Aptamers in Education: Undergraduates Make Aptamers and Acquire 21st Century Skills Along the Way
Abstract
1. Introduction
2. Materials and Methods
2.1. Integration of Aptamer Research into A Teaching Lab
- A fundamental awareness and early experience in scientific research, specifically in the field of aptamer development (oligonucleotide affinity reagent development). This involves an introduction to the terminology, technical concepts, and principles of the research. The learning objectives include:
- Identify a creative, focused, and manageable research question or topic.
- Design a methodology for answering a research question, perusing the project, or small-scale “troubleshooting” tests.
- Demonstrate the understanding of the research implications and its translation to practical applications.
- Teamwork
- Brainstorm troubleshooting and/or problem-solving ideas with other students and/or mentors.
- Make changes to work based on critical analysis of work and on peer review feedback.
- Communication
- Develop and practice scientific writing skills.
- Develop science communication skills, as well as further develop argumentation skills, including the connection between the problem and the solution.
- Data Analysis Experience
- Construct a meaningful figure using research data, which includes appropriate controls and statistics, if appropriate.
- Collect, interpret, evaluate, provide context, and rational conclusions for research data.
- Resilience
- Develop and implement mechanisms to overcome, bypass, and/or wade through setbacks.
- Initiate projects or activities with set deadlines and sometimes incomplete information.
2.2. Anti-CIAP Aptamer Research: in Vitro Aptamer Selection (RNA Aptamers)
2.3. Anti-CIAP Aptamer Research: Sanger Sequencing and Aptamer Binding Assay of Selected Pool
2.4. Creating a Culture of Safety
- In an effort to preserve a culture of safety in the lab, let’s talk about how to protect ourselves in lab. Where do we find personal protective equipment (PPE) in lab and when do we need to wear it?
- Here are a few safety reminders when performing polyacrylamide gel electrophoresis (PAGE): Unpolymerized acrylamide is a neurotoxin and must be kept in the PAGE hood area. Polymerized acrylamide (i.e., solid-gel acrylamide) may be safely disposed of in the regular trash can. If you break a PAGE gel glass, please report it to a mentor and dispose of the glass in the glass waste box. How do we remember these things? Can you create a picture/cartoon, song, or poem to remind us of these safe PAGE practices?
- Sometimes it’s the smallest things. “Hot glass looks like cold glass!” –Alex G. When microwaving agarose solutions, please loosen the lid. Remove clutter from the area to clear a spot to eventually place the hot bottle. And - use a hot pad to remove the bottle from the microwave. Can you think of other "small things" in lab that could be potential safety hazards and what actions can be done to mitigate the hazard potential? Please share one or more idea with your neighbor. You’ll have an opportunity to share your idea with the class.
2.5. Educational Assessment Methods
- Effective Communication: the ability to produce written and oral reports, and make persuasive, evidence-based arguments using appropriate scientific sources and effective figures and graphics.
- Information Literacy: the ability to locate appropriate information, evaluate sources critically, read and interpret primary scientific literature, and synthesize information.
- Computational/Technological Literacy: the ability to organize and interpret data and apply computational skills to solve problems.
- Self-directed Learning: the ability to execute an independent and original project culminating in a product, such as a written document, oral presentation, or physical object; the ability to innovate, create, or conduct original research projects.
- Teamwork: the ability to resolve conflicts, plan, and coordinate group efforts.
3. Results
3.1. Anti-Calf Intestinal Alkaline Phosphatase (CIAP) Aptamer Research Results
3.1.1. Anti-CIAP Aptamer Selection and Sequence Enrichment Results
3.1.2. Anti-CIAP Aptamer Minimized Variant Design, Assay Results, and Dissociation Constants
3.1.3. Anti-CIAP Aptamer Specificity and Activity
3.2. Results of Student Assessment Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A—CIAP Aptamer in vitro Selection Conditions
| Round | RNA:CIAP (pmol:pmol) | Volume of Beads Used (μL) | # of PCR Cycles Necessary to Amplify Selected Pool | Total Wash Volume (mL) | Amount of Recovered Pool (pmol) |
|---|---|---|---|---|---|
| Round 1 | 400:400 | 50.0 | 10 | 2.4 | 2093 |
| Round 2* | 400:400 | 50.0 | 14 | 2.4 | 388 |
| Round 3* | 300:300 | 44.1 | 11 | 2.4 | 301 |
| Round 4* | 120:100 | 14.7 | 15 | 3.2 | 969 |
| Round 5 | 200:100 | 14.7 | 9 | 3.2 | 906 |
| Round 6* | 200:100 | 14.7 | 9 | 4.0 | 298 |
| Round 7* | 200:50 | 7.35 | 9 | 6.0 | 308 |
| Round 8* | 200:50 | 7.35 | 13 | 13.9 | 1241 |
| Round 9* | 800:50 | 7.35 | 13 | 47.2 | 735 |
Appendix B —Early anti-CIAP 32P-labeled RNA Binding Assay Results

Appendix C—Dissociation Constants (Kd) of Anti-CIAP Aptamers

Appendix D—Aptamer Specificity Tests

References
- Zhou, W.; Huang, P.J.J.; Ding, J.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; White, R.R.; Becker, R.C.; Sullenger, B.A. Aptamers as Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 0076. [Google Scholar] [CrossRef]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.B.; Tang, T.H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Autour, A.; Jeng, S.C.Y.; Cawte, A.D.; Abdolahzadeh, A.; Galli, A.; Panchapakesan, S.S.S.; Rueda, D.; Ryckelynck, M.; Unrau, P.J. Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Strum Kenny, S. The Boyer commission on educating undergraduates in the research university, reinventing undergraduate education: A blueprint for America’s research universities. 1998; Stony Brook, NY. Available online: https://files.eric.ed.gov/fulltext/ED424840.pdf (accessed on 24 July 2019).
- American Association for the Advancement of Science. Vision and Change: A Call to Action, Final Report; American Association for the Advancement of Science: Washington, DC, USA, 2011. [Google Scholar]
- Olson, S.; Riordan, D.G. Engage to Excel: Producing One Million Additional College Graduates with Degrees in Science, Technology, Engineering, and Mathematics. Report to the President. Executive Office of the President; Executive Office of the President: Washington, DC, USA, 2012.
- The University of Texas College of Natural Sciences Freshman Research Initiative. Available online: https://cns.utexas.edu/fri (accessed on 8 July 2019).
- University of Maryland, The First-Year Innovation & Research Experience (FIRE). Available online: https://fire.umd.edu (accessed on 8 July 2019).
- Iowa State University, Freshman Research Initiative (FRI). Available online: https://fri.las.iastate.edu/research-streams/ (accessed on 8 July 2019).
- Rodenbusch, S.E.; Hernandez, P.R.; Simmons, S.L.; Dolan, E.L. Early engagement in course-based research increases graduation rates and completion of science, engineering, and mathematics degrees. CBE Life Sci. Educ. 2016, 15, ar20. [Google Scholar] [CrossRef] [PubMed]
- Jordan, T.C.; Burnett, S.H.; Carson, S.; Caruso, S.M.; Clase, K.; DeJong, R.J.; Dennehy, J.J.; Denver, D.R.; Dunbar, D.; Elgin, S.C.R.; et al. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. MBio 2014, 5, e01051-13. [Google Scholar] [CrossRef]
- Lopatto, D.; Alvarez, C.; Barnard, D.; Chandrasekaran, C.; Chung, H.; Du, C.; Eckdahl, T.; Goodman, A.L.; Hauser, C.; Jones, C.J.; et al. Undergraduate research. Genomics Education Partnership. Science 2008, 322, 684–685. [Google Scholar] [CrossRef]
- Bascom-Slack, C.A.; Arnold, A.E.; Strobel, S.A. IBI series winner. Student-directed discovery of the plant microbiome and its products. Science 2012, 338, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Kloser, M.J.; Brownell, S.E.; Shavelson, R.J.; Fukami, T. Effects of a Research-Based Ecology Lab Course: A Study of Nonvolunteer Achievement, Self-Confidence, and Perception of Lab Course Purpose. J. Coll. Sci. Teach. 2013, 42, 72–81. [Google Scholar]
- Walcott, R.L.; Corso, P.S.; Rodenbusch, S.E.; Dolan, E.L. Benefit–Cost Analysis of Undergraduate Education Programs: An Example Analysis of the Freshman Research Initiative. CBE—Life Sci. Educ. 2018, 17, rm1. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, E.; Long, S.R.; Rodenbusch, S.E.; Shear, R.I.; Beckham, J.T.; Procko, K.; DePue, L.; Stevenson, K.J.; Robertus, J.D.; Martin, S.; et al. Teaching through Research: Alignment of Core Chemistry Competencies and Skills within a Multidisciplinary Research Framework. J. Chem. Educ. 2018, 95, 248–258. [Google Scholar] [CrossRef]
- The University of Texas, Aptamer Stream course syllabus, Fall 2018. Available online: https://utdirect.utexas.edu/apps/student/coursedocs/nlogon/download/8866150 (accessed on 23 June 2019).
- Hall, B.; Arshad, S.; Seo, K.; Bowman, C.; Corley, M.; Jhaveri, S.D.; Ellington, A.D. In vitro selection of RNA aptamers to a protein target by filter immobilization. Curr. Protoc. Nucleic Acid Chem. 2010, 40, 9.3.1–9.3.27. [Google Scholar] [CrossRef][Green Version]
- Robertson, M.P.; Ellington, A.D. In vitro selection of nucleoprotein enzymes. Nat. Biotechnol. 2001, 19, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.C.; Ellington, A.D. Automated Selection of Anti-Protein Aptamers. Bioorg. Med. Chem. 2000, 9, 2525–2531. [Google Scholar] [CrossRef]
- Codrea, V.; Hayner, M.; Hall, B.; Jhaveri, S.; Ellington, A.D. In vitro selection of RNA aptamers to a small molecule target. Curr. Protoc. Nucleic. Acid. Chem. 2010, 40, 9.5.1–9.5.23. [Google Scholar] [CrossRef]
- Williams, L.; Wiebe, E.; Yang, K.; Ferzli, M.; Miller, C. In support of paired programming in the introductory computer science course. Comput. Sci. Educ. 2002, 12, 197–212. [Google Scholar] [CrossRef]
- Friday Institute for Educational Innovation. Middle and High School STEM-Student Survey. 2012. Raleigh, NC. Available online: https://miso.fi.ncsu.edu/wp-content/uploads/2013/06/S-STEM_FridayInstitute_DevAndPsychometricProperties_FINAL.pdf (accessed on 24 July 2019).
- Hanauer, D.; Graham, M.; Hatfull, G. A Measure of College Student Persistence in the Sciences (PITS). CBE Life Sci. Educ. 2016, 15, ar54. [Google Scholar] [CrossRef]
- Weston, T.; Laursen, S. The undergraduate research student self-assessment (URSSA): Validation for use in program evaluation. CBE Life Sci. Educ. 2015, 14, ar33. [Google Scholar] [CrossRef] [PubMed]
- Toker, Y.; Ackerman, P.L. Utilizing occupational complexity levels in vocational interest assessments: Assessing interests for STEM areas. J. Vocat. Behav. 2012, 80, 524–544. [Google Scholar] [CrossRef]
- College of Natural Sciences, UT Austin. Strategic Master Plan for CNS Undergraduate Curriculum. 2017. 21st Century Curriculum Implementation Task Force. Available online: https://cns.utexas.edu/teaching-portal/experiential-learning#cns-goals-for-experiential-learning (accessed on 24 July 2019).
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, S.; Pothukuchy, A.; Shroff, R.; Cole, A.W.; Byrom, M.; Ellefson, J.W.; Gollihar, J.D.; Ellington, A.D. Cellular reagents for diagnostics and synthetic biology. PloS ONE 2018, 13, e0201681. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Drolet, D.W.; Jenison, R.D.; Smith, D.E.; Pratt, D.; Hicke, B.J. A high throughput platform for systematic evolution of ligands by exponential enrichment (SELEX). Comb. Chem. High Throughput Screen. 1999, 2, 271–278. [Google Scholar]
- Auchincloss, L.C.; Laursen, S.L.; Branchaw, J.L.; Eagan, K.; Graham, M.; Hanauer, D.I.; Lawrie, G.; McLinn, C.M.; Pelaez, N.; Rowland, S.; et al. Assessment of course-based undergraduate research experiences: a meeting report. CBE—Life Sci. Educ. 2014, 13, 29–40. [Google Scholar] [CrossRef] [PubMed]




| Clone | Sequence |
|---|---|
| c2-4 | GGGUUUACCUAGGUGUAGAUGCUUCUUCACUCCUUAUGAACACGUAGCGCUCAAUCAUCUCUAAUUAAUUCUCAAGUGACGUCUGAACUGCUUCGAA |
| c2-1 | GGGUUUACCUAGGUGUAGAUGCUGAUAGAUUUCCCCUGACUUAGGAGCUCAUUAGAUUUUUAUUGUGUGGGGCAAGUGACGUCUGAACUGCUUCGAA |
| Motif or Clone | % Occurrence (n = 36 clones) | Kd | Fold and Notes | |
|---|---|---|---|---|
| Motif VDH 2.14, 14 nt: GAACUCAACAUAAR | 53% | Not tested |  | VDH 2.14 motif appears in the loop structure (as seen in c2-2) approx. 18 out of 19 clones. Note the presence of an R (A or G). |
| Aptamer Clone 4-3, 97 nt: 5′GGGUUUACCUAGGUGUAGAUGCUGUAUAUAGCGAACUCAACAUAAGGUAUAAUUACAAUUUCUAUACUUCUUCAAGUGACGUCUGAACUGCUUCGAA3′ | 6% | 5 nM | 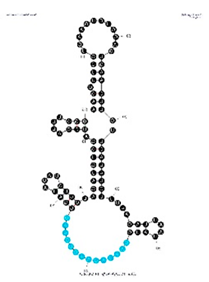 | VDH 2.14 motif appears in the loop structure (as seen in c4-3) approx. 2 out of 2 times. |
| Aptamer Clone 4-9, 97 nt: 5′GGGUUUACCUAGGUGUAGAUGCUUCWAUUGAUAUGUUAUAACUGAACUCAACAUAAGGAUAUGAUGUAUGAUCAAGUGACGUCUGAACUGCUUCGAA3′ | 6% | 9.4 nM |  | VDH 2.14 motif appears in the loop structure (as seen in c4-9) approx. 2 out of 2 times. Note the presence of a W (A or T). |
| Aptamer Clone 3-6, 97 nt: 5′GGGUUUACCUAGGUGUAGAUGCUCUGCCCUUCAGAUUUAUCGAUGACCGUUGAACUCAACAUAAGACCUUCCAAAGUGACGUCUGAACUGCUUCGAA3′ | 3% | 10.8 nM | 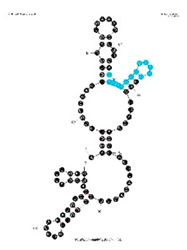 | VDH 2.14 motif is present in the loop and stem structure. |
| Minimized Aptamer Variant 3.1, 55 nt: 5′GGGUAGAUGCUGUAUAUAGCGAACUCAACAUAAGGUAUAAUUACAAUUUCUACCC3′ | N/A | 6.7 nM | 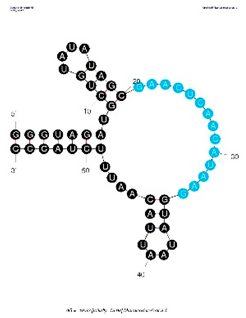 | 3.1 is a minimized variant of aptamer clone 4-3. The triplet GGG was added to 5′ end to improve transcription efficiency and, thus CCC added to 3′ end to participate in the stem structure, contributing to a lower dG. |
| Research-Education Features of Consideration | Aptamer Stream | Replication of Same or Similar CURE |
|---|---|---|
| Student background knowledge | Freshman students, with limited or no experience in aptamer selection laboratory skills or contextual background, are taught both simultaneously in a 1-hr/wk lecture and 6- to 8-hr/wk laboratory work, with additional support mechanisms (e.g., small group meetings, office hours, etc.). | Guided and independent laboratory research opportunities complemented with lecture/contextual background lessons and discussions are suggested. |
| Student interest | The biomedical potential of aptamers (i.e., diagnostic and therapeutic applications) are well-received by the College of Natural Sciences undergraduates, many/most with plans for a career in a health field. | Matching a research experience/CURE with the student interest and/or career prospects is suggested. Surveying and/or consideration of student career interest is encouraged. |
| Student time | Students commit at least 6 hrs/wk in the spring semester and 8 hrs/wk in the fall semester to their aptamer research (~14 wks/semester). Students may schedule their own lab hours M-F 9a-6p. | Consideration of the lab hour flexibility, while providing safe opportunities for training, guidance, and oversight (such as through undergraduate peer mentors) is suggested. |
| Reagents and enzyme | To minimize the cost of reagents and enzymes, reagents are often purchased in bulk or even prepared in lab, if possible (e.g., PCR buffer, plasmid purification reagents, etc.). Enzymes have been provided by other teaching labs, collaborating labs, etc. Additionally, some raw, unpurified enzyme preps have been used, as well [32]. | Consider how enzymes and reagents will be affordably purchased. Undergraduate and new researchers may consume, at times, more reagents than experienced researchers, as the learning curve (such as pipetting errors, solution preparation mistake, etc.) are common. |
| Equipment | Equipment common to most molecular biology labs, with the exception of the speedvac and the plate reader (optional) is sufficient for Aptamer Stream research (see Materials and Methods section). While the speedvac and plate reader are not required, the plate reader is helpful for assay development. | Use caution when obtaining old, well-used equipment, or even flimsy equipment, as novice researchers in their attempts to learn how to use the equipment, may inadvertently damage or induce wear on the equipment. Robust equipment and a maintenance and repair budget is suggested, when possible (such as for micropipette repair/calibration). |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stovall, G.M.; Huynh, V.; Engelman, S.; Ellington, A.D. Aptamers in Education: Undergraduates Make Aptamers and Acquire 21st Century Skills Along the Way. Sensors 2019, 19, 3270. https://doi.org/10.3390/s19153270
Stovall GM, Huynh V, Engelman S, Ellington AD. Aptamers in Education: Undergraduates Make Aptamers and Acquire 21st Century Skills Along the Way. Sensors. 2019; 19(15):3270. https://doi.org/10.3390/s19153270
Chicago/Turabian StyleStovall, Gwendolyn M., Vincent Huynh, Shelly Engelman, and Andrew D. Ellington. 2019. "Aptamers in Education: Undergraduates Make Aptamers and Acquire 21st Century Skills Along the Way" Sensors 19, no. 15: 3270. https://doi.org/10.3390/s19153270
APA StyleStovall, G. M., Huynh, V., Engelman, S., & Ellington, A. D. (2019). Aptamers in Education: Undergraduates Make Aptamers and Acquire 21st Century Skills Along the Way. Sensors, 19(15), 3270. https://doi.org/10.3390/s19153270






