A Review of the Resistance Mechanisms for β-Lactams, Macrolides and Fluoroquinolones among Streptococcus pneumoniae
Abstract
:1. Introduction
2. Etiology
3. Pathogenesis and Immunopathogenesis of Pneumonia
4. Overview of Antibiotic
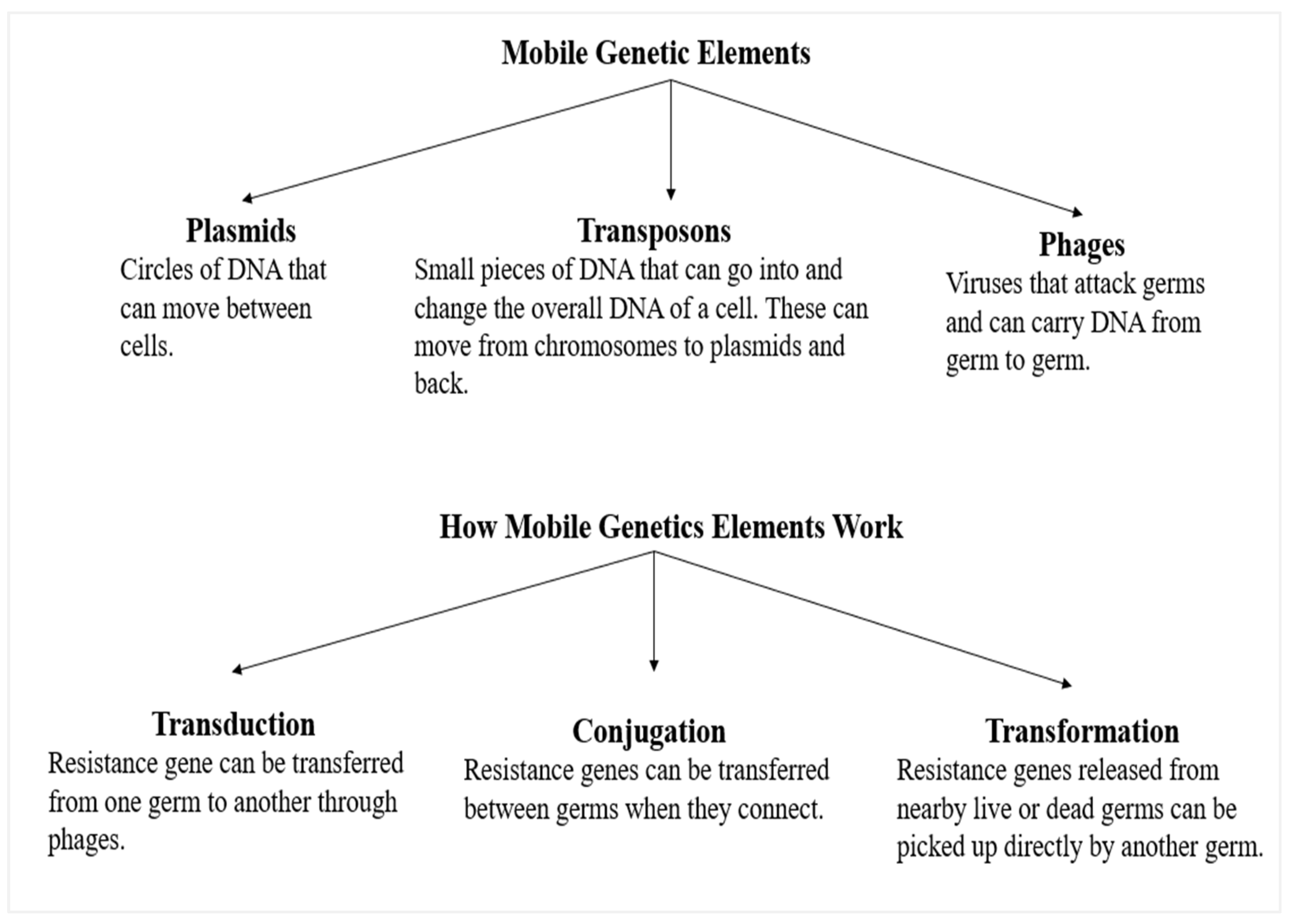
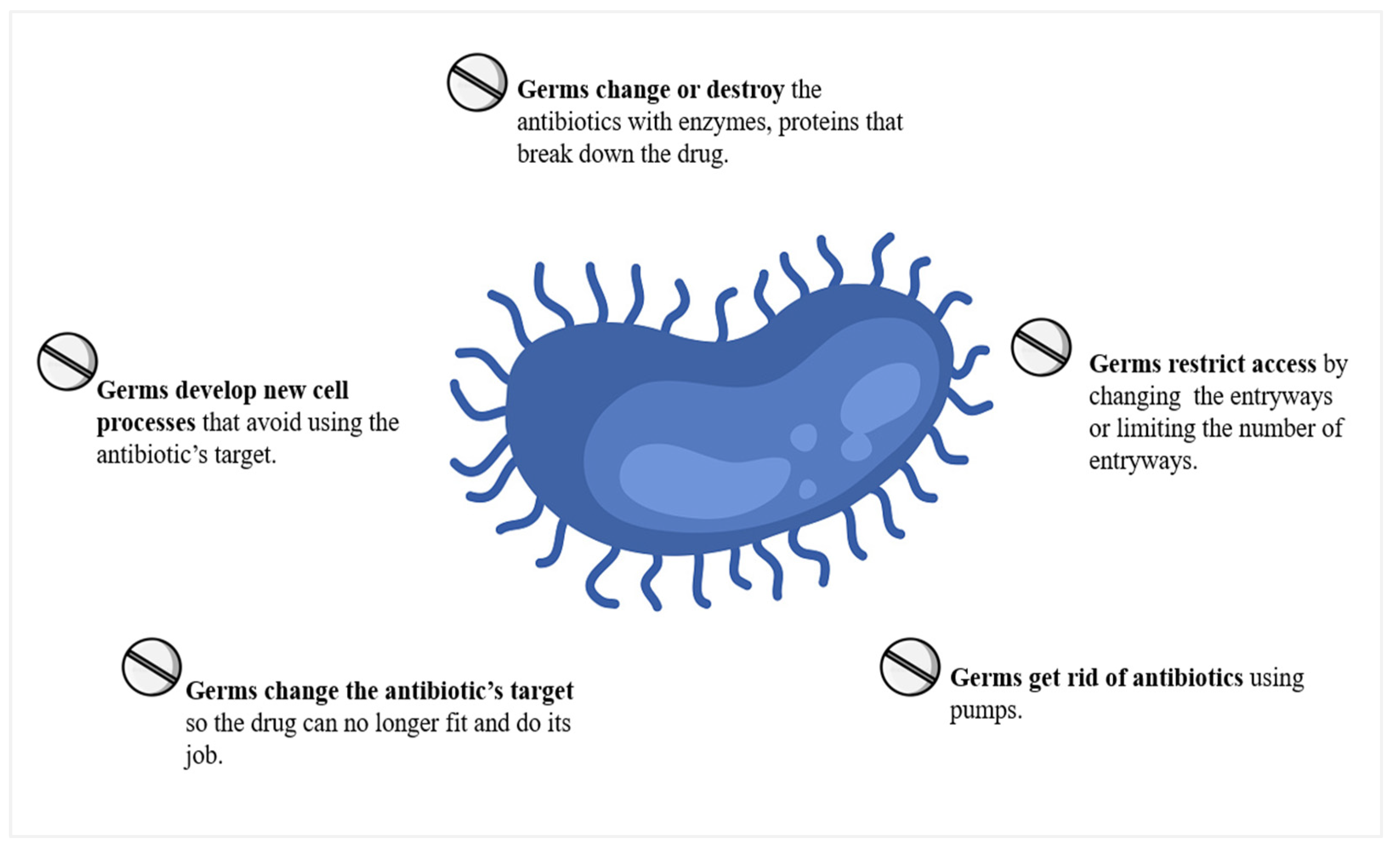
5. Antibiotic Resistance and Its Effects
6. Prevalence of Antimicrobial Resistance in S. pneumoniae
7. Action and Resistance Mechanisms for β-Lactams, Macrolides, and Fluoroquinolones among S. pneumoniae
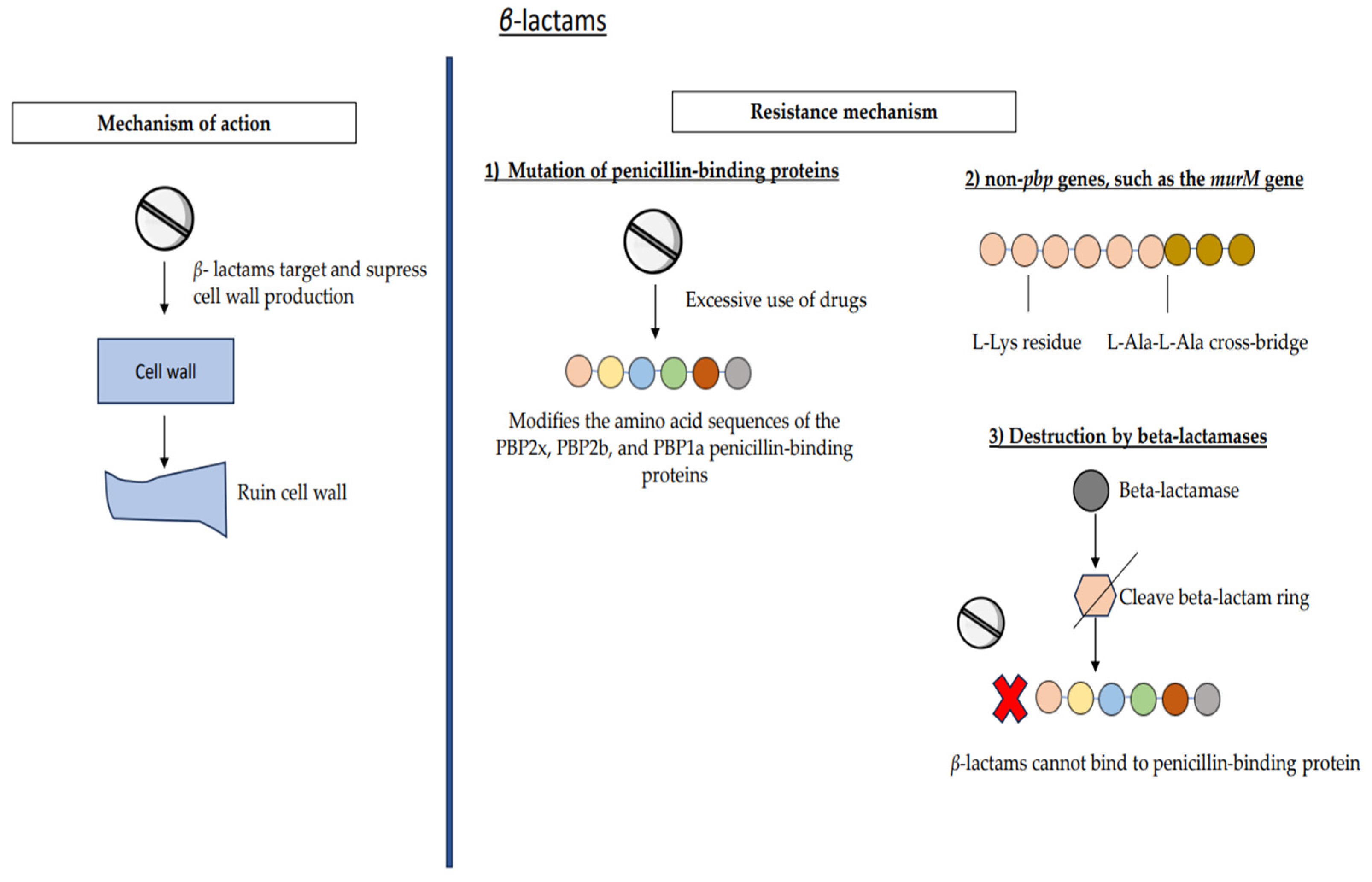
7.1. β-Lactams
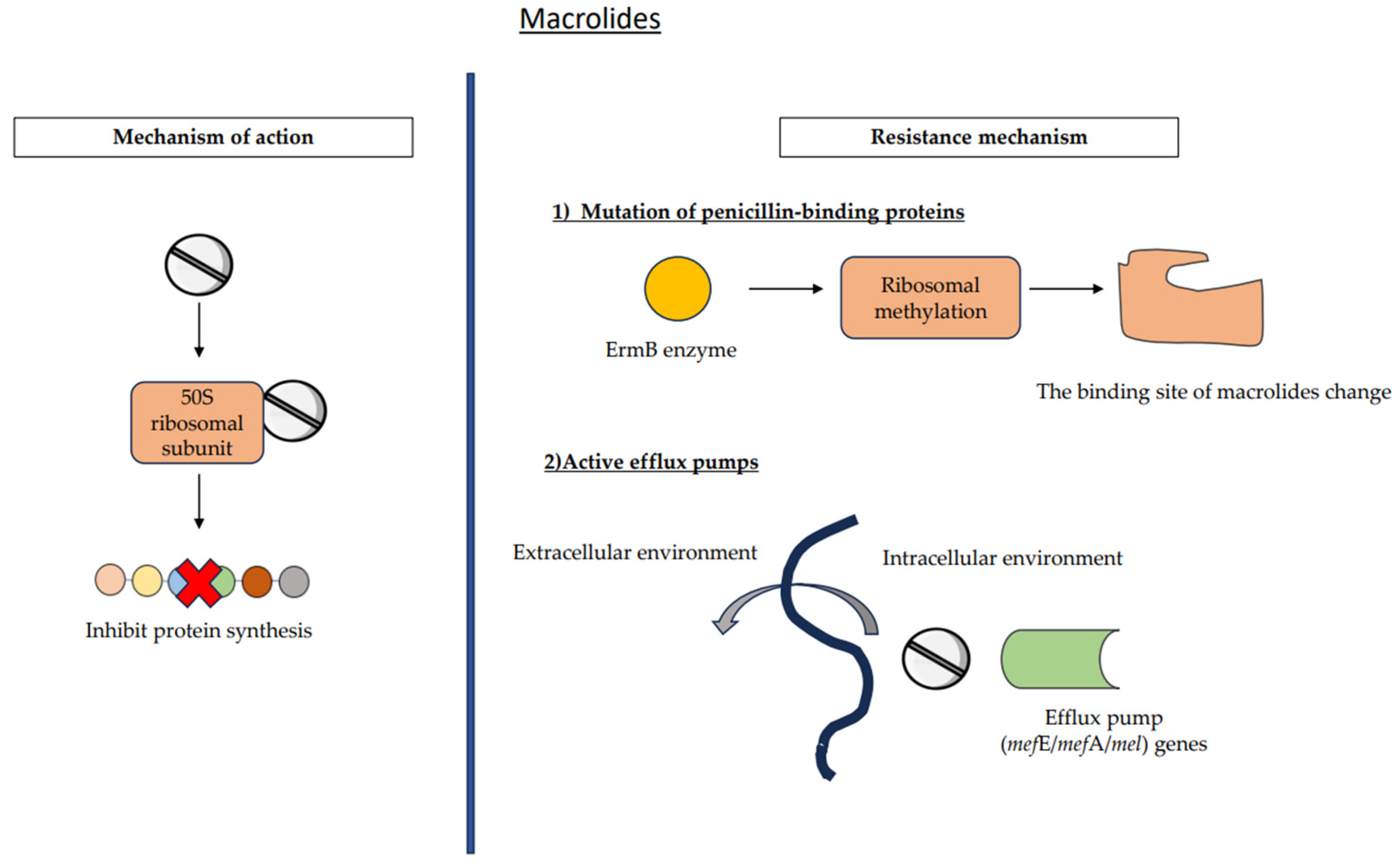
7.2. Macrolides
7.2.1. Ribosomal Alteration
7.2.2. Active Efflux Pumps
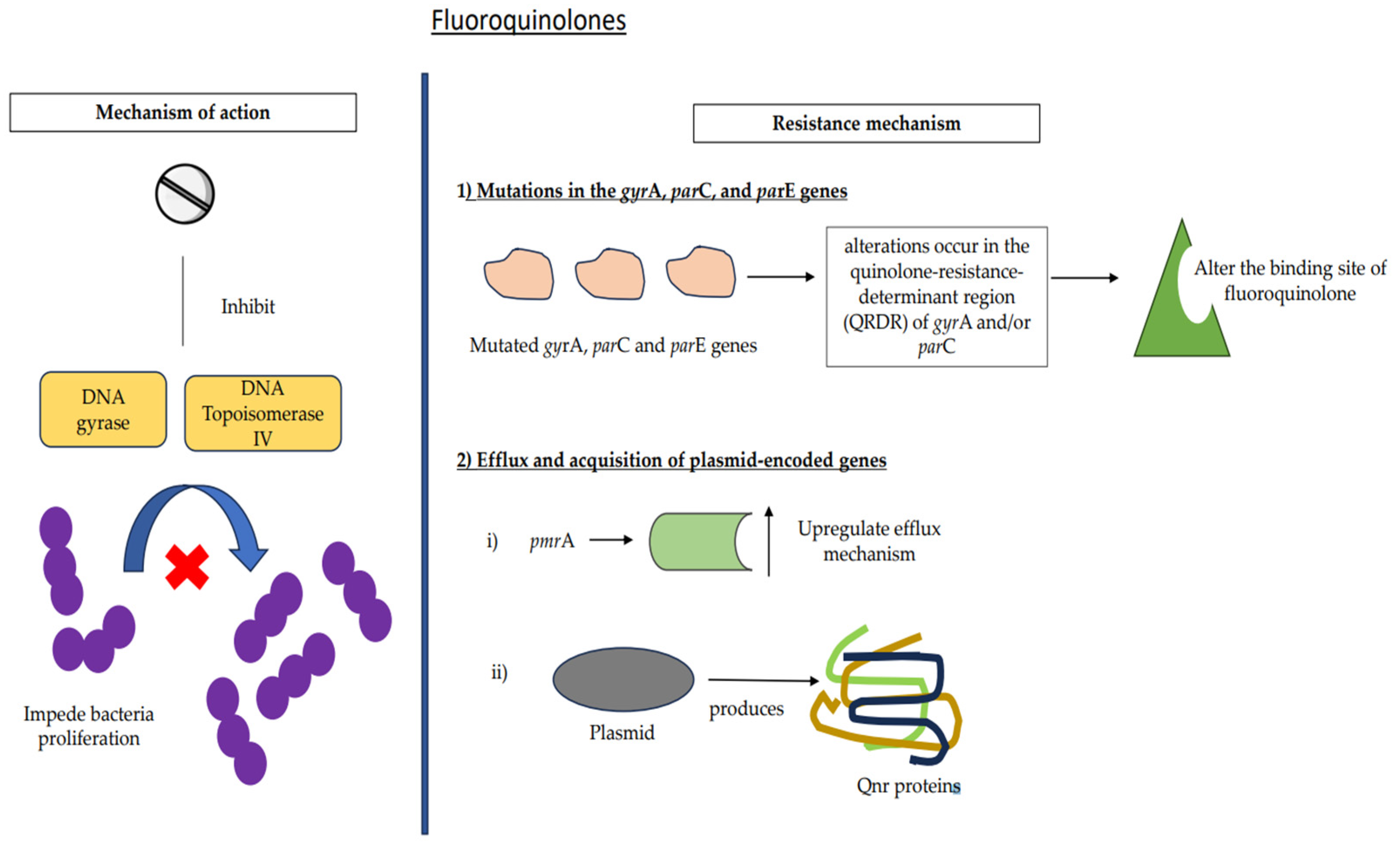
7.3. Fluoroquinolones
7.3.1. Mutations
7.3.2. Efflux and Acquisition of Plasmid-Encoded Genes
8. Current Trends versus New Trends in Terms of Diagnostics
9. Role of Artificial Intelligence in Overcoming AMR
10. Strategies to Curb the Resistance Issue
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Pneumonia. Available online: https://www.who.int/news-room/fact-sheets/detail/pneumonia (accessed on 26 September 2022).
- Lanks, C.W.; Musani, A.I.; Hsia, D.W. Community-Acquired Pneumonia and Hospital-Acquired Pneumonia. Med. Clin. N. Am. 2019, 103, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-H.; Thamlikitkul, V.; Hsueh, P.-R. Clinical and Economic Burden of Community-Acquired Pneumonia amongst Adults in the Asia-Pacific Region. Int. J. Antimicrob. Agents 2011, 38, 108–117. [Google Scholar] [CrossRef]
- Peyrani, P.; Mandell, L.; Torres, A.; Tillotson, G.S. The Burden of Community-Acquired Bacterial Pneumonia in the Era of Antibiotic Resistance. Expert Rev. Respir. Med. 2019, 13, 139–152. [Google Scholar] [CrossRef]
- Weisfelt, M.; van de Beek, D.; Spanjaard, L.; Reitsma, J.B.; de Gans, J. Clinical Features, Complications, and Outcome in Adults with Pneumococcal Meningitis: A Prospective Case Series. Lancet Neurol. 2006, 5, 123–129. [Google Scholar] [CrossRef]
- van de Beek, D.; de Gans, J.; Tunkel, A.R.; Wijdicks, E.F.M. Community-Acquired Bacterial Meningitis in Adults. N. Engl. J. Med. 2006, 354, 44–53. [Google Scholar] [CrossRef]
- Weiser, J.N.; Ferreira, D.M.; Paton, J.C. Streptococcus Pneumoniae: Transmission, Colonization and Invasion. Nat. Rev. Microbiol. 2018, 16, 355–367. [Google Scholar] [CrossRef]
- Cillóniz, C.; Garcia-Vidal, C.; Ceccato, A.; Torres, A. Antimicrobial Resistance Among Streptococcus Pneumoniae. In Antimicrobial Resistance in the 21st Century; Springer International Publishing: Cham, Switzerland, 2018; pp. 13–38. [Google Scholar]
- Xu, L.; Fang, J.; Ou, D.; Xu, J.; Deng, X.; Chi, G.; Feng, H.; Wang, J. Therapeutic Potential of Kaempferol on Streptococcus Pneumoniae Infection. Microbes Infect. 2023, 25, 105058. [Google Scholar] [CrossRef]
- Cherazard, R.; Epstein, M.; Doan, T.-L.; Salim, T.; Bharti, S.; Smith, M.A. Antimicrobial Resistant Streptococcus Pneumoniae: Prevalence, Mechanisms, and Clinical Implications. Am. J. Ther. 2017, 24, e361–e369. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance 2014, Antimicrobial Resistance Global Surveillance Report; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global Antibiotic Consumption 2000 to 2010: An Analysis of National Pharmaceutical Sales Data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Chenoweth, C.; Lynch, J.P. Antimicrobial Resistance: Implications for Managing Respiratory Failure. Curr. Opin. Pulm. Med. 1997, 3, 159–169. [Google Scholar] [CrossRef]
- Doern, G.V.; Pfaller, M.A.; Kugler, K.; Freeman, J.; Jones, R.N. Prevalence of Antimicrobial Resistance Among Respiratory Tract Isolates of Streptococcus Pneumoniae in North America: 1997 Results from the SENTRY Antimicrobial Surveillance Program. Clin. Infect. Dis. 1998, 27, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N.; Low, D.E.; Pfaller, M.A. Epidemiologic Trends in Nosocomial and Community-Acquired Infections Due to Antibiotic-Resistant Gram-Positive Bacteria: The Role of Streptogramins and Other Newer Compounds. Diagn. Microbiol. Infect. Dis. 1999, 33, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Chenoweth, C.E.; Saint, S.; Martinez, F.; Lynch, J.P.; Fendrick, A.M. Antimicrobial Resistance in Streptococcus Pneumoniae: Implications for Patients With Community-Acquired Pneumonia. Mayo Clin. Proc. 2000, 75, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Liñares, J.; Ardanuy, C.; Pallares, R.; Fenoll, A. Changes in Antimicrobial Resistance, Serotypes and Genotypes in Streptococcus Pneumoniae over a 30-Year Period. Clin. Microbiol. Infect. 2010, 16, 402–410. [Google Scholar] [CrossRef]
- Lynch, J.; Zhanel, G. Streptococcus Pneumoniae: Does Antimicrobial Resistance Matter? Semin. Respir. Crit. Care Med. 2009, 30, 210–238. [Google Scholar] [CrossRef]
- Blondeau, J.M. A Review of the Comparative In-Vitro Activities of 12 Antimicrobial Agents, with a Focus on Five New ‘Respiratory Quinolones’. J. Antimicrob. Chemother. 1999, 43, 1–11. [Google Scholar] [CrossRef]
- Wunderink, R.G.; Waterer, G.W. Community-Acquired Pneumonia. N. Engl. J. Med. 2014, 370, 543–551. [Google Scholar] [CrossRef]
- Torres, A.; Peetermans, W.E.; Viegi, G.; Blasi, F. Risk Factors for Community-Acquired Pneumonia in Adults in Europe: A Literature Review. Thorax 2013, 68, 1057–1065. [Google Scholar] [CrossRef]
- Remington, L.T.; Sligl, W.I. Community-Acquired Pneumonia. Curr. Opin. Pulm. Med. 2014, 20, 215–224. [Google Scholar] [CrossRef]
- Mandell, L.A. Community-Acquired Pneumonia: An Overview. Postgrad. Med. 2015, 127, 607–615. [Google Scholar] [CrossRef]
- Reynolds, H.Y. Pulmonary Host Defenses: State of the Art. Chest 1989, 95, 223S–230S. [Google Scholar] [CrossRef]
- Subramanian, K.; Henriques-Normark, B.; Normark, S. Emerging Concepts in the Pathogenesis of the Streptococcus Pneumoniae: From Nasopharyngeal Colonizer to Intracellular Pathogen. Cell. Microbiol. 2019, 21, 13077. [Google Scholar] [CrossRef]
- Roche, A.M.; Richard, A.L.; Rahkola, J.T.; Janoff, E.N.; Weiser, J.N. Antibody Blocks Acquisition of Bacterial Colonization through Agglutination. Mucosal Immunol. 2015, 8, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Weaver, C. Janeway’s Immunobiology-9th Edition; Garland Science: New York, NY, USA, 2017; ISBN 9780815345053. [Google Scholar]
- Chaudhuri, N.; Whyte, M.K.B.; Sabroe, I. Reducing the Toll of Inflammatory Lung Disease. Chest 2007, 131, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhao, L. The Antibacterial Activity of Fluoroquinolone Derivatives: An Update (2018–2021). Eur. J. Med. Chem. 2021, 224, 113741. [Google Scholar] [CrossRef]
- Abushaheen, M.A.; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; Vellappally, S.; et al. Antimicrobial Resistance, Mechanisms and Its Clinical Significance. Disease-a-Month 2020, 66, 100971. [Google Scholar] [CrossRef] [PubMed]
- Brar, R.K.; Jyoti, U.; Patil, R.K.; Patil, H.C. Fluoroquinolone Antibiotics: An Overview. Adesh Univ. J. Med. Sci. Res. 2020, 2, 26. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Alhumaid, S.; Al Mutair, A.; Garout, M.; Abulhamayel, Y.; Halwani, M.A.; Alestad, J.H.; Al Bshabshe, A.; Sulaiman, T.; AlFonaisan, M.K.; et al. Application of Artificial Intelligence in Combating High Antimicrobial Resistance Rates. Antibiotics 2022, 11, 784. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Eljaaly, K.; Alhumaid, S.; Albayat, H.; Al-Adsani, W.; Sabour, A.A.; Alshiekheid, M.A.; Al-Jishi, J.M.; Khamis, F.; Alwarthan, S.; et al. An Overview on Phenotypic and Genotypic Characterisation of Carbapenem-Resistant Enterobacterales. Medicina 2022, 58, 1675. [Google Scholar] [CrossRef]
- Burki, T.K. Superbugs: An Arms Race Against Bacteria. Lancet Respir. Med. 2018, 6, 668. [Google Scholar] [CrossRef]
- Vázquez-Laslop, N.; Mankin, A.S. How Macrolide Antibiotics Work. Trends Biochem. Sci. 2018, 43, 668–684. [Google Scholar] [CrossRef] [PubMed]
- Ojkic, N.; Lilja, E.; Direito, S.; Dawson, A.; Allen, R.J.; Waclaw, B. A Roadblock-and-Kill Mechanism of Action Model for the DNA-Targeting Antibiotic Ciprofloxacin. Antimicrob. Agents Chemother. 2020, 64, e02487-19. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, J.; Yu, Z.; Li, M.; Zhang, W.; Sun, H. Epidemiological Characteristics and Antibiotic Resistance Mechanisms of Streptococcus Pneumoniae: An Updated Review. Microbiol. Res. 2023, 266, 127221. [Google Scholar] [CrossRef]
- Feldman, C.; Anderson, R. Recent Advances in the Epidemiology and Prevention of Streptococcus Pneumoniae Infections. F1000Research 2020, 9, 338. [Google Scholar] [CrossRef] [PubMed]
- Larsson, M.; Nguyen, H.Q.; Olson, L.; Tran, T.K.; Nguyen, T.V.; Nguyen, C.T.K. Multi-drug Resistance in Streptococcus Pneumoniae among Children in Rural Vietnam More than Doubled from 1999 to 2014. Acta Paediatr. 2021, 110, 1916–1923. [Google Scholar] [CrossRef] [PubMed]
- Aliberti, S.; Cook, G.S.; Babu, B.L.; Reyes, L.F.; Rodriguez, A.H.; Sanz, F.; Soni, N.J.; Anzueto, A.; Faverio, P.; Sadud, R.F.; et al. International Prevalence and Risk Factors Evaluation for Drug-Resistant Streptococcus Pneumoniae Pneumonia. J. Infect. 2019, 79, 300–311. [Google Scholar] [CrossRef]
- Appelbaum, P.C. Resistance among Streptococcus Pneumoniae: Implications for Drug Selection. Clin. Infect. Dis. 2002, 34, 1613–1620. [Google Scholar] [CrossRef]
- Jaber, Z.K.; Al-Deresawi, M.S.; Matrood, A.A. Mutation in Ompk35 Are Evolution Pathway in Antibiotic Resistance. HIV Nurs. 2022, 23, 830–835. [Google Scholar]
- Willcox, D.; Chappell, B.G.N.; Hogg, K.F.; Calleja, J.; Smalley, A.P.; Gaunt, M.J. A General Catalytic β-C–H Carbonylation of Aliphatic Amines to β-Lactams. Science 2016, 354, 851–857. [Google Scholar] [CrossRef]
- Karampela, I.; Dalamaga, M. Could Respiratory Fluoroquinolones, Levofloxacin and Moxifloxacin, Prove to Be Beneficial as an Adjunct Treatment in COVID-19? Arch. Med. Res. 2020, 51, 741–742. [Google Scholar] [CrossRef]
- Mathy, V.; Grohs, P.; Compain, F. In Vitro Activity of β-Lactams in Combination with Avibactam against Multidrug-Resistant Pseudomonas Aeruginosa, Stenotrophomonas Maltophilia and Achromobacter Xylosoxidans Isolates from Patients with Cystic Fibrosis. J. Med. Microbiol. 2018, 67, 1217–1220. [Google Scholar] [CrossRef] [PubMed]
- Massip, C.; Mathieu, C.; Gaudru, C.; Miaut, V.; Floch, P.; Oswald, E.; Segonds, C.; Guet-Revillet, H. In Vitro Activity of Seven β-Lactams Including Ceftolozane/Tazobactam and Ceftazidime/Avibactam against Burkholderia Cepacia Complex, Burkholderia Gladioli and Other Non-Fermentative Gram-Negative Bacilli Isolated from Cystic Fibrosis Patients. J. Antimicrob. Chemother. 2019, 74, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Hansman, D.; Bullen, M.M. A Resistant Pneumococcus. Lancet 1967, 2, 264–265. [Google Scholar] [CrossRef]
- Weinstein, M.P.; Klugman, K.P.; Jones, R.N. Rationale for Revised Penicillin Susceptibility Breakpoints versus Streptococcus Pneumoniae: Coping with Antimicrobial Susceptibility in an Era of Resistance. Clin. Infect. Dis. 2009, 48, 1596–1600. [Google Scholar] [CrossRef] [PubMed]
- Chi, F.; Nolte, O.; Bergmann, C.; Ip, M.; Hakenbeck, R. Crossing the Barrier: Evolution and Spread of a Major Class of Mosaic Pbp2x in Streptococcus Pneumoniae, S. Mitis and S. Oralis. Int. J. Med. Microbiol. 2007, 297, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Hakenbeck, R.; Brückner, R.; Denapaite, D.; Maurer, P. Molecular Mechanisms of β-Lactam Resistance in Streptococcus Pneumoniae. Future Microbiol. 2012, 7, 395–410. [Google Scholar] [CrossRef]
- Jensen, A.; Valdórsson, O.; Frimodt-Møller, N.; Hollingshead, S.; Kilian, M. Commensal Streptococci Serve as a Reservoir for β-Lactam Resistance Genes in Streptococcus Pneumoniae. Antimicrob. Agents Chemother. 2015, 59, 3529–3540. [Google Scholar] [CrossRef]
- Reichmann, P.; König, A.; Liñares, J.; Alcaide, F.; Tenover, F.C.; McDougal, L.; Swidsinski, S.; Hakenbeck, R. A Global Gene Pool for High-Level Cephalosporin Resistance in Commensal Streptococcus Species and Streptococcus Pneumoniae. J. Infect. Dis. 1997, 176, 1001–1012. [Google Scholar] [CrossRef]
- Filipe, S.R.; Severina, E.; Tomasz, A. Distribution of the Mosaic Structured MurM Genes among Natural Populations of Streptococcus Pneumoniae. J. Bacteriol. 2000, 182, 6798–6805. [Google Scholar] [CrossRef]
- Filipe, S.R.; Severina, E.; Tomasz, A. The Role of MurMN Operon in Penicillin Resistance and Antibiotic Tolerance of Streptococcus Pneumoniae. Microb. Drug Resist. 2001, 7, 303–316. [Google Scholar] [CrossRef]
- Rodríguez-Villodres, Á.; Gil-Marqués, M.L.; Álvarez-Marín, R.; Bonnin, R.A.; Pachón-Ibáñez, M.E.; Aguilar-Guisado, M.; Naas, T.; Aznar, J.; Pachón, J.; Lepe, J.A.; et al. Extended-Spectrum Resistance to β-Lactams/β-Lactamase Inhibitors (ESRI) Evolved from Low-Level Resistant Escherichia Coli. J. Antimicrob. Chemother. 2020, 75, 77–85. [Google Scholar] [CrossRef]
- Bergman, M.; Huikko, S.; Huovinen, P.; Paakkari, P.; Seppälä, H. Macrolide and Azithromycin Use Are Linked to Increased Macrolide Resistance in Streptococcus Pneumoniae. Antimicrob. Agents Chemother. 2006, 50, 3646–3650. [Google Scholar] [CrossRef] [PubMed]
- Malhotra-Kumar, S.; Lammens, C.; Coenen, S.; Van Herck, K.; Goossens, H. Effect of Azithromycin and Clarithromycin Therapy on Pharyngeal Carriage of Macrolide-Resistant Streptococci in Healthy Volunteers: A Randomised, Double-Blind, Placebo-Controlled Study. Lancet 2007, 369, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.; Chalmers, J.D. The Immunomodulatory Effects of Macrolide Antibiotics in Respiratory Disease. Pulm. Pharmacol. Ther. 2021, 71, 102095. [Google Scholar] [CrossRef] [PubMed]
- Gaynor, M.; Mankin, A. Macrolide Antibiotics: Binding Site, Mechanism of Action, Resistance. Curr. Top. Med. Chem. 2003, 3, 949–960. [Google Scholar] [CrossRef]
- Patel, P.H.; Hashmi, M.F. Macrolides; StatPearls Publishing: St. Petersburg, FL, USA, 2022. [Google Scholar]
- Heidary, M.; Ebrahimi Samangani, A.; Kargari, A.; Kiani Nejad, A.; Yashmi, I.; Motahar, M.; Taki, E.; Khoshnood, S. Mechanism of Action, Resistance, Synergism, and Clinical Implications of Azithromycin. J. Clin. Lab. Anal. 2022, 36, e24427. [Google Scholar] [CrossRef]
- Schroeder, M.R.; Stephens, D.S. Macrolide Resistance in Streptococcus Pneumoniae. Front. Cell. Infect. Microbiol. 2016, 6, 98. [Google Scholar] [CrossRef]
- Shortridge, V.D.; Doern, G.V.; Brueggemann, A.B.; Beyer, J.M.; Flamm, R.K. Prevalence of Macrolide Resistance Mechanisms in Streptococcus Pneumoniae Isolates from a Multicenter Antibiotic Resistance Surveillance Study Conducted in the United States in 1994–1995. Clin. Infect. Dis. 1999, 29, 1186–1188. [Google Scholar] [CrossRef]
- Chancey, S.T.; Zhou, X.; Zähner, D.; Stephens, D.S. Induction of Efflux-Mediated Macrolide Resistance in Streptococcus Pneumoniae. Antimicrob. Agents Chemother. 2011, 55, 3413–3422. [Google Scholar] [CrossRef]
- He, W.; Jiang, K.; Qiu, H.; Liao, L.; Wang, S. 16-Membered Ring Macrolides and Erythromycin Induce ErmB Expression by Different Mechanisms. BMC Microbiol. 2022, 22, 152. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, K.; Du, X.; Lu, Y.; Liao, L.; He, Z.; He, W. Translational Attenuation Mechanism of ErmB Induction by Erythromycin Is Dependent on Two Leader Peptides. Front. Microbiol. 2021, 12, 690744. [Google Scholar] [CrossRef]
- Arenz, S.; Ramu, H.; Gupta, P.; Berninghausen, O.; Beckmann, R.; Vázquez-Laslop, N.; Mankin, A.S.; Wilson, D.N. Molecular Basis for Erythromycin-Dependent Ribosome Stalling during Translation of the ErmBL Leader Peptide. Nat. Commun. 2014, 5, 3501. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.-H.; Kwon, A.-R.; Yoon, E.-J.; Shim, M.-J.; Choi, E.-C. Translational Attenuation and MRNA Stabilization as Mechanisms of Erm (B) Induction by Erythromycin. Antimicrob. Agents Chemother. 2008, 52, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Liu, B.; Klepacki, D.; Gupta, V.; Schulten, K.; Mankin, A.S.; Vázquez-Laslop, N. Nascent Peptide Assists the Ribosome in Recognizing Chemically Distinct Small Molecules. Nat. Chem. Biol. 2016, 12, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gupta, V.; Pathania, R. Efflux Pump Inhibitors for Bacterial Pathogens: From Bench to Bedside. Indian J. Med. Res. 2019, 149, 129. [Google Scholar] [CrossRef]
- Chancey, S.T.; Zähner, D.; Stephens, D.S. Acquired Inducible Antimicrobial Resistance in Gram-Positive Bacteria. Future Microbiol. 2012, 7, 959–978. [Google Scholar] [CrossRef]
- Chancey, S.T.; Bai, X.; Kumar, N.; Drabek, E.F.; Daugherty, S.C.; Colon, T.; Ott, S.; Sengamalay, N.; Sadzewicz, L.; Tallon, L.J.; et al. Transcriptional Attenuation Controls Macrolide Inducible Efflux and Resistance in Streptococcus Pneumoniae and in Other Gram-Positive Bacteria Containing Mef/Mel(Msr(D)) Elements. PLoS ONE 2015, 10, e0116254. [Google Scholar] [CrossRef]
- Ambrose, K.D.; Nisbet, R.; Stephens, D.S. Macrolide Efflux in Streptococcus Pneumoniae Is Mediated by a Dual Efflux Pump (Mel and Mef) and Is Erythromycin Inducible. Antimicrob. Agents Chemother. 2005, 49, 4203–4209. [Google Scholar] [CrossRef]
- Zähner, D.; Zhou, X.; Chancey, S.T.; Pohl, J.; Shafer, W.M.; Stephens, D.S. Human Antimicrobial Peptide LL-37 Induces MefE/Mel-Mediated Macrolide Resistance in Streptococcus Pneumoniae. Antimicrob. Agents Chemother. 2010, 54, 3516–3519. [Google Scholar] [CrossRef]
- Ezelarab, H.A.A.; Abbas, S.H.; Hassan, H.A.; Abuo-Rahma, G.E.-D.A. Recent Updates of Fluoroquinolones as Antibacterial Agents. Arch. Pharm. 2018, 351, 1800141. [Google Scholar] [CrossRef]
- Gupta, V.; Datta, P. Next-Generation Strategy for Treating Drug Resistant Bacteria: Antibiotic Hybrids. Indian J. Med. Res. 2019, 149, 97. [Google Scholar] [CrossRef] [PubMed]
- Khondker, A.; Bider, R.-C.; Passos-Gastaldo, I.; Wright, G.D.; Rheinstädter, M.C. Membrane Interactions of Non-Membrane Targeting Antibiotics: The Case of Aminoglycosides, Macrolides, and Fluoroquinolones. Biochim. Biophys. Acta-Biomembr. 2021, 1863, 183448. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.C.; Goyal, R.; Sharma, A.; Sharma, D.; Saini, N.; Rajak, H.; Sharma, S.; Thakur, V.K. Insights on Fluoroquinolones in Cancer Therapy: Chemistry and Recent Developments. Mater. Today Chem. 2020, 17, 100296. [Google Scholar] [CrossRef]
- Ferreira, M.; Gameiro, P. Fluoroquinolone-Transition Metal Complexes: A Strategy to Overcome Bacterial Resistance. Microorganisms 2021, 9, 1506. [Google Scholar] [CrossRef] [PubMed]
- Blondeau, J.M. Fluoroquinolones: Mechanism of Action, Classification, and Development of Resistance. Surv. Ophthalmol. 2004, 49, S73–S78. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhao, S.-J.; Lv, Z.-S.; Gao, F.; Wang, Y.; Zhang, F.; Bai, L.; Deng, J.-L. Fluoroquinolone-Isatin Hybrids and Their Biological Activities. Eur. J. Med. Chem. 2019, 162, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-H.; Kim, E.Y.; Kim, Y.-J. Systemic Use of Fluoroquinolone in Children. Korean J. Pediatr. 2013, 56, 196. [Google Scholar] [CrossRef]
- Bradley, J.S.; Jackson, M.A. The Use of Systemic and Topical Fluoroquinolones. Pediatrics 2011, 128, e1034–e1045. [Google Scholar] [CrossRef]
- Jones, R.N.; Jacobs, M.R.; Sader, H.S. Evolving Trends in Streptococcus Pneumoniae Resistance: Implications for Therapy of Community-Acquired Bacterial Pneumonia. Int. J. Antimicrob. Agents 2010, 36, 197–204. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Topoisomerase Inhibitors: Fluoroquinolone Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025320. [Google Scholar] [CrossRef]
- Cornick, J.E.; Bentley, S.D. Streptococcus Pneumoniae: The Evolution of Antimicrobial Resistance to Beta-Lactams, Fluoroquinolones and Macrolides. Microbes Infect. 2012, 14, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.J. Dual Activity of Fluoroquinolones against Streptococcus Pneumoniae: The Facts behind the Claims. J. Antimicrob. Chemother. 2002, 49, 893–895. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C. Emerging Mechanisms of Fluoroquinolone Resistance. Available online: https://www.medscape.com/viewarticle/414418_3 (accessed on 29 November 2022).
- Anahtar, M.N.; Yang, J.H.; Kanjilal, S. Applications of Machine Learning to the Problem of Antimicrobial Resistance: An Emerging Model for Translational Research. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Leanse, L.G.; Feng, Y. Artificial Intelligence and Machine Learning Assisted Drug Delivery for Effective Treatment of Infectious Diseases. Adv. Drug Deliv. Rev. 2021, 178, 113922. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Deng, S.; Zhang, L. A Review of Artificial Intelligence Applications for Antimicrobial Resistance. Biosaf. Health 2021, 3, 22–31. [Google Scholar] [CrossRef]
- Lau, H.J.; Lim, C.H.; Foo, S.C.; Tan, H.S. The Role of Artificial Intelligence in the Battle against Antimicrobial-Resistant Bacteria. Curr. Genet. 2021, 67, 421–429. [Google Scholar] [CrossRef]
- Sharma, P.; Sethi, G.; Tripathi, M.K.; Rana, S.; Singh, H.; Kaur, P. Role of Nature-Inspired Intelligence in Genomic Diagnosis of Antimicrobial Resistance. Stud. Comput. Intell. 2023, 1066, 223–245. [Google Scholar] [CrossRef]
- Iskandar, K.; Molinier, L.; Hallit, S.; Sartelli, M.; Hardcastle, T.C.; Haque, M.; Lugova, H.; Dhingra, S.; Sharma, P.; Islam, S.; et al. Surveillance of Antimicrobial Resistance in Low- and Middle-Income Countries: A Scattered Picture. Antimicrob. Resist. Infect. Control 2021, 10, 63. [Google Scholar] [CrossRef]
- Brandileone, M.-C.C.; Almeida, S.C.G.; Bokermann, S.; Minamisava, R.; Berezin, E.N.; Harrison, L.H.; Andrade, A.-L. Dynamics of Antimicrobial Resistance of Streptococcus Pneumoniae Following PCV10 Introduction in Brazil: Nationwide Surveillance from 2007 to 2019. Vaccine 2021, 39, 3207–3215. [Google Scholar] [CrossRef]
- de Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year Due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- Kuk, A.C.Y.; Hao, A.; Guan, Z.; Lee, S.-Y. Visualizing Conformation Transitions of the Lipid II Flippase MurJ. Nat. Commun. 2019, 10, 1736. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mollo, A.; Kahne, D.; Ruiz, N. The Bacterial Cell Wall: From Lipid II Flipping to Polymerization. Chem. Rev. 2022, 122, 8884–8910. [Google Scholar] [CrossRef] [PubMed]
- Sham, L.-T.; Butler, E.K.; Lebar, M.D.; Kahne, D.; Bernhardt, T.G.; Ruiz, N. MurJ Is the Flippase of Lipid-Linked Precursors for Peptidoglycan Biogenesis. Science 2014, 345, 220–222. [Google Scholar] [CrossRef] [PubMed]

| Antimicrobial Classes | Resistance Mechanism | Resistance Prevalence | |
|---|---|---|---|
| Beta-lactams | (1) Mutation of penicillin-binding proteins (2) Non-pbp genes, murM gene (3) Destruction by beta-lactamases | Penicillin | Penicillin G: 13.8% |
| Penicillin V: 41.8% | |||
| Cephalosporins: <1–29.9% | |||
| Cefuroxime: 29.9% | |||
| Ceftriaxone: 11.7% | |||
| Ceftraroline: 0–<1% | |||
| Macrolides | (1) Ribosomal alteration (2) Active efflux pumps | 20–40% | |
| Fluoroquinolones | (1) Mutations in gyrA, parC, and parE regions (2) Efflux pumps (3) Acquisition of plasmid-encoded genes | <1–2% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahari, N.I.N.; Engku Abd Rahman, E.N.S.; Irekeola, A.A.; Ahmed, N.; Rabaan, A.A.; Alotaibi, J.; Alqahtani, S.A.; Halawi, M.Y.; Alamri, I.A.; Almogbel, M.S.; et al. A Review of the Resistance Mechanisms for β-Lactams, Macrolides and Fluoroquinolones among Streptococcus pneumoniae. Medicina 2023, 59, 1927. https://doi.org/10.3390/medicina59111927
Zahari NIN, Engku Abd Rahman ENS, Irekeola AA, Ahmed N, Rabaan AA, Alotaibi J, Alqahtani SA, Halawi MY, Alamri IA, Almogbel MS, et al. A Review of the Resistance Mechanisms for β-Lactams, Macrolides and Fluoroquinolones among Streptococcus pneumoniae. Medicina. 2023; 59(11):1927. https://doi.org/10.3390/medicina59111927
Chicago/Turabian StyleZahari, Nurul Izzaty Najwa, Engku Nur Syafirah Engku Abd Rahman, Ahmad Adebayo Irekeola, Naveed Ahmed, Ali A. Rabaan, Jawaher Alotaibi, Shayea A. Alqahtani, Mohammed Y. Halawi, Ibrahim Ateeq Alamri, Mohammed S. Almogbel, and et al. 2023. "A Review of the Resistance Mechanisms for β-Lactams, Macrolides and Fluoroquinolones among Streptococcus pneumoniae" Medicina 59, no. 11: 1927. https://doi.org/10.3390/medicina59111927










