction
2. Materials and Methods
2.1. DMF Device Design and Fabrication
2.2. DMF Control System
2.3. Temperature Control System
2.4. Benchtop LAMP Amplification of c-Myc
2.5. LAMP Reaction Optimization for On-Chip Integration
2.6. DMF-LAMP Amplification of c-Myc (On-Chip)
3. Results and Discussion
3.1. DMF Device Characterization and Performance
3.2. Temperature Control Evaluation on the DMF Platform
3.3. Benchtop LAMP Optimization for On-Chip Integration
3.4. DMF-LAMP
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Coelho, B.J.; Veigas, B.; Fortunato, E.; Martins, R.; Águas, H.; Igreja, R.; Baptista, P.V. Digital Microfluidics for Nucleic Acid Amplification. Sensors 2017, 17, 1495. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Ng, A.H.C.; Fobel, R.; Wheeler, A.R. Digital Microfluidics. Annu. Rev. Anal. Chem. 2012, 5, 413–440. [Google Scholar] [CrossRef] [PubMed]
- Millington, D.S.; Sista, R.; Eckhardt, A.; Rouse, J.; Bali, D.; Goldberg, R.; Cotten, M.; Buckley, R.; Pamula, V. Digital Microfluidics: A Future Technology in the Newborn Screening Laboratory? Semin. Perinatol. 2010, 34, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Seyrig, G.; Tourlousse, D.M.; Stedtfeld, R.D.; Tiedje, J.M.; Hashsham, S.A. A CCD-based fluorescence imaging system for real-time loop-mediated isothermal amplification-based rapid and sensitive detection of waterborne pathogens on microchips. Biomed. Microdevices 2011, 13, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Norian, H.S.; Shepard, K.; Kymissis, J.; Field, R. An integrated CMOS quantitative-polymerase-chain-reaction lab-on-chip for point-of-care diagnostics. Lab Chip 2014, 14, 4076–4084. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, S.; Valiadi, M.; Tsaloglou, M.; Parry-jones, L.; Jacobs, A.; Watson, R.; Turner, C.; Amos, R.; Hadwen, B.; Buse, J.; et al. Rapid and sensitive detection of antibiotic resistance on a programmable digital microfluidic platform. Lab Chip 2015, 15, 3065–3075. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Lee, G.-B.; Huang, F.-C.; Chen, Y.-Y.; Lin, J.-L. Integrated polymerase chain reaction chips utilizing digital microfluidics. Biomed. Microdevices 2006, 8, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Rouse, J.L.; Eckhardt, A.E.; Srinivasan, V.; Pamula, V.K.; Schell, W.A.; Benton, J.L.; Mitchell, T.G.; Pollack, M.G. Multiplexed real-time polymerase chain reaction on a digital microfluidic platform. Anal. Chem. 2010, 82, 2310–2316. [Google Scholar] [CrossRef] [PubMed]
- Fouillet, Y.; Jary, D.; Brachet, A.; Berthier, J.; Blervaque, R.; Davous, L.; Roux, J.; Achard, J.; Peponnet, C. EWOD digital microfluidics for Lab on a Chip. In Proceedings of the ASME 4th International Conference on Nanochannels, Microchannels and Minichannels, Limerick, Ireland, 19–21 June 2006; pp. 1255–1264. [Google Scholar]
- Berthier, J.; Mourier, V.; Sarrut, N.; Jary, D.; Fouillet, Y.; Pouteau, P.; Caillat, P.; Peponnet, C. Some examples of micro-devices for biotechnology developed at the Department of Technologies for Life Sciences and Healthcare of the LETI. Int. J. Nanotechnol. 2010, 7, 802–818. [Google Scholar] [CrossRef]
- Kühnemund, M.; Witters, D.; Nilsson, M.; Lammertyn, J. Circle-to-circle amplification on a digital microfluidic chip for amplified single molecule detection. Lab Chip 2014, 14, 2983–2992. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The Use of Loop-Mediated Isothermal Amplification (TB-LAMP) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance; GPS Publishing: Geneva, Switzerland, 2016. [Google Scholar]
- Veigas, B.; Pinto, J.; Vinhas, R.; Calmeiro, T.; Martins, R.; Fortunato, E.; Baptista, P.V. Quantitative real-time monitoring of RCA amplification of cancer biomarkers mediated by a flexible ion sensitive platform. Biosens. Bioelectron. 2017, 91, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-M.; Chang, W.-H.; Wang, C.-H.; Wang, J.-H.; Mai, J.; Lee, G.-B. Nucleic acid amplification using microfluidic systems. Lab Chip 2013, 13, 1225–1242. [Google Scholar] [CrossRef] [PubMed]
- Bunz, F. Chapter 5- Cancer Gene Pathways; C-MYC is a Downstream Effector of Multiple Cancer Gene Pathways. In Principles of Cancer Genetics; Springer: New York, NY, USA, 2008; pp. 201–205. [Google Scholar]
- Cenci, T.; Martini, M.; Montano, N.; Alessandris, Q.G.; Falchetti, M.L.; Annibali, D.; Savino, M.; Bianchi, F.; Pierconti, F.; Nasi, S.; et al. Prognostic relevance of c-Myc and BMI1 expression in patients with glioblastoma. Am. J. Clin. Pathol. 2012, 138, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Giroux-Leprieur, E.; Wislez, M.; Hu, M.; Zhang, Y.; Shi, H.; Du, K.; Wang, L. Human RNA polymerase II associated factor 1 complex promotes tumorigenesis by activating c-MYC transcription in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2015, 465, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Mashraei, Y.; Sivashankar, S.; Buttner, U.; Salama, K.N. Integration of Fractal Biosensor in a Digital Microfluidic Platform. IEEE Sens. J. 2016, 16, 8775–8783. [Google Scholar] [CrossRef]
- Xu, X.; Sun, L.; Chen, L.; Zhou, Z.; Xiao, J.; Zhang, Y. Electrowetting on dielectric device with crescent electrodes for reliable and low-voltage droplet manipulation. Biomicrofluidics 2014, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dak, P.; Ebrahimi, A.; Swaminathan, V.; Duarte-Guevara, C.; Bashir, R.; Alam, M.A. Droplet-based biosensing for lab-on-a-chip, open microfluidics platforms. Biosensors 2016, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Fobel, R.; Fobel, C.; Wheeler, A.R. DropBot: An open-source digital microfluidic control system with precise control of electrostatic driving force and instantaneous drop velocity measurement. Appl. Phys. Lett. 2013, 102, 1–5. [Google Scholar] [CrossRef]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Veigas, B.; Branquinho, R.; Pinto, J.V.; Wojcik, P.J.; Martins, R.; Fortunato, E.; Baptista, P.V. Ion sensing (EIS) real-time quantitative monitorization of isothermal DNA amplification. Biosens. Bioelectron. 2014, 52, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Stedtfeld, R.D.; Tourlousse, D.M.; Seyrig, G.; Stedtfeld, T.M.; Kronlein, M.; Price, S.; Ahmad, F.; Gulari, E.; Tiedje, J.M.; Hashsham, S. Gene-Z: A device for point of care genetic testing using a smartphone. Lab Chip 2012, 12, 1454–1462. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Shen, F.; McCalla, S.E.; Kreutz, J.E.; Karymov, M.A.; Ismagilov, R.F. Mechanistic evaluation of the pros and cons of digital RT-LAMP for HIV-1 viral load quantification on a microfluidic device and improved efficiency via a two-step digital protocol. Anal. Chem. 2013, 85, 1540–1546. [Google Scholar] [CrossRef] [PubMed]
- Schoepp, N.G.; Schlappi, T.S.; Curtis, M.S.; Butkovich, S.S.; Miller, S.; Humphries, R.M.; Ismagilov, R.F. Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci. Transl. Med. 2017, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, M.C.; Spoto, G. Integration of isothermal amplification methods in microfluidic devices: Recent advances. Biosens. Bioelectron. 2017, 90, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Jebrail, M.J.; Sinha, A.; Vellucci, S.; Renzi, R.F.; Ambriz, C.; Gondhalekar, C.; Schoeniger, J.S.; Patel, K.D.; Branda, S.S. World-to-Digital-Microfluidic Interface Enabling Extraction and Purification of RNA from Human Whole Blood. Anal. Chem. 2014, 86, 3856–3862. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coelho, B.J.; Veigas, B.; Águas, H.; Fortunato, E.; Martins, R.; Baptista, P.V.; Igreja, R. A Digital Microfluidics Platform for Loop-Mediated Isothermal Amplification Detection. Sensors 2017, 17, 2616. https://doi.org/10.3390/s17112616
Coelho BJ, Veigas B, Águas H, Fortunato E, Martins R, Baptista PV, Igreja R. A Digital Microfluidics Platform for Loop-Mediated Isothermal Amplification Detection. Sensors. 2017; 17(11):2616. https://doi.org/10.3390/s17112616
Chicago/Turabian StyleCoelho, Beatriz Jorge, Bruno Veigas, Hugo Águas, Elvira Fortunato, Rodrigo Martins, Pedro Viana Baptista, and Rui Igreja. 2017. "A Digital Microfluidics Platform for Loop-Mediated Isothermal Amplification Detection" Sensors 17, no. 11: 2616. https://doi.org/10.3390/s17112616