Composition and Quantitation of Microalgal Lipids by ERETIC 1H NMR Method
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sample Preparation

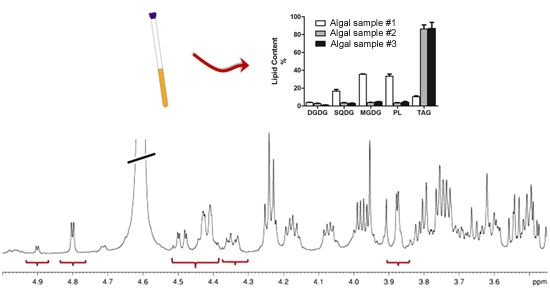
2.2. NMR Methodology
| S | Chemical Shift (ppm) | n | Chemical Assignment | Lipid Class |
|---|---|---|---|---|
| 1 | 4.34 | 2 | methylene protons of glycerol | TAG |
| 2 | 4.53–4.38 | 1 | methylene protons of glycerol | PL + GL |
| 3 | 3.88 | 1 | methine proton at C4 of galactose | MGDG |
| 4 | 4.90 | 1 | anomeric proton of galactose | DGDG |
| 5 | 4.80 | 1 | anomeric proton of sulfoquinovoside | SQDG |
| 6 | 2.35 a | 2 | methylene protons α to carboxy group | TFA |
| 7 | 2.06 b | 4 | allylic protons | UFA |

| Integrated Signal | TW | NS | CYC | ||||
|---|---|---|---|---|---|---|---|
| AS | C (µmol) | AS | C (µmol) | AS | C (µmol) | ||
| TAG | 1 | 0.198 | 23.0 | 27.43 | 140.0 | 52.04 | 530.0 |
| PL + GL | 2 | 1.055 | 192.4 | 6.913 | 22.2 | 3.139 | 79.8 |
| MGDG | 3 | 0.277 | 75.0 | 0.946 | 6.4 | 0.726 | 28.4 |
| DGDG | 4 | 0.033 | 40.0 | 1.087 | 4.6 | 0.757 | 18.0 |
| SQDG | 5 | 0.184 | 9.2 | 1.113 | 5.6 | 0.229 | 7.0 |
| TFA | 6 | 2.642 | 453.8 | 109.8 | 464.4 | 166.7 | 1749.6 |
| UFA | 7 | 5.083 | 436.5 | 149.5 | 316.3 | 176.8 | 927.7 |
2.3. Lipid NMR Analysis
| SQDG | DGDG | MGDG | PL | TAG | FFA | UFA | SFA | |
|---|---|---|---|---|---|---|---|---|
| TW | 9.2 ± 1.4 | 40.0 ± 9.0 | 75.0 ± 9.0 | 68.2 ± 16.8 | 23.0 ± 1.6 | 35.6 ± 15.0 | 436.5 ± 32.7 | 17.3 ± 4.9 |
| NS | 5.6 ± 0.8 | 4.6 ± 1.4 | 6.4 ± 0.8 | 5.6 ± 0.8 | 141.8 ± 22.0 | 55.4 ± 5.4 | 316.3 ± 44.0 | 148.1 ± 24.8 |
| CYC | 7.0 ± 3.4 | 18.0·± 5.6 | 28.4 ± 4.8 | 26.4 ± 11.0 | 530.0 ± 93.0 | 147.6 ± 29.4 | 927.7 ± 121.2 | 821.9 ± 130.2 |

3. Experimental Section
3.1. General
3.2. Algal Culturing
3.3. Lipid Extraction
3.4. NMR Analysis
3.5. Protocol for General Analysis of Fatty Acid-Based Lipids in Microalgae
- Lyophilize frozen sample.
- Cover dry sample with 5 mL methanol and kept at 4 °C for 1 min.
- Add 20 µg (24.4 × 10−3 µmol) internal standard [(4-chlorophenyl)-trihexadecylsilane] for each mg of dry sample.
- Add 10 mL chloroform.
- Homogenized and incubate with shaking for 5 min.
- Centrifuge (3750 rpm) for 5 min at room temperature.
- Transfer supernatant to a fresh tube.
- Suspend solid residue in 15 mL chloroform:methanol (2:1 v/v) and incubate with shaking for 2 min.
- Repeat steps 6 and 7.
- To combined supernatants from steps 7 and 9, add 7 mL deionized water.
- Vortex, centrifuge and discard the upper phase.
- Recover lower phase (organic extract) with a Pasteur pipette and transfer to a glass rotary evaporator flask.
- Remove solvent and dry sample under vacuum at room temperature.
- Dissolve dry extract in 700 µL CD3OD/CDCl3 (1:1 v/v).
- Transfer to a fresh NMR tube.
- Acquire 1H NMR spectrum (ns = 1, ds = 0, spectral with 14 ppm, O1P = 5).
- Integrate diagnostic signal (Figure 2) with the “Process—Integrate” function of the Bruker Top Spin software or corresponding programs of other manufacturers.
- Calculate concentration (µmol/L) of each lipid class by the ERETIC function of the same software.
- suspend algal pellet in 5 mL of 0.5 M ammonium formate solution.
- centrifuge the sample (3600 rpm) for 5 min at room temperature.
- remove the supernatant.
- repeat twice the above steps.
4. Conclusions
Conflict of Interest
References
- Radwan, S.S. Sources of C20-polyunsaturated fatty acids for biotechnological use. Appl. Microbiol. Biotechnol. 1991, 35, 421–430. [Google Scholar] [CrossRef]
- Alvarez, A.M.R.; Spolaore, P.; Joannis-Cassan, C.; Duran, E.; Isambert, A. Commercial applications of microalgae. J. Biosci. Bioeng. 2006, 101, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Atalah, E.; Hernández Cruz, C.M.; Izquierdo, M.S.; Rosenlund, G.; Caballero, M.J.; Valencia, A.; Robaina, L. Two Microalgae Crypthecodinium cohnii and Phaeodactylum tricornutum as alternative source of essential fatty acids in starter feeds for seabream (Sparus aurata). Aquaculture 2007, 270, 178–185. [Google Scholar] [CrossRef]
- Kassis, N.M.; Beamer, S.K.; Matak, K.E.; Tou, J.C.; Jaczynski, J. Nutritional Composition of novel nutraceutical egg products developed with omega-3-rich oils. LWT-Food Sci. Technol. 2010, 43, 1204–1212. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef]
- Lam, M.K.; Lee, K.T. Microalgae biofuels: A critical review of issues, problems and the way forward. Biotechnol. Adv. 2012, 30, 673–690. [Google Scholar] [CrossRef]
- Mata, T.M.; Martins, A.A.; Caetano, N.S. Microalgae for biodiesel production and other applications: a review. Renew. Sust. Energ. Rev. 2010, 14, 217–232. [Google Scholar] [CrossRef]
- Wijffels, R.H.; Barbosa, M.J. An outlook on microalgal biofuels. Science 2010, 329, 796–799. [Google Scholar] [CrossRef]
- Fukuda, H.; Kondo, A.; Noda, H. Biodiesel fuel production by transesterification of oils. J. Biosci. Bioeng. 2001, 92, 405–416. [Google Scholar]
- Hernandez, E. Lipids, Pharmaceutical and Cosmetic Use. In Kirk-Othmer Ecyclopedia of Chemical Technology; Wiley: New York, NY, USA, 2012. [Google Scholar]
- Shifrin, N.S.; Chisholm, S.W. Phytoplankton lipids: interspecific differences and effects of nitrate sulfate and light-dark cycles. J. Phycol. 1981, 17, 374–384. [Google Scholar] [CrossRef]
- Stuart, A.S.; Matthew, P.D.; John, S.D.; Irmtraud, H.; Christopher, J.; Lea-Smith, D.J.; Alison, G.S. Biodiesel from algae: Challenges and prospects. Curr. Opin. Biotechnol. 2010, 21, 277–286. [Google Scholar]
- Han, Y.; Wen, Q.; Chen, Z.; Li, P. Review of Methods Used for Microalgal Lipid-Content Analysis. Energ. Procedia 2011, 12, 944–950. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Soham, C.; Sancharini, D.; Ramkrishna, S. Rapid and precise estimation of biodiesel by high performance thin layer chromatography. Appl. Energy 2011, 88, 5188–5192. [Google Scholar] [CrossRef]
- Wang, R.; Song, B.; Zhou, W.; Zhang, Y.; Hu, D.; Bhadury, P.S.; Yang, S. A facile and feasible method to evaluate and control the quality of Jatropha curcus L. seed oil for biodiesel feedstock: Gas chromatographic fingerprint. Appl. Energy 2011, 88, 2064–2070. [Google Scholar] [CrossRef]
- Franz, A.K.; Danielewicz, M.A.; Wong, D.M.; Anderson, L.A.; Boothe, J.R. Phenotypic Screening with Oleaginous Microalgae Reveals Modulators of Lipid Productivity. ACS Chem. Biol. 2013, 8, 1053–1062. [Google Scholar] [CrossRef]
- Jones, J.; Manning, S.; Montoya, M.; Keller, K.; Poenie, M. Extraction of Algal Lipids and Their Analysis by HPLC and Mass Spectrometry. J. Am. Oil Chem. Soc. 2012, 89, 1371–1381. [Google Scholar]
- Cooksey, K.E.; Guckert, J.B.; Williams, S.A.; Callis, P.R. Fluorimetric determination of the neutral lipid-content of microalgal cells using Nile red. J. Microbiol. Methods 1987, 6, 333–345. [Google Scholar] [CrossRef]
- Elsey, D.; Jameson, D.; Raleigh, B.; Cooney, M.J. Fluorescent measurement of microalgal neutral lipids. J. Microbiol. Methods 2007, 68, 639–642. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, C.; Song, L.; Sommerfeld, M.; Hu, Q. A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J. Microbiol. Methods 2009, 77, 41–47. [Google Scholar] [CrossRef]
- Knothe, G. Analyzing Biodiesel: Standards and Other Methods. J. Am. Oil Chem. Soc. 2006, 83, 823–833. [Google Scholar] [CrossRef]
- Zagonel, G.F.; Zamora, P.P.; Ramos, L.P. Multivariate monitoring of soybean oil ethanolysis by FTIR. Talanta 2004, 63, 1021–1025. [Google Scholar] [CrossRef]
- Neto, P.R.C.; Caro, M.S.B.; Mazzuco, L.M.; Nascimento, M.G. Quantification of soybean oil ethanolysis with 1H NMR. J. Am. Oil Chem. Soc. 2004, 81, 1111–1114. [Google Scholar] [CrossRef]
- Chongkhong, S.; Tongurai, C.; Chetpattananondh, P.; Bunyakan, C. Biodiesel production by esterification of palm fatty acid distillate. Biomass Bioenerg. 2007, 31, 563–568. [Google Scholar] [CrossRef]
- Ferreira Ghesti, G.; Lemos de Macedo, J.; Sabioni Resck, I.; Alves Dias, J.; Loureiro Dias, S.C. FT-Raman Spectroscopy Quantification of Biodiesel in a Progressive Soybean Oil Transesterification Reaction and Its Correlation with 1H NMR Spectroscopy Methods. Energ. Fuels 2007, 21, 2475–2480. [Google Scholar] [CrossRef]
- Casas, A.; Ramos, M.J.; Pérez, A.; Simon, A.; Lucas-Torres, C.; Moreno, A. Rapid quantitative determination by 13C NMR of the composition of acetylglycerol mixtures as byproduct in biodiesel synthesis. Fuel 2012, 92, 180–186. [Google Scholar] [CrossRef]
- Ciubota-Rosie, C.; Macoveanu, M.; Fernàndez, C.M.; Ramos, M.J.; Pérez, A.; Moreno, A. Sinapis alba seed as a prospective biodiesel source. Biomass Bioenerg. 2013, 53, 83–90. [Google Scholar]
- Brown, M.R.; Farmer, C.L. Riboflavin content of six species of microalgae used in mariculture. J. Appl. Phycol. 1994, 6, 61–65. [Google Scholar] [CrossRef]
- Wikfors, G.H.; Ferris, G.E.; Smith, B.C. The relationship between gross biochemical composition of cultured algal foods and growth of the hard clam, Mercenaria mercenaria (L.). Aquacolture 1992, 108, 135–154. [Google Scholar] [CrossRef]
- Zhu, C.J.; Lee, Y.K. Determination of biomass dry weight of marine microalgae. J. Appl. Phycol. 1997, 9, 189–194. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Akoka, S.; Barantin, L.; Trierweiler, M. Concentration Measurement by Proton NMR Using the ERETIC Method. Anal. Chem. 1999, 71, 2554–2557. [Google Scholar] [CrossRef]
- Annarao, S.; Sidhu, O.P.; Roy, R.; Tuli, R.; Khetrapal, C.L. Lipid profiling of developing Jatropha curcas L. seeds using 1H NMR spectroscopy. Bioresour. Technol. 2008, 99, 9032–9035. [Google Scholar] [CrossRef]
- Pahl, S.D.; Lewis, D.M.; Chen, F.; King, K.D. Growth dynamics and the proximate biochemical composition and fatty acid profile of the heterotrophically grown diatom Cyclotella cryptica. J. Appl. Phycol. 2010, 22, 165–171. [Google Scholar] [CrossRef] [Green Version]
- Prartono, T.; Kawaroe, M.; Katili, V. Fatty Acid Composition of Three Diatom Species Skeletonema costatum, Thalassiosira sp. and Chaetoceros gracilis. Int. J. Environ. Bioenerg. 2013, 6, 28–43. [Google Scholar]
- Mohammady, N.G. Characterization of the fatty acid composition of Nannochloropsis salina as a determinant of biodiesel properties. J. Biosci. 2011, 66, 328–332. [Google Scholar]
- Cutignano, A.; d’Ippolito, G.; Romano, G.; Lamari, N.; Cimino, G.; Febbraio, F.; Nucci, R.; Fontana, A. Chloroplastic glycolipids fuel aldehyde biosynthesis in the marine diatom Thalassiosira rotula. ChemBioChem 2006, 7, 450–456. [Google Scholar] [CrossRef]
- Manzo, E.; Ciavatta, M.L.; Pagano, D.; Fontana, A. Synthetic strategy for the preparation of bioactive galactoglycerolipids. Chem. J. Mold. 2011, 6, 27–29. [Google Scholar]
- Manzo, E.; Ciavatta, M.L.; Pagano, D.; Fontana, A. An efficient and versatile chemical synthesis of bioactive glycoglycerolipids. Tetrahedron Lett. 2012, 53, 879–881. [Google Scholar] [CrossRef]
- Manzo, E.; Tramice, A.; Pagano, D.; Trincone, A.; Fontana, A. Chemoenzymatic preparation of α-6-sulfoquinovosyl-1,2-O-diacylglycerols. Tetrahedron 2012, 68, 10169–10175. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Ryther, J.H. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Merchant, T.E.; Lass, J.H.; Meneses, P.; Greiner, J.V.; Glonek, T. Human crystalline lens phospholipid analysis with age. Invest. Ophthalmol. Vis. Sci. 1991, 32, 549–555. [Google Scholar]
- Huang, L.; Grami, V.; Marrero, Y.; Tang, D.; Yappert, M.C.; Rasi, V.; Borchman, D. Human Lens Phospholipid Changes with Age and Cataract. Invest. Ophthalmol. Vis. Sci. 2005, 46, 1682–1688. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nuzzo, G.; Gallo, C.; D'Ippolito, G.; Cutignano, A.; Sardo, A.; Fontana, A. Composition and Quantitation of Microalgal Lipids by ERETIC 1H NMR Method. Mar. Drugs 2013, 11, 3742-3753. https://doi.org/10.3390/md11103742
Nuzzo G, Gallo C, D'Ippolito G, Cutignano A, Sardo A, Fontana A. Composition and Quantitation of Microalgal Lipids by ERETIC 1H NMR Method. Marine Drugs. 2013; 11(10):3742-3753. https://doi.org/10.3390/md11103742
Chicago/Turabian StyleNuzzo, Genoveffa, Carmela Gallo, Giuliana D'Ippolito, Adele Cutignano, Angela Sardo, and Angelo Fontana. 2013. "Composition and Quantitation of Microalgal Lipids by ERETIC 1H NMR Method" Marine Drugs 11, no. 10: 3742-3753. https://doi.org/10.3390/md11103742
APA StyleNuzzo, G., Gallo, C., D'Ippolito, G., Cutignano, A., Sardo, A., & Fontana, A. (2013). Composition and Quantitation of Microalgal Lipids by ERETIC 1H NMR Method. Marine Drugs, 11(10), 3742-3753. https://doi.org/10.3390/md11103742