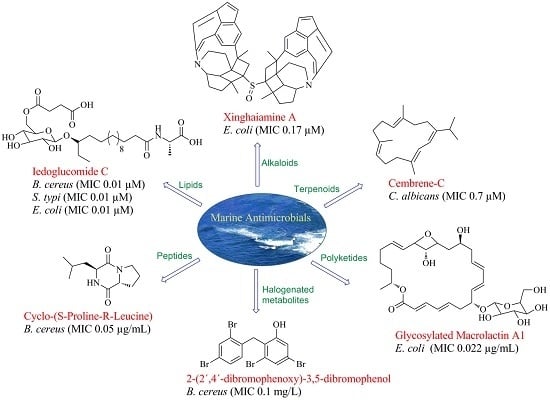
Current Status and Future Prospects of Marine Natural Products (MNPs) as Antimicrobials
Abstract
:1. Introduction
2. Marine Natural Products (MNPs)
3. Chemical Entities in the Preclinical Antimicrobial Pipeline
4. Alkaloids
5. Terpenoids
6. Lipids
7. Peptides
8. Halogenated Compounds
9. Polyketides
10. Isocoumarins
11. Nucleosides
12. Miscellaneous Compounds
13. Synthetic Interventions
14. Genome Mining of Marine Microorganisms—The Future of Antimicrobial Discovery
15. Concluding Remarks and Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- O’Neill, J. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014, 1–16. [Google Scholar] [CrossRef]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 2016, 7, 1728. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, R.; Ibrahim, S.A. Natural products as antimicrobial agents. Food Control 2014, 46, 412–429. [Google Scholar] [CrossRef]
- Fenical, W. New pharmaceuticals from marine organisms. Trends Biotechnol. 1997, 15, 339–341. [Google Scholar] [CrossRef]
- Shen, B. A new golden age of natural products drug discovery. Cell 2015, 163, 1297–1300. [Google Scholar] [CrossRef] [PubMed]
- Montaser, R.; Luesch, H. Marine natural products: A new wave of drugs? Future 2011, 3, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Patridge, E.; Gareiss, P.; Kinch, M.S.; Hoyer, D. An analysis of fda-approved drugs: Natural products and their derivatives. Drug Discov. Today 2016, 21, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.T. Natural products and pharma 2011: Strategic changes spur new opportunities. Nat. Prod. Rep. 2011, 28, 1783–1789. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Margulis, L.; Chapman, M.J. Kingdoms and Domains: An illustrated Guide to the Phyla of Life on Earth; Elsevier Science, Marine Biological Laboratory: Woods Hole, MA, USA, 2009. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef] [Green Version]
- Mayer, A.M.; Nguyen, M.; Newman, D.J.; Glaser, K.B. The marine pharmacology and pharmaceuticals pipeline in 2015. FASEB J. 2016, 30, 932.7. [Google Scholar]
- Mayer, A.M.; Nguyen, M.; Kalwajtys, P.; Kerns, H.; Newman, D.J.; Glaser, K.B. The marine pharmacology and pharmaceuticals pipeline in 2016. FASEB J. 2017, 31, 18.1. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef] [PubMed]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Falaise, C.; François, C.; Travers, M.-A.; Morga, B.; Haure, J.; Tremblay, R.; Turcotte, F.; Pasetto, P.; Gastineau, R.; Hardivillier, Y. Antimicrobial compounds from eukaryotic microalgae against human pathogens and diseases in aquaculture. Mar. Drugs 2016, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Drugs and drug candidates from marine sources: An assessment of the current “state of play”. Planta Med. 2016, 82, 775–789. [Google Scholar] [CrossRef]
- Mehbub, M.F.; Perkins, M.V.; Zhang, W.; Franco, C.M. New marine natural products from sponges (porifera) of the order dictyoceratida (2001 to 2012); a promising source for drug discovery, exploration and future prospects. Biotechnol. Adv. 2016, 34, 473–491. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Agrawal, S.; Adholeya, A. The pharmacological potential of nonribosomal peptides from marine sponge and tunicates. Front. Pharmacol. 2016, 7, 333. [Google Scholar] [CrossRef]
- Moreno-González, R.; Rodríguez-Mozaz, S.; Huerta, B.; Barceló, D.; León, V. Do pharmaceuticals bioaccumulate in marine molluscs and fish from a coastal lagoon? Environ. Res. 2016, 146, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Anjum, K.; Abbas, S.Q.; Shah, S.A.A.; Akhter, N.; Batool, S.; ul Hassan, S.S. Marine sponges as a drug treasure. Biomol. Ther. 2016, 24, 347–362. [Google Scholar] [CrossRef]
- Putz, A.; Proksch, P. Chemical Defence in Marine Ecosystems. In Functions and Biotechnology of Plant Secondary Metabolites, 2nd ed.; Wink, M., Ed.; Wiley-Blackwell: Oxford, UK, 2010. [Google Scholar] [CrossRef]
- Mayer, A.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2009–2011: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2013, 11, 2510–2573. [Google Scholar]
- Mayer, A.M.; Rodríguez, A.D.; Berlinck, R.G.; Fusetani, N. Marine pharmacology in 2007–2008: Marine compounds with antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous system, and other miscellaneous mechanisms of action. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2011, 153, 191–222. [Google Scholar] [PubMed]
- Huang, T.; Lin, S. Microbial natural products: A promising source for drug discovery. J. Appl. Microbiol. Biochem. 2017, 2, 1–3. [Google Scholar]
- Pangestuti, R.; Kim, S.-K. Bioactive peptide of marine origin for the prevention and treatment of non-communicable diseases. Mar. Drugs 2017, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Keffer, J.L.; Huecas, S.; Hammill, J.T.; Wipf, P.; Andreu, J.M.; Bewley, C.A. Chrysophaentins are competitive inhibitors of ftsz and inhibit Z-ring formation in live bacteria. Bioorg. Med. Chem. 2013, 21, 5673–5678. [Google Scholar] [CrossRef]
- Plaza, A.; Keffer, J.L.; Bifulco, G.; Lloyd, J.R.; Bewley, C.A. Chrysophaentins A–H, antibacterial bisdiarylbutene macrocycles that inhibit the bacterial cell division protein ftsz. J. Am. Chem. Soc. 2010, 132, 9069–9077. [Google Scholar] [CrossRef]
- Kwan, J.C.; Meickle, T.; Ladwa, D.; Teplitski, M.; Paul, V.; Luesch, H. Lyngbyoic acid, a “tagged” fatty acid from a marine cyanobacterium, disrupts quorum sensing in Pseudomonas aeruginosa. Mol. BioSyst. 2011, 7, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- DiGirolamo, J.A.; Li, X.-C.; Jacob, M.R.; Clark, A.M.; Ferreira, D. Reversal of fluconazole resistance by sulfated sterols from the marine sponge Topsentia sp. J. Nat. Prod. 2009, 72, 1524–1528. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.-H.; Li, X.-M.; Liu, Y.; Wang, B.-G. Polyoxygenated dihydropyrano [2,3-c]pyrrole-4,5-dione derivatives from the marine mangrove-derived endophytic fungus Penicillium brocae MA-231 and their antimicrobial activity. Chin. Chem. Lett. 2015, 26, 610–612. [Google Scholar] [CrossRef]
- Du, F.-Y.; Li, X.; Li, X.-M.; Zhu, L.-W.; Wang, B.-G. Indolediketopiperazine alkaloids from Eurotium cristatum EN-220, an endophytic fungus isolated from the marine alga Sargassum thunbergii. Mar. Drugs 2017, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Du, F.-Y.; Li, X.-M.; Li, C.-S.; Shang, Z.; Wang, B.-G. Cristatumins A–D, new indole alkaloids from the marine-derived endophytic fungus Eurotium cristatum EN-220. Bioorg. Med. Chem. Lett. 2012, 22, 4650–4653. [Google Scholar] [CrossRef]
- Youssef, D.T.; Shaala, L.A.; Alshali, K.Z. Bioactive hydantoin alkaloids from the red sea marine sponge Hemimycale arabica. Mar. Drugs 2015, 13, 6609–6619. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mándi, A.; Li, X.-M.; Meng, L.-H.; Kurtán, T.; Wang, B.-G. Peniciadametizine A, a dithiodiketopiperazine with a unique spiro [furan-2,7′-pyrazino [1,2-b][1,2] oxazine] skeleton, and a related analogue, peniciadametizine B, from the marine sponge-derived fungus Penicillium adametzioides. Mar. Drugs 2015, 13, 3640–3652. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.-H.; Zhang, P.; Li, X.-M.; Wang, B.-G. Penicibrocazines A–E, five new sulfide diketopiperazines from the marine-derived endophytic fungus Penicillium brocae. Mar. Drugs 2015, 13, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Sun, S.; Ference, C.; Zhu, W.; Zhou, N.; Zhang, Y.; Zhou, K. A potent antimicrobial compound isolated from Clathria cervicornis. Bioorg. Med. Chem. Lett. 2015, 25, 67–69. [Google Scholar] [CrossRef]
- Jiao, W.; Zhang, F.; Zhao, X.; Hu, J.; Suh, J.-W. A novel alkaloid from marine-derived actinomycete Streptomyces xinghaiensis with broad-spectrum antibacterial and cytotoxic activities. PLoS ONE 2013, 8, e75994. [Google Scholar] [CrossRef] [PubMed]
- Youssef, D.T.; Shaala, L.A.; Asfour, H.Z. Bioactive compounds from the red sea marine sponge Hyrtios species. Mar. Drugs 2013, 11, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Hamann, M.T.; Zou, Y.; Zhang, M.-Y.; Gong, X.-B.; Xiao, J.-R.; Chen, W.-S.; Lin, H.-W. Antimicrobial metabolites from the paracel islands sponge Agelas mauritiana. J. Nat. Prod. 2012, 75, 774–778. [Google Scholar] [CrossRef]
- Kubota, T.; Nakamura, K.; Kurimoto, S.-I.; Sakai, K.; Fromont, J.; Gonoi, T.; Kobayashi, J.I. Zamamidine D, a manzamine alkaloid from an okinawan Amphimedon sp. Marine sponge. J. Nat. Prod. 2017. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, X.-M.; Meng, L.-H.; Jiang, W.-L.; Xu, G.-M.; Huang, C.-G.; Wang, B.-G. Bisthiodiketopiperazines and acorane sesquiterpenes produced by the marine-derived fungus Penicillium adametzioides AS-53 on different culture media. J. Nat. Prod. 2015, 78, 1294–1299. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.J.; Tang, X.L.; Qin, G.F.; Sun, Y.T.; Li, L.; de Voogd, N.J.; Li, P.L.; Li, G.Q. Pyrrole derivatives and diterpene alkaloids from the south china sea sponge Agelas nakamurai. Chem. Biodivers. 2017. [Google Scholar] [CrossRef]
- Asiri, I.A.; Badr, J.M.; Youssef, D.T. Penicillivinacine, antimigratory diketopiperazine alkaloid from the marine-derived fungus Penicillium vinaceum. Phytochem. Lett. 2015, 13, 53–58. [Google Scholar] [CrossRef]
- Gao, X.; Lu, Y.; Xing, Y.; Ma, Y.; Lu, J.; Bao, W.; Wang, Y.; Xi, T. A novel anticancer and antifungus phenazine derivative from a marine actinomycete BM-17. Microbiol. Res. 2012, 167, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, K.; Garcia Hernandez, J.E.; Harper, M.K.; Carroll, A.; Motti, C.A.; Awaya, J.; Nguyen, H.-Y.; Wright, A.D. Puupehenol, a potent antioxidant antimicrobial meroterpenoid from a hawaiian deep-water Dactylospongia sp. Sponge. J. Nat. Prod. 2015, 78, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.-H.; Li, X.-M.; Liu, Y.; Wang, B.-G. Penicibilaenes A and B, sesquiterpenes with a tricyclo [6.3. 1.01, 5] dodecane skeleton from the marine isolate of Penicillium bilaiae MA-267. Org. Lett. 2014, 16, 6052–6055. [Google Scholar] [CrossRef]
- Wang, J.-F.; Lin, X.-P.; Qin, C.; Liao, S.-R.; Wan, J.-T.; Zhang, T.-Y.; Liu, J.; Fredimoses, M.; Chen, H.; Yang, B. Antimicrobial and antiviral sesquiterpenoids from sponge-associated fungus, Aspergillus sydowii ZSDS1-F6. J. Antibiot. 2014, 67, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Alarif, W.M.; Al-Lihaibi, S.S.; Ayyad, S.-E.N.; Abdel-Rhman, M.H.; Badria, F.A. Laurene-type sesquiterpenes from the red sea red alga Laurencia obtusa as potential antitumor—Antimicrobial agents. Eur. J. Med. Chem. 2012, 55, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.B.; Jensen, P.R.; Fenical, W. Cytotoxic and antimicrobial napyradiomycins from two marine-derived Streptomyces strains. Eur. J. Org. Chem. 2013, 2013, 3751–3757. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Mándi, A.; Li, S.; Chen, Y.; Zhang, W.; Tian, X.; Zhang, H.; Li, H.; Zhang, W.; Zhang, S. N–N-coupled indolo-sesquiterpene atropo-diastereomers from a marine-derived actinomycete. Eur. J. Org. Chem. 2012, 2012, 5256–5262. [Google Scholar] [CrossRef]
- Al-Footy, K.O.; Alarif, W.M.; Asiri, F.; Aly, M.M.; Ayyad, S.-E.N. Rare pyrane-based cembranoids from the red sea soft coral Sarcophyton trocheliophorum as potential antimicrobial—Antitumor agents. Med. Chem. Res. 2015, 24, 505–512. [Google Scholar] [CrossRef]
- Kumar, R.; Subramani, R.; Aalbersberg, W. Three bioactive sesquiterpene quinones from the fijian marine sponge of the genus Hippospongia. Nat. Prod. Res. 2013, 27, 1488–1491. [Google Scholar] [CrossRef] [PubMed]
- Murshid, S.S.; Badr, J.M.; Youssef, D.T. Penicillosides a and b: New cerebrosides from the marine-derived fungus Penicillium species. Rev. Bras. Farmacogn. 2016, 26, 29–33. [Google Scholar] [CrossRef]
- Tareq, F.S.; Kim, J.H.; Lee, M.A.; Lee, H.-S.; Lee, Y.-J.; Lee, J.S.; Shin, H.J. Ieodoglucomides A and B from a marine-derived bacterium Bacillus licheniformis. Org. Lett. 2012, 14, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Tareq, F.S.; Lee, H.-S.; Lee, Y.-J.; Lee, J.S.; Shin, H.J. Ieodoglucomide C and ieodoglycolipid, new glycolipids from a marine-derived bacterium Bacillus Licheniformis 09IDYM23. Lipids 2015, 50, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Tareq, F.S.; Lee, M.A.; Lee, H.-S.; Lee, Y.-J.; Lee, J.S.; Hasan, C.M.; Islam, M.T.; Shin, H.J. Gageotetrins A–C, noncytotoxic antimicrobial linear lipopeptides from a marine bacterium Bacillus subtilis. Org. Lett. 2014, 16, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Mondol, M.A.M.; Shin, H.J. Antibacterial and antiyeast compounds from marine-derived bacteria. Mar. Drugs 2014, 12, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Mondol, M.A.M.; Tareq, F.S.; Kim, J.H.; Lee, M.A.; Lee, H.-S.; Lee, J.S.; Lee, Y.-J.; Shin, H.J. New antimicrobial compounds from a marine-derived Bacillus sp. J. Antibiot. 2013, 66, 89–95. [Google Scholar] [CrossRef]
- Lu, Y.; Zhao, M. Two highly acetylated sterols from the marine sponge Dysidea sp. Z. Naturforschung B 2017, 72, 49–52. [Google Scholar] [CrossRef]
- Lima, B.D.A.; de Lira, S.P.; Kossuga, M.H.; Gonçalves, R.B.; Berlinck, R.G.; Kamiya, R.U. Halistanol sulfate A and rodriguesines A and B are antimicrobial and antibiofilm agents against the cariogenic bacterium Streptococcus mutans. Rev. Bras. Farmacogn. 2014, 24, 651–659. [Google Scholar] [CrossRef]
- Kalinovskaya, N.I.; Romanenko, L.A.; Kalinovsky, A.I. Antibacterial low-molecular-weight compounds produced by the marine bacterium Rheinheimera japonica KMM 9513T. Anieke Van Leeuwenhoek 2017, 110, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Cuong, P.V.; Cuc, N.T.K.; Quyen, V.T.; Binh, P.T.; Kiem, P.; Nam, N.H.; Dat, N.T. Antimicrobial constituents from the Bacillus megaterium IC isolated from marine sponge Haliclona oculata. Nat. Prod. Sci. 2015, 21, 202–205. [Google Scholar]
- Du, F.-Y.; Zhang, P.; Li, X.-M.; Li, C.-S.; Cui, C.-M.; Wang, B.-G. Cyclohexadepsipeptides of the isaridin class from the marine-derived fungus Beauveria felina EN-135. J. Nat. Prod. 2014, 77, 1164–1169. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, Q.; Liu, X.; Chen, Y.; Zhang, Y.; Sun, A.; Zhang, W.; Zhang, J.; Ju, J. Cyclic hexapeptides from the deep south china sea-derived Streptomyces scopuliridis SCSIO ZJ46 active against pathogenic gram-positive bacteria. J. Nat. Prod. 2014, 77, 1937–1941. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Canning, C.B.; Bhargava, K.; Sun, X.; Zhu, W.; Zhou, N.; Zhang, Y.; Zhou, K. Polybrominated diphenyl ethers with potent and broad spectrum antimicrobial activity from the marine sponge Dysidea. Bioorg. Med. Chem. Lett. 2015, 25, 2181–2183. [Google Scholar] [CrossRef] [PubMed]
- Gotsbacher, M.P.; Karuso, P. New antimicrobial bromotyrosine analogues from the sponge Pseudoceratina purpurea and its predator Tylodina corticalis. Mar. Drugs 2015, 13, 1389–1409. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Alves, C.; Horta, A.; Pinteus, S.; Silva, J.; Culioli, G.; Thomas, O.P.; Pedrosa, R. Antitumor and antimicrobial potential of bromoditerpenes isolated from the red alga, Sphaerococcus coronopifolius. Mar. Drugs 2015, 13, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, J.; Tang, K.; Shi, X.; Wang, S.; Zhu, W.-M.; Zhang, X.-H. Purification and characterization of antibacterial compounds of Pseudoalteromonas flavipulchra JG1. Microbiology 2012, 158, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Kusama, T.; Takahashi-Nakaguchi, A.; Gonoi, T.; Fromont, J.; Kobayashi, J.I. Nagelamides X–Z, dimeric bromopyrrole alkaloids from a marine sponge agelas sp. Org. Lett. 2013, 15, 3262–3265. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Davis, R.A.; Feng, Y.; Sykes, M.L.; Shelper, T.; Avery, V.M.; Camp, D.; Quinn, R.J. Ianthelliformisamines A–C, antibacterial bromotyrosine-derived metabolites from the marine sponge Suberea ianthelliformis. J. Nat. Prod. 2012, 75, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xie, X.; Chen, L.; Yan, S.; Ye, X.; Anjum, K.; Huang, H.; Lian, X.; Zhang, Z. Bioactive polycyclic quinones from marine streptomyces sp. 182SMLY. Mar. Drugs 2016, 14, 10. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wiese, J.; Labes, A.; Kramer, A.; Schmaljohann, R.; Imhoff, J.F. Lindgomycin, an unusual antibiotic polyketide from a marine fungus of the lindgomycetaceae. Mar. Drugs 2015, 13, 4617–4632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gushiken, M.; Kagiyama, I.; Kato, H.; Kuwana, T.; Losung, F.; Mangindaan, R.E.; De Voogd, N.J.; Tsukamoto, S. Manadodioxans A–E: Polyketide endoperoxides from the marine sponge Plakortis bergquistae. J. Nat. Med. 2015, 69, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Thilakan, B.; Chakraborty, R.D.; Raola, V.K.; Joy, M. O-heterocyclic derivatives with antibacterial properties from marine bacterium Bacillus subtilis associated with seaweed, Sargassum myriocystum. Appl. Microbiol. Biotechnol. 2017, 101, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Ye, X.; Yu, S.; Lian, X.-Y.; Zhang, Z. New capoamycin-type antibiotics and polyene acids from marine Streptomyces fradiae PTZ0025. Mar. Drugs 2012, 10, 2388–2402. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.; Piggott, A.M.; Khalil, Z.; Bernhardt, P.V.; Capon, R.J. Heronamycin a: A new benzothiazine ansamycin from an australian marine-derived streptomyces sp. Tetrahedron Lett. 2012, 53, 1063–1065. [Google Scholar] [CrossRef]
- Chakraborty, K.; Thilakan, B.; Raola, V.K.; Joy, M. Antibacterial polyketides from Bacillus amyloliquefaciens associated with edible red seaweed Laurenciae papillosa. Food Chem. 2017, 218, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Du, F.Y.; Li, X.M.; Song, J.Y.; Li, C.S.; Wang, B.G. Anthraquinone derivatives and an orsellinic acid ester from the marine alga-derived endophytic fungus Eurotium cristatum EN-220. Helv. Chim. Acta 2014, 97, 973–978. [Google Scholar] [CrossRef]
- Xu, R.; Li, X.-M.; Wang, B.-G. Penicisimpins A-C, three new dihydroisocoumarins from Penicillium simplicissimum MA-332, a marine fungus derived from the rhizosphere of the mangrove plant Bruguiera sexangula var. Rhynchopetala. Phytochem. Lett. 2016, 17, 114–118. [Google Scholar] [CrossRef]
- Aksoy, S.Ç.; Uzel, A.; Bedir, E. Cytosine-type nucleosides from marine-derived Streptomyces rochei 06CM016. J. Antibiot. 2016, 69, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Shaala, L.A.; Youssef, D.T. Identification and bioactivity of compounds from the fungus Penicillium sp. CYE-87 isolated from a marine tunicate. Mar. Drugs 2015, 13, 1698–1709. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Ren, B.; Chen, C.; Yu, K.; Liu, X.; Zhang, Y.; Yang, N.; He, H.; Liu, X.; Dai, H. Three new sterigmatocystin analogues from marine-derived fungus Aspergillus versicolor MF359. Appl. Microbiol. Biotechnol. 2014, 98, 3753–3758. [Google Scholar] [CrossRef] [PubMed]
- Ambavane, V.; Tokdar, P.; Parab, R.; Sreekumar, E.; Mahajan, G.; Mishra, P.D.; D’Souza, L.; Ranadive, P. Caerulomycin A—An antifungal compound isolated from marine actinomycetes. Adv. Microbiol. 2014, 4, 567–578. [Google Scholar] [CrossRef]
- Wu, B.; Oesker, V.; Wiese, J.; Schmaljohann, R.; Imhoff, J.F. Two new antibiotic pyridones produced by a marine fungus, Trichoderma sp. strain MF106. Mar. Drugs 2014, 12, 1208–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasari, V.R.R.K.; Muthyala, M.K.K.; Nikku, M.Y.; Donthireddy, S.R.R. Novel pyridinium compound from marine actinomycete, Amycolatopsis alba var. Nov. DVR D4 showing antimicrobial and cytotoxic activities in vitro. Microbiol. Res. 2012, 167, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhao, S.; Wei, J.; Fang, N.; Yin, L.; Sun, J. Porric acid D from marine-derived fungus Alternaria sp. isolated from bohai sea. Chem. Nat. Compd. 2012, 47, 893–895. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Y.; Liu, L.; Han, Z.; Lai, P.Y.; Guo, X.; Zhang, X.; Lin, W.; Qian, P.-Y. Five new amicoumacins isolated from a marine-derived bacterium Bacillus subtilis. Mar. Drugs 2012, 10, 319–328. [Google Scholar] [CrossRef]
- Eltamany, E.E.; Abdelmohsen, U.R.; Ibrahim, A.K.; Hassanean, H.A.; Hentschel, U.; Ahmed, S.A. New antibacterial xanthone from the marine sponge-derived Micrococcus sp. EG45. Bioorg. Med. Chem. Lett. 2014, 24, 4939–4942. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Opportunities for natural products in 21 st century antibiotic discovery. Nat. Prod. Rep. 2017, 34, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Liu, J.; Xu, S.; Zhu, Z.; Xu, J. The structural modification of natural products for novel drug discovery. Expert Opin. Drug Discov. 2017, 12, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Majik, M.S.; Rodrigues, C.; Mascarenhas, S.; D’Souza, L. Design and synthesis of marine natural product-based 1H-indole-2, 3-dione scaffold as a new antifouling/antibacterial agent against fouling bacteria. Bioorg. Chem. 2014, 54, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Pang, H.D.; Fan, J.; Ge, F.; Yang, X.X.; Liu, Q.Y.; Liao, X.J.; Xu, S.H. In vitro and in vivo antineoplastic activity of a novel bromopyrrole and its potential mechanism of action. Br. J. Pharmacol. 2010, 159, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Tasdemir, D.; Topaloglu, B.; Perozzo, R.; Brun, R.; O’Neill, R.; Carballeira, N.M.; Zhang, X.; Tonge, P.J.; Linden, A.; Rüedi, P. Marine natural products from the turkish sponge Agelas oroides that inhibit the enoyl reductases from Plasmodium falciparum, Mycobacterium tuberculosis and Escherichia coli. Bioorg. Med. Chem. 2007, 15, 6834–6845. [Google Scholar] [CrossRef] [PubMed]
- Schillaci, D.; Petruso, S.; Cascioferro, S.; Raimondi, M.; Haagensen, J.A.J.; Molin, S. In vitro anti-gram-positive and antistaphylococcal biofilm activity of newly halogenated pyrroles related to pyrrolomycins. Int. J. Antimicrob. Agents 2008, 31, 380–382. [Google Scholar] [CrossRef] [PubMed]
- Fattorusso, E.; Taglialatela-Scafati, O. Modern Alkaloids: Structure, Isolation, Synthesis, and Biology, 1st ed.; John Wiley & Sons: Chichester, UK, 2008; p. 271. [Google Scholar]
- Rane, R.A.; Gutte, S.D.; Sahu, N.U. Synthesis and evaluation of novel 1, 3, 4-oxadiazole derivatives of marine bromopyrrole alkaloids as antimicrobial agent. Bioorg. Med. Chem. Lett. 2012, 22, 6429–6432. [Google Scholar] [CrossRef] [PubMed]
- Rane, R.A.; Sahu, N.U.; Shah, C.P. Synthesis and antibiofilm activity of marine natural product-based 4-thiazolidinones derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 7131–7134. [Google Scholar] [CrossRef] [PubMed]
- Rane, R.A.; Sahu, N.U.; Shah, C.P.; Shah, N.K. Design, synthesis and antistaphylococcal activity of marine pyrrole alkaloid derivatives. J. Enzym. Inhib. Med. 2014, 29, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Zidar, N.; Montalvão, S.; Hodnik, Ž.; Nawrot, D.A.; Žula, A.; Ilaš, J.; Kikelj, D.; Tammela, P.; Mašič, L.P. Antimicrobial activity of the marine alkaloids, clathrodin and oroidin, and their synthetic analogues. Mar. Drugs 2014, 12, 940–963. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-C.; Li, X.-M.; Gloer, J.B.; Wang, B.-G. First total syntheses and antimicrobial evaluation of penicimonoterpene, a marine-derived monoterpenoid, and its various derivatives. Mar. Drugs 2014, 12, 3352–3370. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.D.; Chu, M.; Oza, U.; Rajgarhia, V. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007, 24, 1225–1244. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resource Coordinators. Database resources of the national center for biotechnology information. In Nucleic Acids Res.; 2017; 45, pp. D12–D17. [Google Scholar]
- Genomes OnLine Database (GOLD). Available online: https://gold.Jgi.Doe.Gov/ (accessed on 20 July 2017).
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L.; Ravel, J. Coelichelin. A new peptide siderophore encoded by the Streptomyces coelicolor genome: Structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol. Lett. 2000, 187, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L. Exploitation of the streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 2014, 41, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, K.; Kotowska, M.; Chater, K.F.; Kuczek, K.; Takano, E. A cryptic type I polyketide synthase (CPK) gene cluster in Streptomyces coelicolor A3(2). Arch. Microbiol. 2007, 187, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.Q.; Wang, J.F.; Hao, Y.Y.; Wang, Y. Recent advances in the discovery and development of marine microbial natural products. Mar. Drugs 2013, 11, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Atlas of Biosynthetic Gene Clusters. Available online: https://img.Jgi.Doe.Gov/abc (accessed on 20 July 2017).
- The Secondary Metabolite Bioinformatics Forum. Available online: http://www.Secondarymetabolites.Org/ (accessed on 20 July 2017).
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Muller, R.; Wohlleben, W.; et al. Antismash 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- De Jong, A.; van Hijum, S.A.; Bijlsma, J.J.; Kok, J.; Kuipers, O.P. Bagel: A web-based bacteriocin genome mining tool. Nucleic Acids Res. 2006, 34, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Dejong, C.A.; Rees, P.N.; Johnston, C.W.; Li, H.; Webster, A.L.; Wyatt, M.A.; Magarvey, N.A. Genomes to natural products prediction informatics for secondary metabolomes (PRISM). Nucleic Acids Res. 2015, 43, 9645–9662. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Zouhir, A.; Le Lay, C.; Ben Hamida, J.; Fliss, I. Bactibase second release: A database and tool platform for bacteriocin characterization. BMC Microbiol. 2010, 10, 22. [Google Scholar] [CrossRef]
- Conway, K.R.; Boddy, C.N. Clustermine360: A database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 2013, 41, D402–D407. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Wieland Brown, L.C.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. Antismash 2.0—A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. Antismash: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. Antismash 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Kautsar, S.A.; Suarez Duran, H.G.; Blin, K.; Osbourn, A.; Medema, M.H. Plantismash: Automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, W55–W63. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Morales, P.; Kopp, J.F.; Martinez-Guerrero, C.; Yanez-Guerra, L.A.; Selem-Mojica, N.; Ramos-Aboites, H.; Feldmann, J.; Barona-Gomez, F. Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model streptomycetes. Genome Biol. Evol. 2016, 8, 1906–1916. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, S.; Zhu, Y.; Chen, Y.; Chen, Y.; Zhang, H.; Zhang, G.; Tian, X.; Pan, Y.; Zhang, S.; et al. Heronamides D-F, polyketide macrolactams from the deep-sea-derived streptomyces sp. SCSIO 03032. J. Nat. Prod. 2014, 77, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, W.; Chen, Y.; Yuan, C.; Zhang, H.; Zhang, G.; Ma, L.; Zhang, Q.; Tian, X.; Zhang, S.; et al. Characterization of heronamide biosynthesis reveals a tailoring hydroxylase and indicates migrated double bonds. ChemBioChem 2015, 16, 2086–2093. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Song, Y.; Qin, X.; Zhang, X.; Sun, A.; Ju, J. Identification of the biosynthetic gene cluster for the anti-infective desotamides and production of a new analogue in a heterologous host. J. Nat. Prod. 2015, 78, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Chen, Q.; Chen, S.; Xu, B.; Ju, J.; Wang, H. Discovery and biosynthesis of mathermycin from marine-derived marinactinospora thermotolerans scsio 00652. Appl. Environ. Microbiol. 2017. [Google Scholar] [CrossRef]
- Viegelmann, C.; Margassery, L.M.; Kennedy, J.; Zhang, T.; O’Brien, C.; O’Gara, F.; Morrissey, J.P.; Dobson, A.D.; Edrada-Ebel, R. Metabolomic profiling and genomic study of a marine sponge-associated streptomyces sp. Mar. Drugs 2014, 12, 3323–3351. [Google Scholar] [CrossRef]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Ruckert, C.; Braig, S.; Zahler, S.; et al. New natural products identified by combined genomics-metabolomics profiling of marine streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, B.; Li, H.; Xie, Y.; Li, Q.; Qin, X.; Zhang, X.; Ju, J. Biosynthesis of the anti-infective marformycins featuring pre-NRPS assembly line N-formylation and O-methylation and post-assembly line C-hydroxylation chemistries. Org. Lett. 2015, 17, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.N.; Wang, W.; Spanogiannopoulos, P.; Waglechner, N.; King, A.M.; Medina, R.; Wright, G.D. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 2013, 31, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.N.; Waglechner, N.; Wright, G.D. Antibiotic resistance-mediated isolation of scaffold-specific natural product producers. Nat. Protoc. 2014, 9, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, J.; Millan-Aguinaga, N.; Zhang, J.J.; O’Neill, E.C.; Ugalde, J.A.; Jensen, P.R.; Mantovani, S.M.; Moore, B.S. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 2015, 10, 2841–2849. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.H.; Ahuja, M.; Chiang, Y.M.; Oakley, C.E.; Moore, S.; Yoon, O.; Hajovsky, H.; Bok, J.W.; Keller, N.P.; Wang, C.C.; et al. Resistance gene-guided genome mining: Serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic pathway for fellutamide B, a proteasome inhibitor. ACS Chem. Biol. 2016, 11, 2275–2284. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.W.; Skinnider, M.A.; Dejong, C.A.; Rees, P.N.; Chen, G.M.; Walker, C.G.; French, S.; Brown, E.D.; Berdy, J.; Liu, D.Y.; et al. Assembly and clustering of natural antibiotics guides target identification. Nat. Chem. Biol. 2016, 12, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Tracanna, V.; de Jong, A.; Medema, M.H.; Kuipers, O.P. Mining prokaryotes for antimicrobial compounds: From diversity to function. FEMS Microbiol. Rev. 2017, 41, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.M.; Hodgson, J.E. The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘super-cluster’ in three streptomyces. FEMS Microbiol. Lett. 1993, 110, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Trefzer, A.; Kovalchuk, A.; van den Berg, M.; Muller, U.; Heijne, W.; Wu, L.; Alam, M.T.; Ronning, C.M.; Nierman, W.C.; et al. The sequence of a 1.8-MB bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol. Evol. 2010, 2, 212–224. [Google Scholar] [CrossRef]
- Alanjary, M.; Kronmiller, B.; Adamek, M.; Blin, K.; Weber, T.; Huson, D.; Philmus, B.; Ziemert, N. The antibiotic resistant target seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 2017, 45, W42–W48. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. Card 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. Arg-annot. A new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
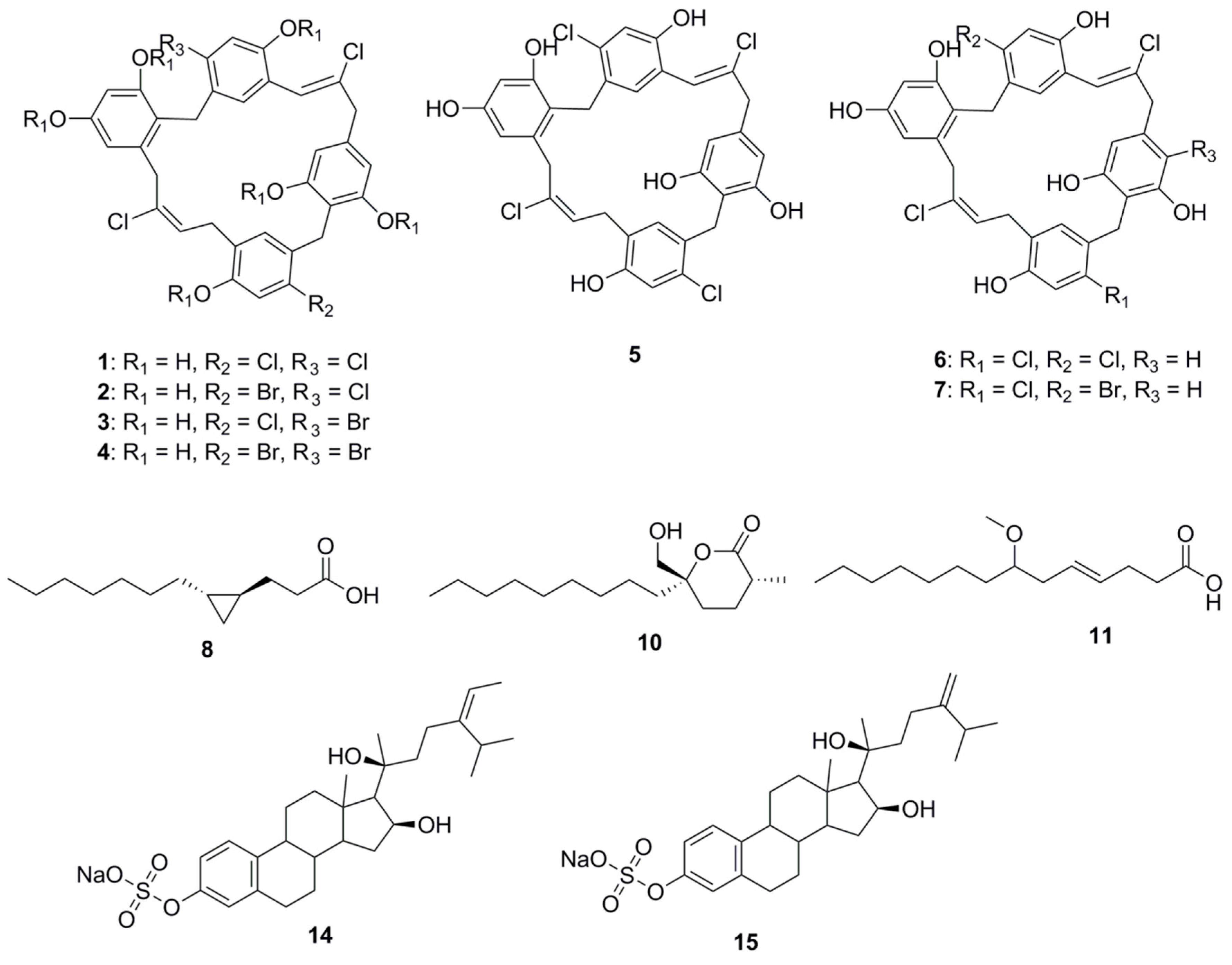
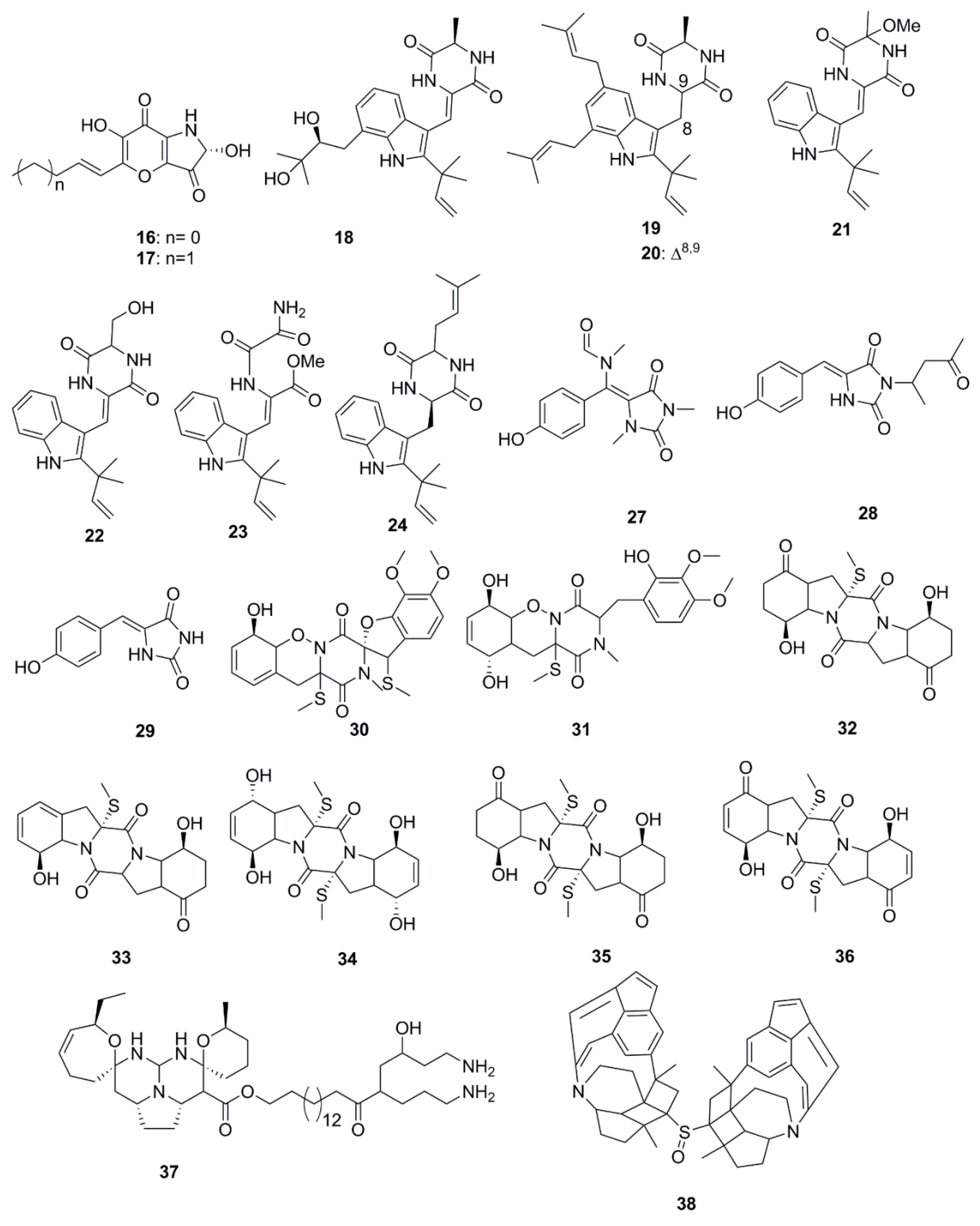
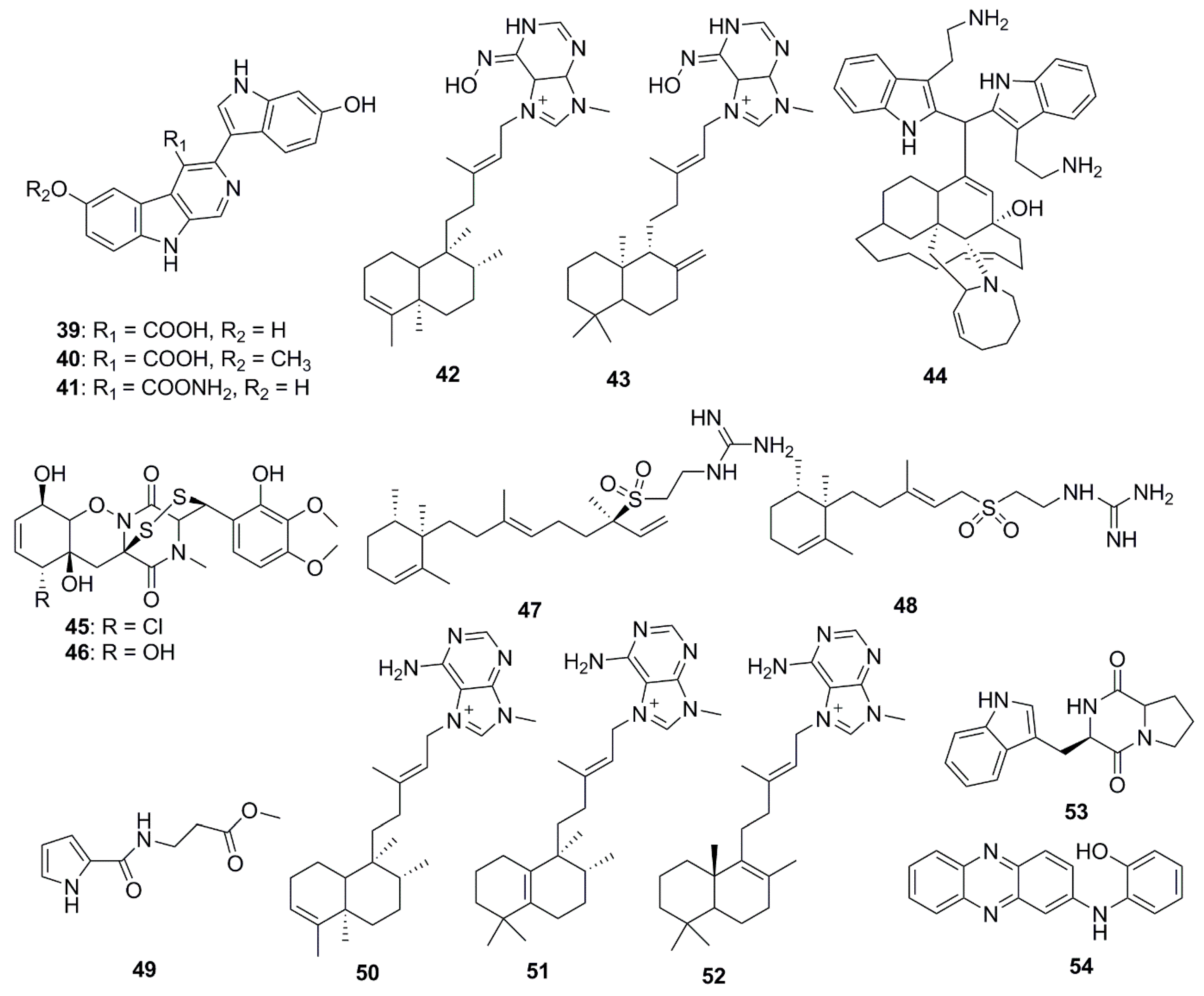


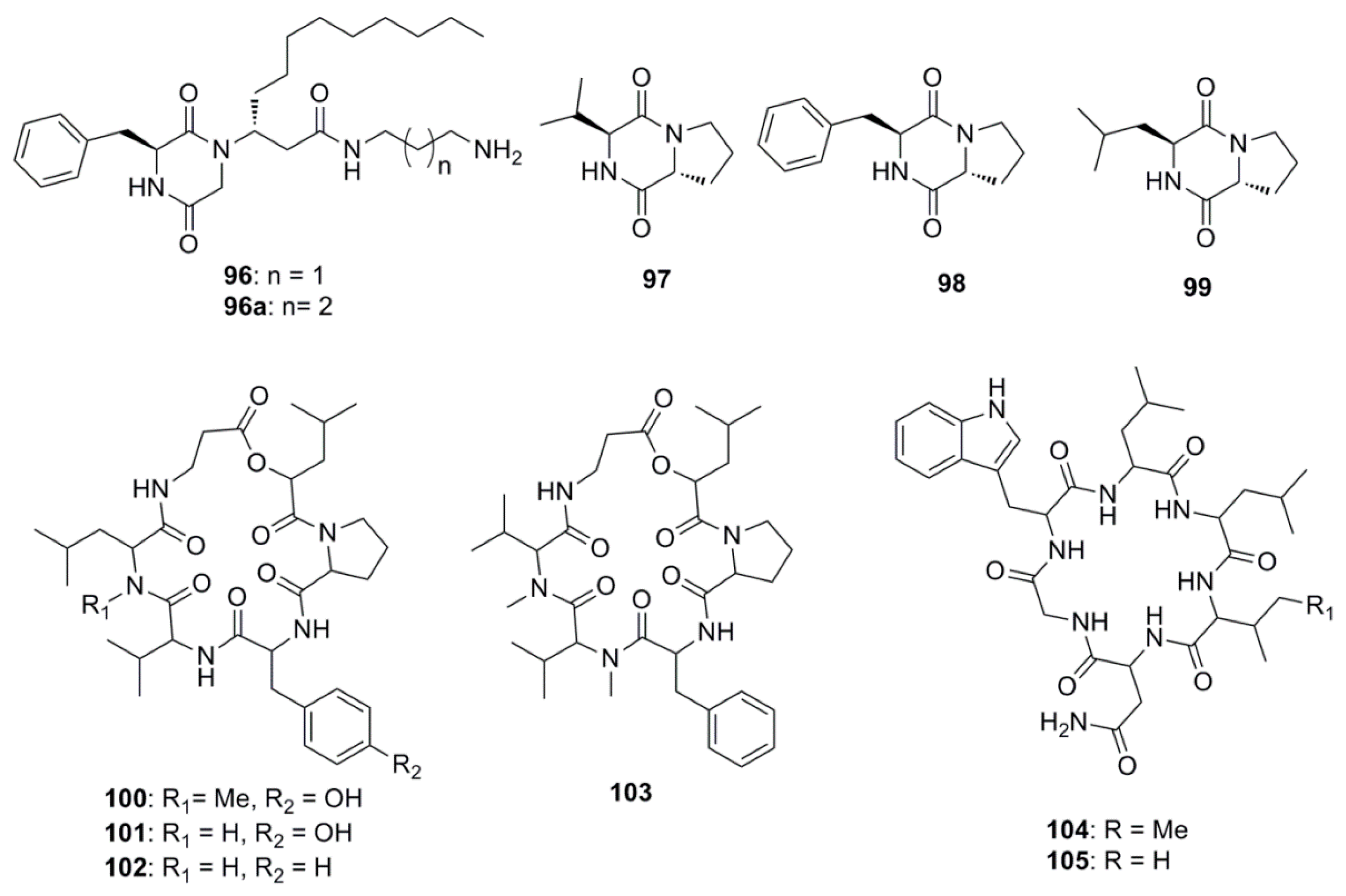
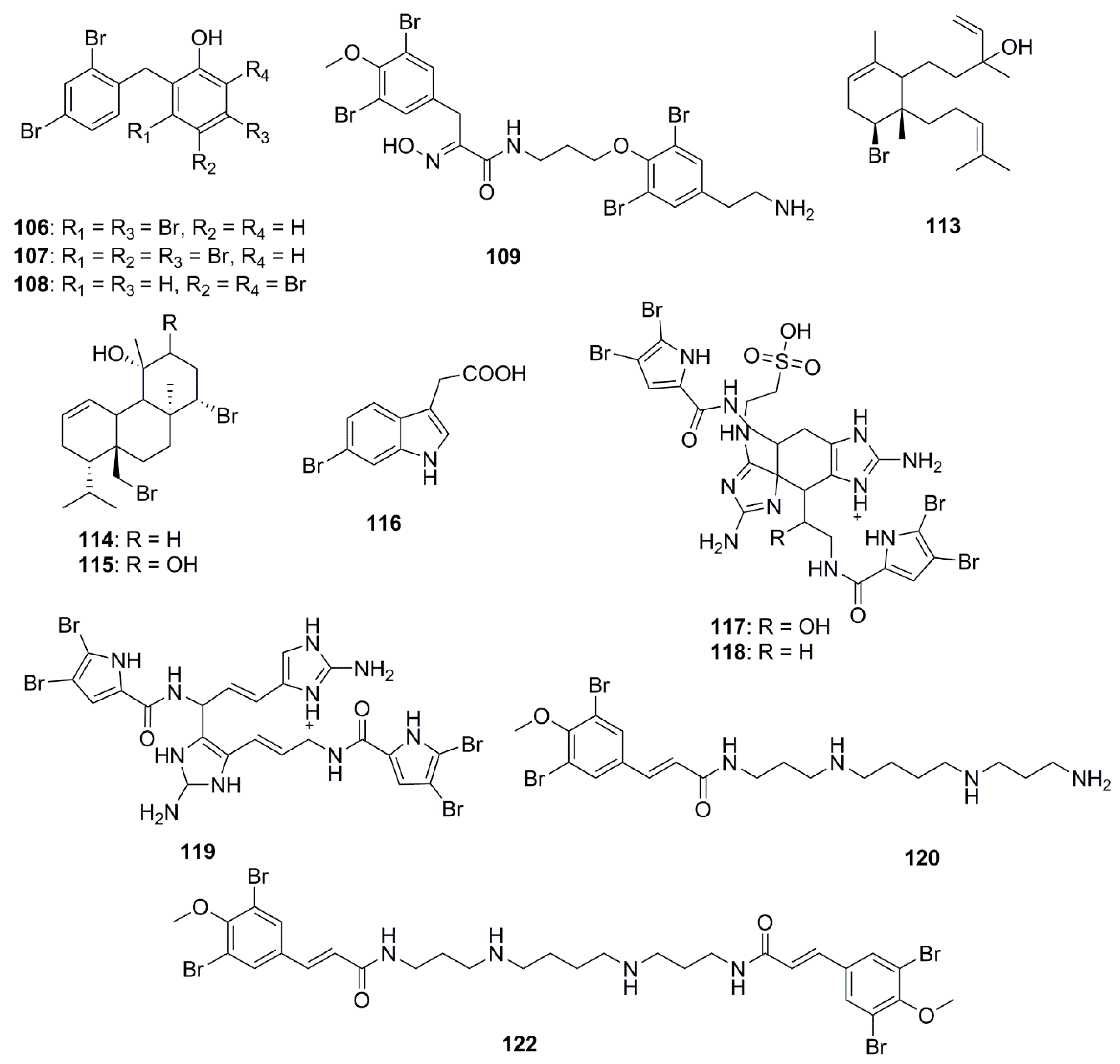
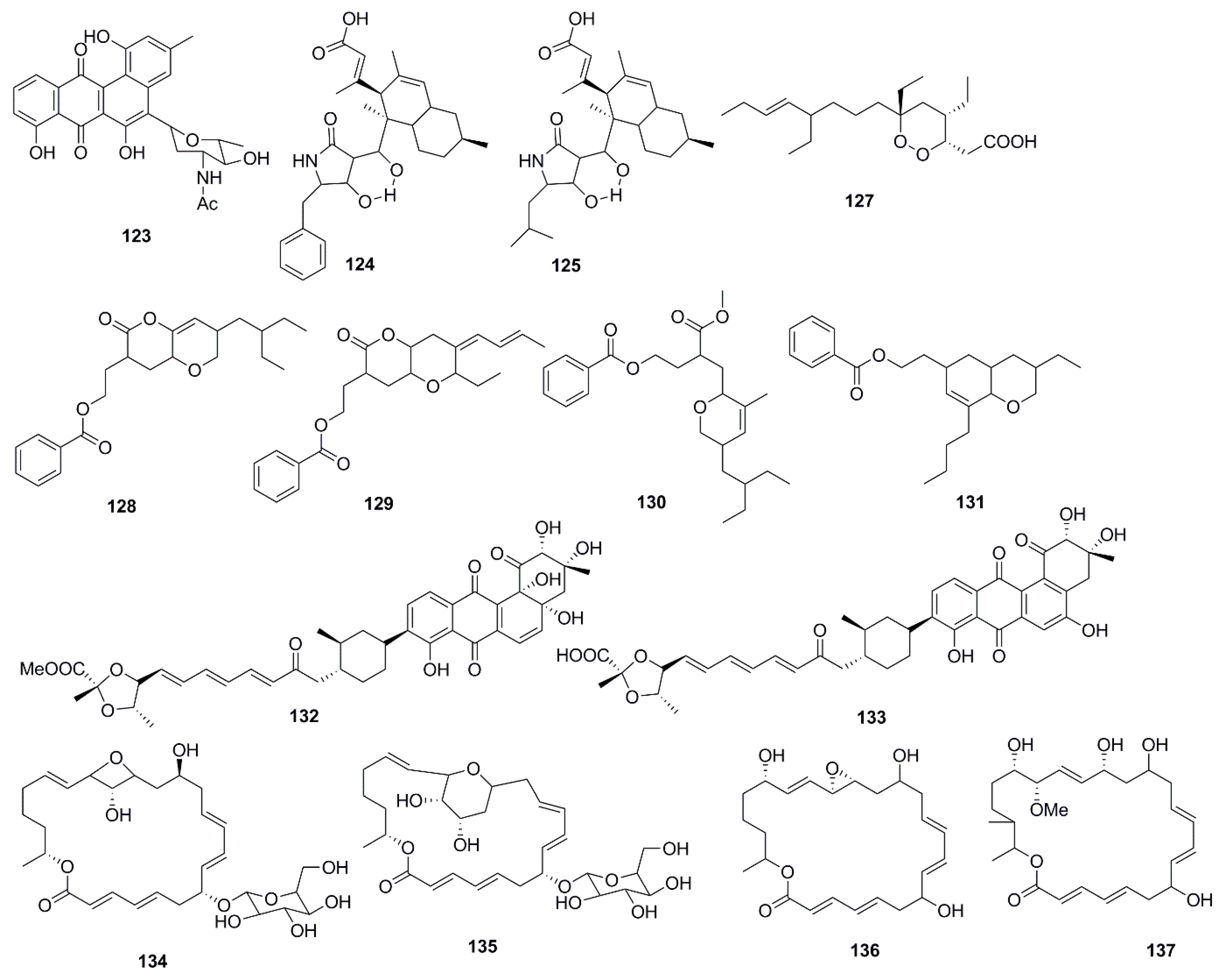
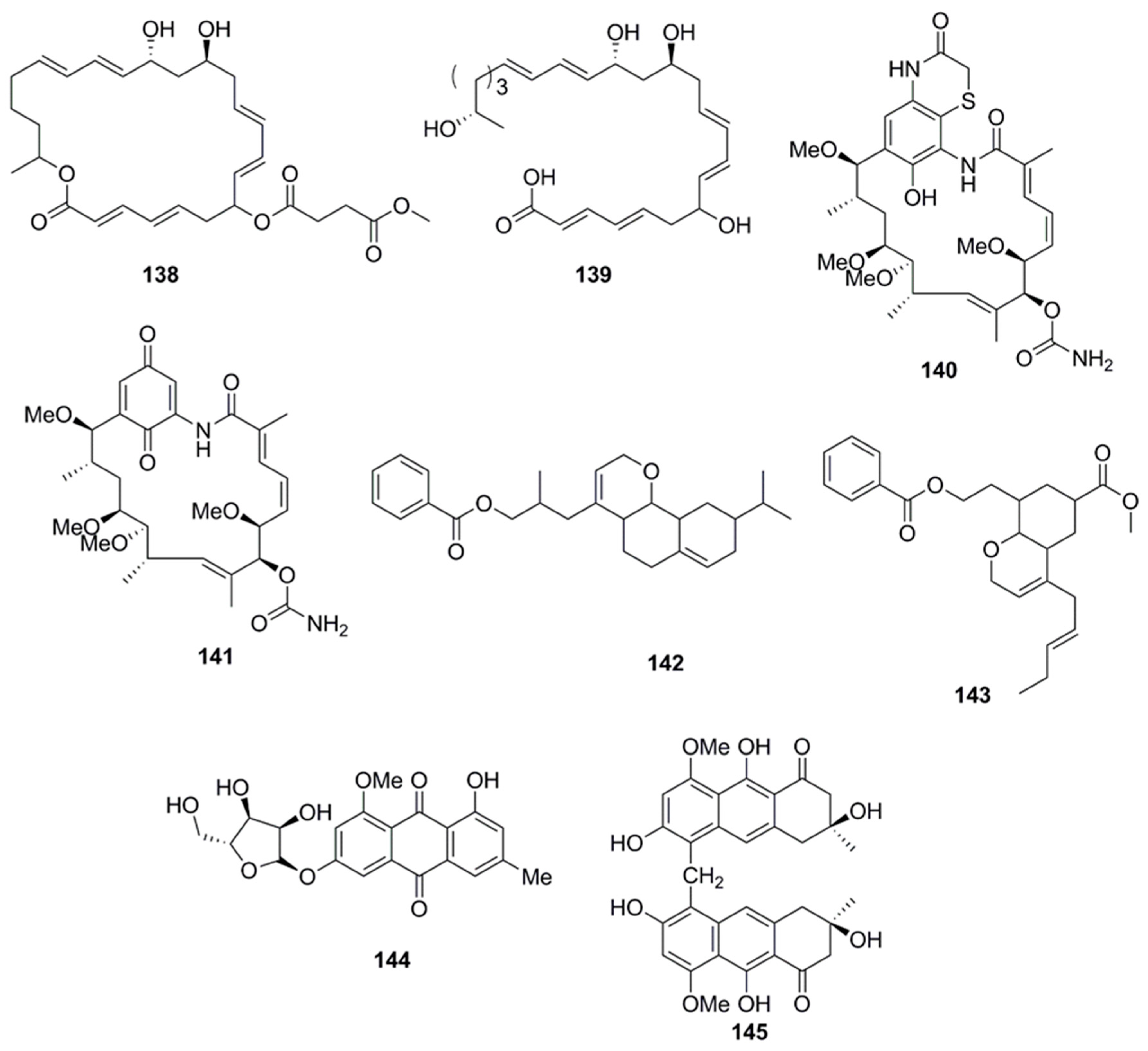
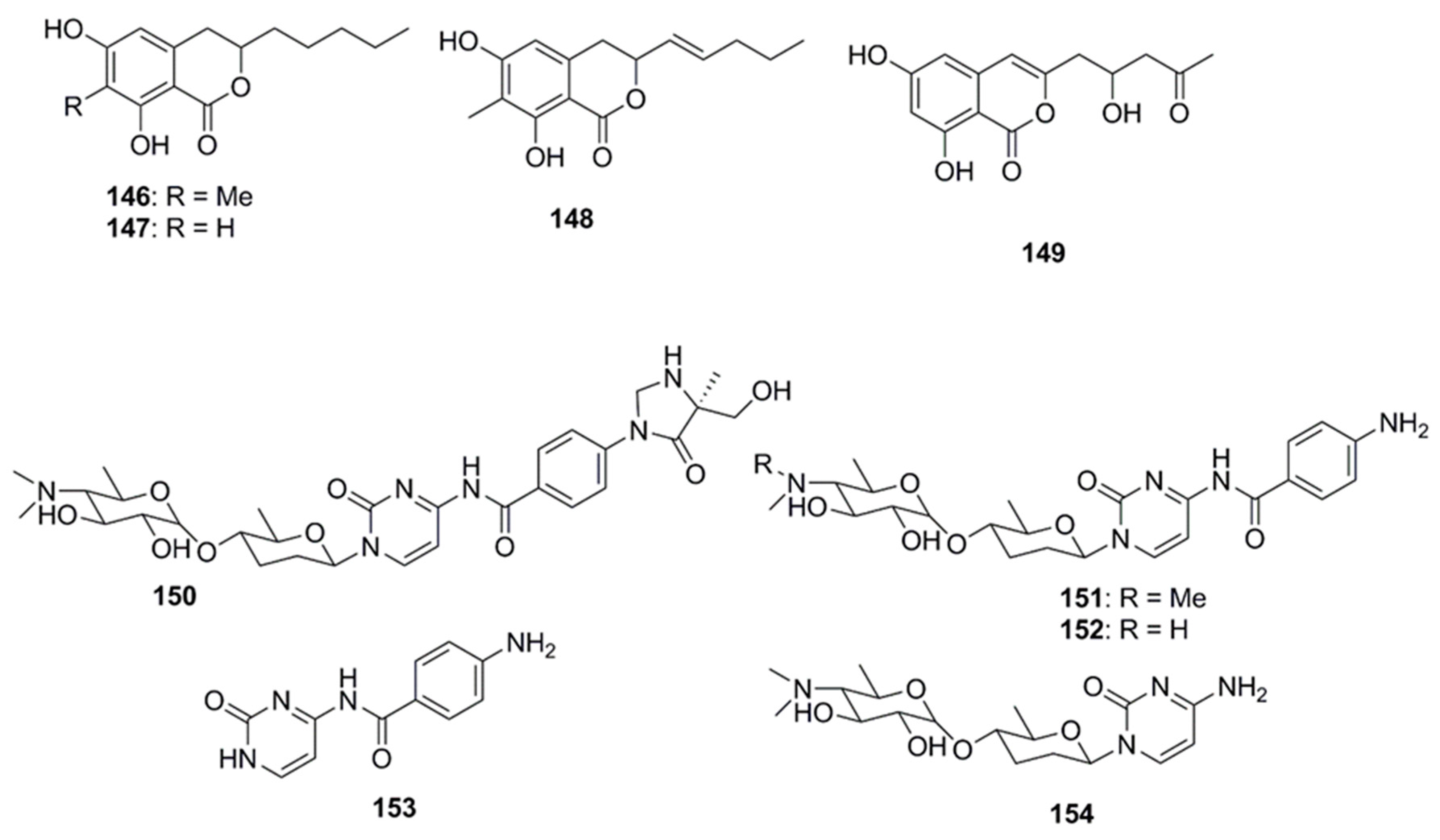
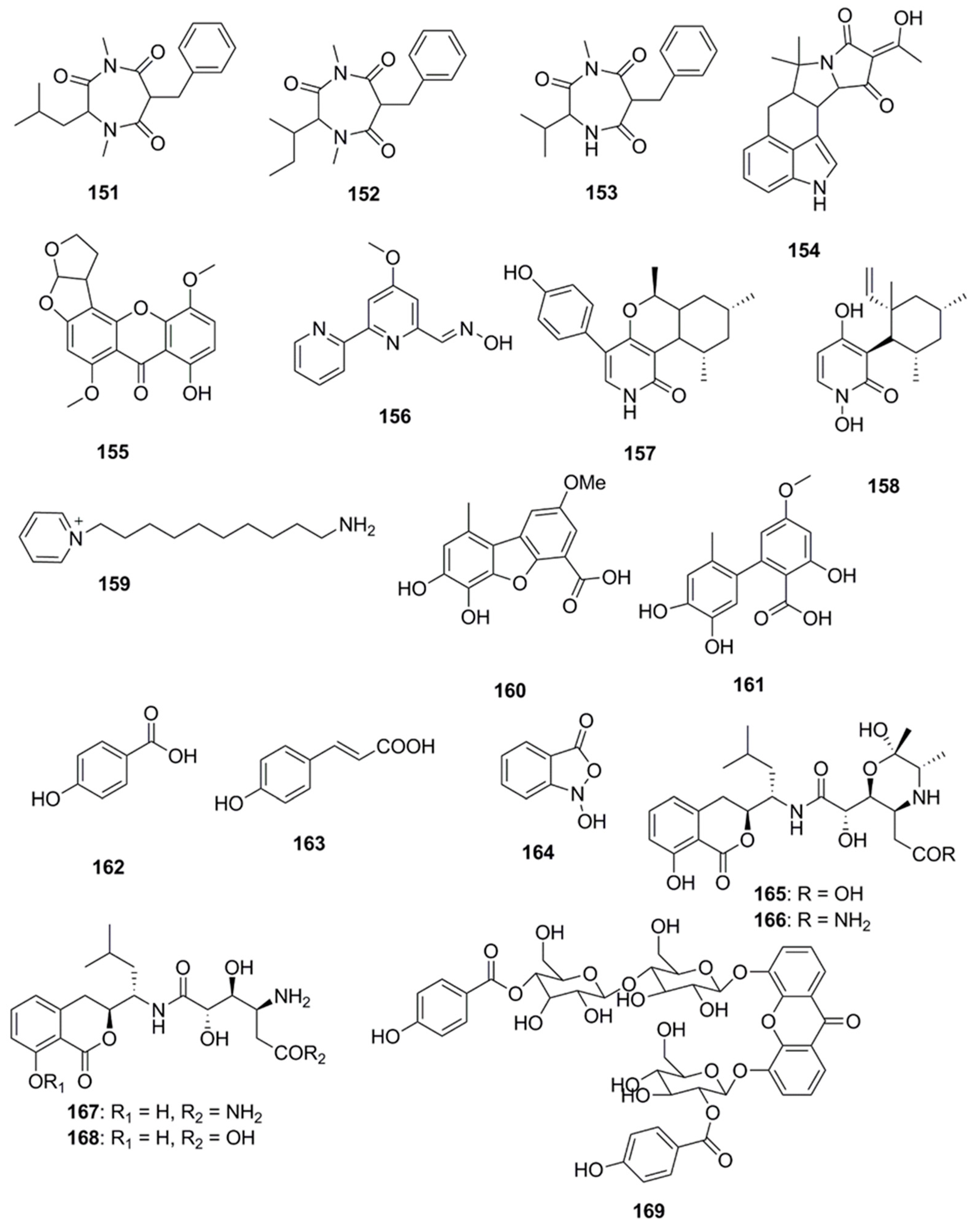
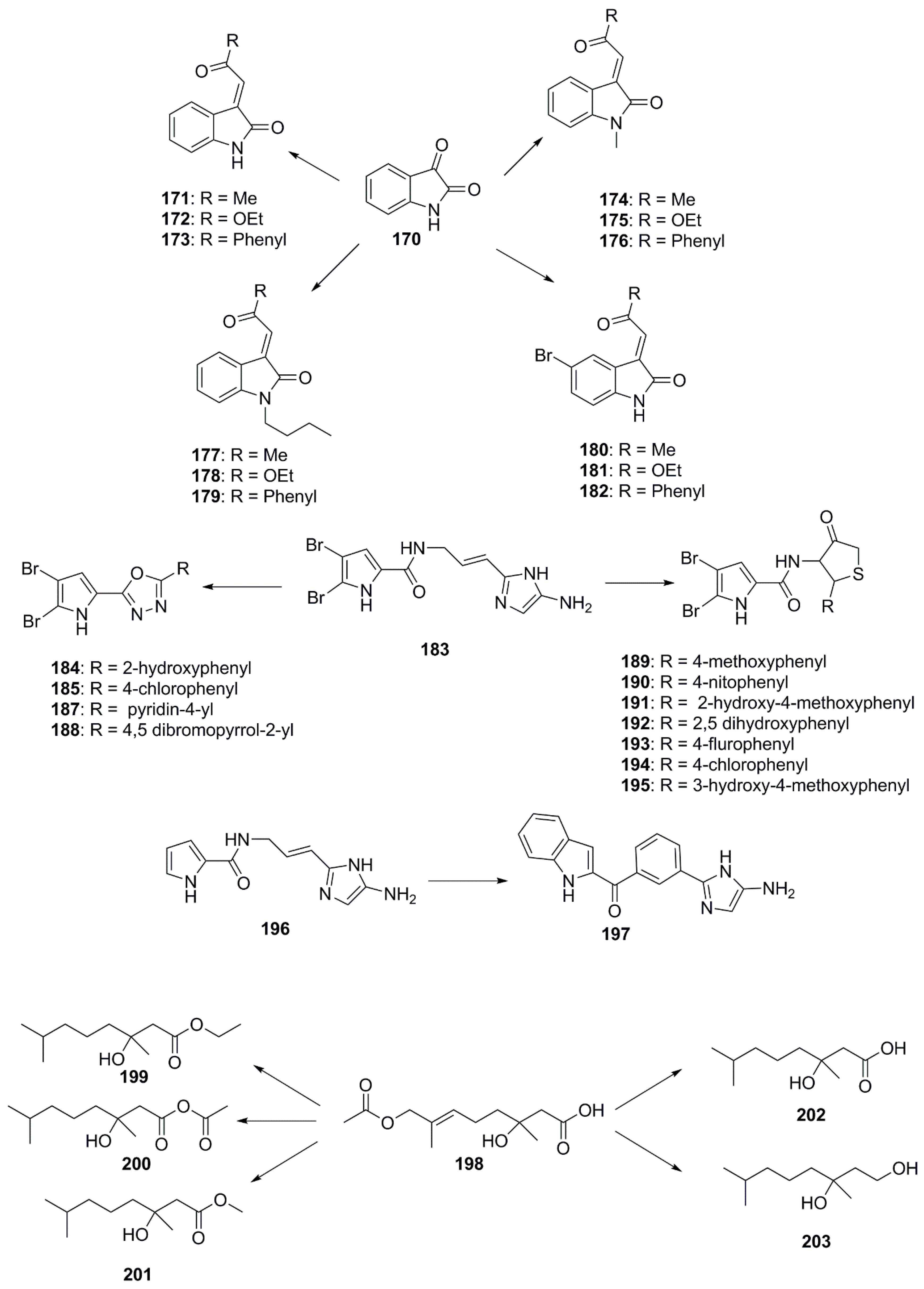
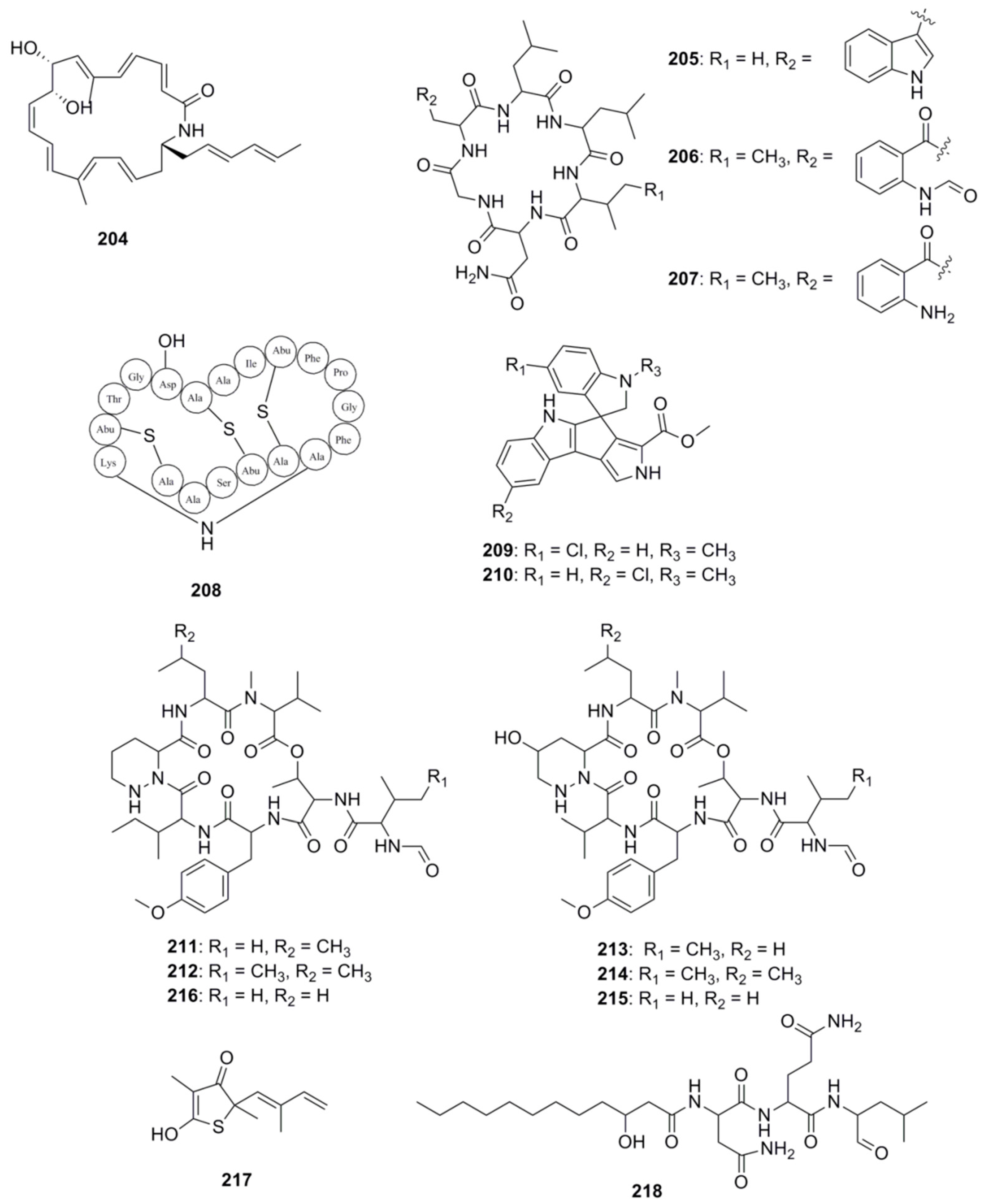
| Compound | Source | Activity Against Pathogen |
|---|---|---|
| Alkaloids | ||
| Pyranonigrin A (16) | Penicillium brocae MA-231 | S. aureus (MIC 0.5 µg/mL) |
| V. harveyi (MIC 0.5 µg/mL) | ||
| V. parahaemolyticus (MIC 0.5 µg/mL) | ||
| A. brassicae (MIC 0.5 µg/mL) | ||
| C. gloeosprioide (MIC 0.5 µg/mL) | ||
| Pyranonigrin F (17) | P. brocae MA-231 | S. aureus (MIC 0.5 µg/mL) |
| V. harveyi (MIC 0.5 µg/mL) | ||
| Vibrio parahaemolyticus (MIC 0.5 µg/mL) | ||
| Alternaria brassicae (MIC 0.5 µg/mL) | ||
| C. gloeosprioide (MIC 0.5 µg/mL) | ||
| Rubrumazine B (18) | E. cristatum EN-220 | Magnaporthe grisea (MIC 64 µg/mL) |
| Echinulin (19) | E. cristatum EN-220 | S. aureus (MIC 256 µg/mL) |
| Dehydroechinulin (20) | E.cristatum EN-220 | S. aureus (MIC 256 µg/mL) |
| Variecolorin H (21) | E. cristatum EN-220 | S. aureus (MIC 256 µg/mL) |
| Cristatumin A (22) | E.cristatum | E. coli (MIC 64 µg/mL) |
| S. aureus (MIC 8 µg/mL) | ||
| Cristatumin D (23) | E. cristatum | S. aureus (Zone of inhibition 8 mm at 100 µg/disk) |
| Tardioxopiperazine A (24) | E. cristatum | E. coli (MIC 64 µg/mL) |
| S. aureus (MIC 8 µg/mL) | ||
| Hemimycalin A (27) | Hemimycale arabica | E. coli (Inhibition zone 18 mm at 100 µg/disk) |
| C. albicans (Inhibition zone 22 mm at 100 µg/disk) | ||
| Hemimycalin B (28) | H. arabica | E. coli (Inhibition zone 10 mm at 100 µg/disk) |
| C. albicans (Inhibition zone 14 mm at 100 µg/disk) | ||
| (Z)-5-(4-hydroxybenzylidene)imidazolidine-2,4-dione (29) | H. arabica | E. coli (Inhibition zone 20 mm at 100 µg/disk) |
| C. albicans (Inhibition zone 20 mm at 100 µg/disk) | ||
| Peniciadametizine A (30) | Penicillium adametzioides | A. brassicae (MIC 4 µg/mL) |
| Peniciadametizine B (31) | P. adametzioides | A. brassicae (MIC 32 µg/mL) |
| Penicibrocazine B (33) | P. brocae | S. aureus (MIC 32 µg/mL) |
| G.graminis (MIC 0.25 µg/mL) | ||
| Penicibrocazine C (34) | P. brocae | S. aureus (MIC 0.25 µg/mL) |
| M. luteus (MIC 0.25 µg/mL) | ||
| Penicibrocazine D (35) | P. brocae | S. aureus (MIC 8 µg/mL) |
| G.graminis (MIC 8 µg/mL) | ||
| Penicibrocazine E (36) | P. brocae | G. graminis (MIC 0.25 µg/mL) |
| Crambescidin 800 (37) | Clathria cervicornis | A. baumannii (MIC 2 µg/mL) |
| K. pneumonia (MIC 1 µg/mL) | ||
| P. aeruginosa. (MIC 1 µg/mL) | ||
| Xinghaiamine A (38) | Streptomyces xinghaiensis | S. aureus (MIC 0.69 µM) |
| B. subtilis (MIC 0.35 µM) | ||
| E. coli (MIC 0.17 µM) | ||
| A. baumanii (MIC 2.76 µM) | ||
| P. aeruginosa (MIC 11 µM) | ||
| MRSA 5301 (MIC 5.52 µM) | ||
| MRSA 5438 (MIC 2.76 µM) | ||
| MRSA 5885 (MIC 5.52 µM) | ||
| Hyrtioerectine D (39) | Hyrtios sp. | C. albicans (Zone of inhibition 17 mm at 100 µg/disk) |
| S. aureus (Zone of inhibition 20 mm at 100 µg/disk) | ||
| P. aeruginosa (Zone of inhibition 9 mm at 100 µg/disk) | ||
| Hyrtioerectine E (40) | Hyrtios sp. | C. albicans (Zone of inhibition 19 mm at 100 µg/disk) |
| S. aureus (Zone of inhibition 10 mm at 100 µg/disk) | ||
| P. aeruginosa (Zone of inhibition 9 mm at 100 µg/disk) | ||
| Hyrtioerectine F (41) | Hyrtios sp. | C. albicans (Zone of inhibition 14 mm at 100 µg/disk) |
| S. aureus (Zone of inhibition 16 mm at 100 µg/disk) | ||
| P. aeruginosa (Zone of inhibition 9 mm at 100 µg/disk) | ||
| Ageloxime B (42) | Agelas mauritiana | C. neoformans (MIC 5 µg/mL) |
| S. aureus (MIC 7 µg/mL) | ||
| Ageloxime D (43) | A. mauritiana | C. neoformans (MIC 6 µg/mL) |
| Zamamidine D (44) | Amphimedon sp. | E. coli (MIC 32 µg/mL) |
| S. aureus (MIC 8 µg/mL) | ||
| B. subtilis (MIC 8 µg/mL) | ||
| M. luteus (MIC 8 µg/mL) | ||
| A. niger (MIC 16 µg/mL) | ||
| T. mentagrophytes (MIC 8 µg/mL) | ||
| C. albicans (MIC 16 µg/mL) | ||
| C. neoformans (MIC 2 µg/mL) | ||
| Adametizine A (45) | P. adametzioides AS-53 | S.aureus (MIC 8 µg/mL) |
| A. hydrophilia (MIC 8 µg/mL) | ||
| V. harveyi (MIC 32 µg/mL) | ||
| V. parahaemolyticus (MIC 8 µg/mL) | ||
| G. graminis (MIC 16 µg/mL) | ||
| Adametizine B (46) | P.adametzioides AS-53 | S. aureus (MIC 64 µg/mL) |
| Iso-Agelasidine B (47) | A. nakamurai | C. albicans (MIC 2.34 µg/mL) |
| (−)-Agelasidine C (48) | A. nakamurai | C. albicans (MIC 2.34 µg/mL) |
| Iso-agelasine C (49) | A. nakamurai | S. aureus (MIC 75 µg/mL) |
| E. coli (MIC 150 µg/mL) | ||
| P. vulgaris (MIC 19 µg/mL) | ||
| C. albicans (MIC 5 µg/mL) | ||
| Agelasine B (50) | A. nakamurai | P. vulgaris (MIC 19 µg/mL) |
| C. albicans (MIC 2 µg/mL) | ||
| Agelasine J (51) | A. nakamurai | S. aureus (MIC 75 µg/mL) |
| E. coli (MIC 75 µg/mL) | ||
| P. vulgaris (MIC 9 µg/mL) | ||
| C. albicans (MIC 0.6 µg/mL) | ||
| Nemoechine G (52) | A. nakamurai | S. aureus (MIC 150 µg/mL) |
| E. coli (MIC 75 µg/mL) | ||
| P. vulgaris (MIC 9 µg/mL) | ||
| C. albicans (MIC 0.6 µg/mL) | ||
| Brevianamide F (53) | Penicillium vinaceum | C. albicans (Zone of inhibition 25 mm at 100 µg/disk) |
| S. aureus (Zone of inhibition 19 mm at 100 µg/disk) | ||
| N-(2-hydroxyphenyl)-2-phenazinamine (54) | Nocardia dassonvillei | C. albicans (MIC of 64 µg/mL) |
| Terpenoids | ||
| Puupehenol (55) | Dactylospongia sp. | S. aureus (Zone of inhibition 4 mm at 10 µg/disk) |
| B. cereus (Zone of inhibition 4 mm at 10 µg/disk) | ||
| Puupehenone (56) | Dactylospongia sp. | S. aureus (Zone of inhibition 3 mm at 10 µg/disk) |
| B. cereus (Zone of inhibition 3 mm at 10 µg/disk) | ||
| Penicibilaene A (57) | Penicillium bilaiae | C. gloeosporioides (MIC 1 µg/mL) |
| Penicibilaene B (58) | P. bilaiae | C. gloeosporioides (MIC 0.1 µg/mL) |
| Aspergillusene A (59) | Aspergillus sydowii | K. pneumonia (MIC 21 µM) |
| A. hydrophila (MIC 4.3 µM) | ||
| (Z)-5-(Hydroxymenthyl)-2-(6′)-methylhept-2′-en-2′-yl)-phenol (60) | A. sydowii | K. pneumonia (MIC 11 µM) |
| Sydonic acid (61) | A. sydowii | E. faecalis (MIC 19 µM) |
| 12-hydroxy isolaurene (62) | Laurencia obtuse | B. subtilis (MIC 46 µg/mL) |
| S. aureus (MIC 52 µg/mL) | ||
| 8,11-dihydro-12-hydroxy isolaurene (63) | L. obtuse | C. albicans (MIC 120 µg/mL) |
| A. fumigatus (MIC 200 µg/mL) | ||
| B. subtilis (MIC 39 µg/mL) | ||
| S. aureus (MIC 31 µg/mL) | ||
| Isolauraldehyde (64) | L. obtuse | C. albicans (MIC 70 µg/mL) |
| A. fumigatus (MIC 100 µg/mL) | ||
| B. subtilis (MIC 35 µg/mL) | ||
| S. aureus (MIC 27 µg/mL) | ||
| Napyradiomycin 1 (65) | Streptomyces strain | MRSA (MIC 16 µg/mL) |
| Napyradiomycin 2 (66) | Streptomyces strain | MRSA (MIC 64 µg/mL) |
| Napyradiomycin B2 (67) | Streptomyces strain | MRSA (MIC 32–64 µg/mL) |
| Napyradiomycin B3 (68) | Streptomyces strain | MRSA (MIC 2 µg/mL) |
| Napyradiomycin B4 (69) | Streptomyces strain | MRSA (MIC 32 µg/mL) |
| Dixiamycins A (70) | Streptomyces sp. | E. coli (MIC 8 µg/mL) |
| S. aureus (MIC 8 µg/mL) | ||
| B. subtilis (MIC 16 µg/mL) | ||
| B. thuringensis (MIC 4 µg/mL) | ||
| Dixiamycins B (71) | Streptomyces sp. | E. coli (MIC 8 µg/mL) |
| S. aureus (MIC 16 µg/mL) | ||
| B. subtilis (MIC 16 µg/mL) | ||
| B. thuringensis (MIC 8 µg/mL) | ||
| Oxiamycin (72) | Streptomyces sp. | E. coli (MIC 64 µg/mL) |
| S. aureus (MIC 128 µg/mL) | ||
| Chloroxiamycin (73) | Streptomyces sp. | E. coli (MIC 64 µg/mL) |
| S. aureus (MIC 64 µg/mL) | ||
| B. thuringensis (MIC 64 µg/mL) | ||
| Xiamycin A (74) | Streptomyces sp. | E. coli (MIC 64 µg/mL) |
| S. aureus (MIC 64 µg/mL) | ||
| Sarcotrocheliol acetate (75) | Sarcophyton trocheliophorum | S. aureus (MIC 1.7 µM) |
| MRSA (MIC 1.7 µM) | ||
| Acinetobacter pp. (MIC 4.3 µM) | ||
| Sarcotrocheliol (76) | S. trocheliophorum | S. aureus (MIC 1.5 µM) |
| MRSA (MIC 3.0 µM) | ||
| Acinetobacter sp. (MIC 3.0 µM) | ||
| Cembrene-C (77) | S. trocheliophorum | C. albicans (MIC 0.7 µM) |
| A. flavus (MIC 0.7 µM) | ||
| Sarcophine (78) | S. trocheliophorum | S. aureus (MIC 9.4 µM) |
| MRSA (MIC 9.4 µM) | ||
| Acinetobacter sp. (MIC 9.4 µM) | ||
| Palustrol (79) | S. trocheliophorum | S. aureus (MIC 6.6 µM) |
| MRSA (MIC 6.6 µM) | ||
| Acinetobacter sp. (MIC 11.1 µM) | ||
| epi-Ilimaquinone (80) | Hippospongia sp. | MRSA (MIC 63 µg/mL) |
| S. aureus (MIC 31 µg/mL) | ||
| Vancomycin resistant E. faecium (MIC 16 µg/mL) | ||
| Amphotericin-resistant C. albicans (MIC 125 µg/mL) | ||
| Penicillosides A (81) | Penicillium sp. | C. albicans (inhibition zone 23 mm at 100 µg/disk) |
| Penicillosides B (82) | Penicillium sp. | S. aureus (inhibition zone 19 mm at 100 µg/disk) |
| E. coli (inhibition zone 20 mm at 100 µg/disk) | ||
| Ieodoglucomide A (83) | Bacillus licheniformis | S. aureus (MIC 8 µg/mL) |
| B. subtilis (MIC 16 µg/mL) | ||
| B. cereus (MIC 16 µg/mL) | ||
| S. typi (MIC 16 µg/mL) | ||
| E. coli (MIC 8 µg/mL) | ||
| P. aeruginosa (MIC 8 µg/mL) | ||
| A. niger (MIC 32 µg/mL) | ||
| C. albicans (MIC 32 µg/mL) | ||
| Ieodoglucomide B (84) | B. licheniformis | S. aureus (MIC 8 µg/mL) |
| B. subtilis (MIC 16µg/mL) | ||
| B. cereus (MIC 8 µg/mL) | ||
| S. typi (MIC 16 µg/mL) | ||
| E. coli (MIC 16µg/mL) | ||
| P. aeruginosa (MIC 8 µg/mL) | ||
| A. niger (MIC 32 µg/mL) | ||
| C. albicans (MIC 16 µg/mL) | ||
| Iedoglucomide C (85) | Bacillus licheniformis | S. aureus (MIC 0.03 µM) |
| B. subtilis (MIC 0.03 µM) | ||
| B. cereus (MIC 0.01 µM) | ||
| S. typi (MIC 0.01 µM) | ||
| E. coli (MIC 0.01 µM) | ||
| P. aeruginosa (MIC 0.01 µM) | ||
| A. niger (MIC 0.05 µM) | ||
| R. solani (MIC 0.05 µM) | ||
| C. acutatum (MIC 0.03 µM) | ||
| B. cenerea (MIC 0.03 µM) | ||
| C. albicans (MIC 0.03 µM) | ||
| Iedoglycoloipd (86) | B. licheniformis | S. aureus (MIC 0.03 µM) |
| B. subtilis (MIC 0.05 µM) | ||
| B. cereus (MIC 0.03 µM) | ||
| S. typi (MIC 0.05 µM) | ||
| E. coli (MIC 0.03 µM) | ||
| P. aeruginosa (MIC 0.03 µM) | ||
| A. niger (MIC 0.03 µM) | ||
| R. solani (MIC 0.05 µM) | ||
| C. acutatum (MIC 0.03 µM) | ||
| B. cenerea (MIC 0.03 µM) | ||
| C. albicans (MIC 0.05 µM) | ||
| Gageotetrin A (87) | Bacillus subtilis | C. acutatum (MIC 0.03 µM) |
| B. cinera. (MIC 0.03 µM) | ||
| S. aureus (MIC 0.03 µM) | ||
| B. subtilis (MIC 0.03 µM) | ||
| Gageotetrin B (88) | B. subtilis | C. acutatum (MIC 0.01 µM) |
| B. cinera. (MIC 0.01 µM) | ||
| S. aureus (MIC 0.04 µM) | ||
| B. subtilis (MIC 0.02 µM) | ||
| Gageotetrin C (89) | B. subtilis | C. acutatum (MIC 0.02 µM) |
| B. cinera (MIC 0.01 µM) | ||
| S. aureus (MIC 0.04 µM) | ||
| B. subtilis (MIC 0.04 µM) | ||
| Lauramide diethanolamine (90) | Streptomyces sp. | B. subtilis (MIC 0.055 µg/mL) |
| E. coli (MIC 0.055 µg/mL) | ||
| P. aeruginosa (MIC 0.011 µg/mL) | ||
| S. aureus (MIC 0.011 µg/mL) | ||
| S. cerevisiae (MIC 0.022 µg/mL) | ||
| Linieodolide A (91) | Bacillus sp. | B. subtilis (MIC 8 µg/mL) |
| E. coli (MIC 8 µg/mL) | ||
| S. cerevisiae (MIC 32 µg/mL) | ||
| Linieodolide B (92) | Bacillus sp. | B. subtilis (MIC 64 µg/mL) |
| E. coli (MIC 64 µg/mL) | ||
| S. cerevisiae (MIC 128 µg/mL) | ||
| Dysiroid A (93) | Dysidea sp. | S. aureus ATCC 29213 (MIC 4 µg/mL) |
| S. aureus ATCC 43300(MIC 8 µg/mL) | ||
| E. faecalis ATCC 29212(MIC 4 µg/mL) | ||
| B. licheniformis ATCC 10716 (MIC 16 µg/mL) | ||
| Dysiroid B (94) | Dysidea sp. | S. aureus ATCC 29213 (MIC 4 µg/mL) |
| S. aureus ATCC 43300(MIC 4 µg/mL) | ||
| E. faecalis ATCC 29212(MIC 4 µg/mL) | ||
| B. licheniformis ATCC 10716 (MIC 8 µg/mL) | ||
| Halistanol sulfate A (95) | Petromica ciocalyptoides | S. mutans clinical isolate (MIC 15 µg/mL) |
| S. mutans UA159 (MIC 15 µg/mL) | ||
| Peptides | ||
| Rodriguesines A and B (96 and 96a) | Didemnum sp. | S. mutans clinical isolate (MIC 31 µg/mL) |
| S. mutans UA159 (MIC 62 µg/mL) | ||
| Cyclo-(l-valyl-d-proline) (97) | Rheinheimera japonica | S. aureus (Zone of inhibition 17 mm at 0.5 mg/mL) |
| V. parahaemolyticus (MIC 0.05 µg/mL) | ||
| V. vulnificus (MIC 5 µg/mL) | ||
| M. luteus (MIC 5 µg/mL) | ||
| Cyclo-(l-phenylalanyl-d-proline (98) | R. japonica | S. aureus (Zone of inhibition 15 mm at 0.5 mg/mL) |
| Cyclo-(S-Pro-R-Leu) (99) | Haliclona oculata | V. parahaemolyticus (MIC 0.5 µg/mL) |
| V. vulnificus (MIC 5 µg/mL) | ||
| B. cereus (MIC 0.05 µg/mL) | ||
| Isaridin G (100) | Beauveria felina EN-135 | E. coli (MIC 64 µg/mL) |
| Desmethylisaridin G (101) | B. felina EN-135 | E. coli (MIC 64 µg/mL) |
| Desmethylisaridin C1(102) | B. felina EN-135 | E. coli (MIC 8 µg/mL) |
| Isaridin E (103) | B. felina EN-135 | E. coli (MIC 16 µg/mL) |
| Desotamide (104) | Streptomyces scopuliridis SCSIO ZJ46 | S. pnuemoniae (MIC 13 µg/mL) |
| S. aureus (MIC 16 µg/mL) | ||
| MRSE (MIC 32 µg/mL) | ||
| Desotamide B (105) | S. scopuliridis SCSIO ZJ46 | S. pnuemoniae (MIC 13 µg/mL) |
| S. aureus (MIC 16 µg/mL) | ||
| MRSE (MIC 32 µg/mL) | ||
| Halogenated Compounds | ||
| 2-(2′,4′-dibromophenoxy)-3,5-dibromophenol (106) | Dysidea granulosa | MSSA (MIC 1 mg/L) |
| L. monocytogenes (MIC 2 mg/L) | ||
| B. cereus (MIC 0.1 mg/L) | ||
| C. diffiile (MIC 4 mg/L) | ||
| MRSA (MIC 0.1 mg/L) | ||
| Salmonella sp. (MIC 1 mg/L) | ||
| E. coli O157:H7 (MIC 8 mg/L) | ||
| Pseudomonas (MIC 4 mg/L) | ||
| K. pneumoniae (MIC 0.1 mg/L) | ||
| N. gonorrhoeae (MIC 2 mg/L) | ||
| A. baumannii (MIC 16 mg/L) | ||
| C. jejuni (MIC 5 mg/L) | ||
| 2-(2′,4′-dibromophenoxy)-3,4,5-tribromophenol (107) | Dysidea granulosa | L. monocytogenes (MIC 0.1 mg/L) |
| C. diffiile (MIC 10 mg/L) | ||
| MRSA (MIC 0.1 mg/L) | ||
| Salmonella sp. (MIC 10 mg/L) | ||
| C. jejuni (MIC 5 mg/L) | ||
| 2-(2′,4′-dibromophenoxy)-4,6-dibromophenol (108) | Dysidea sp. | L. monocytogenes (MIC 1 mg/L) |
| B. cereus (MIC 5 mg/L) | ||
| S. pneumoniae (MIC 5 mg/L) | ||
| C. diffiile (MIC 10 mg/L) | ||
| MRSA (MIC 1 mg/L) | ||
| Salmonella sp. (MIC 10 mg/L) | ||
| E. coli O157:H7 (MIC 10 mg/L) | ||
| K. pneumoniae (MIC 5 mg/L) | ||
| C. jejuni (MIC 5 mg/L) | ||
| Aplysamine 8 (109) | Pseudoceratina purpurea | E. coli (MIC 125 µg/mL) |
| S. aureus (MIC 31 µg/mL) | ||
| Sphaerodactylomelol (113) | Sphaerococcus coronopifolius | S. aureus (MIC 96 μM) |
| Bromosphaerol (114) | Sphaerococcus coronopifolius | S. aureus (MIC 22 µM) |
| 12R-hydroxybromosphaerol (115) | Sphaerococcus coronopifolius | S. aureus (MIC 6 μM) |
| 6-bromoindolyl-3-acetic acid (116) | Pseudoalteromonas flavipulchra | V. anguillarum (MIC 0.25 mg/mL) |
| Nagelamide X (117) | Agelas sp. | S. aureus (MIC 8 μg/mL) |
| M. luteus (MIC 8 µg/mL) | ||
| A. niger (IC50 32 µg/mL) | ||
| T. mentagrophytes (IC50 16 µg/mL) | ||
| C. albicans (IC50 2 µg/mL) | ||
| Nagelamide Y (118) | Agelas sp. | C. albicans (IC50 2 µg/mL) |
| Nagelamide Z (119) | Agelas sp. | S. aureus (MIC 16 µg/mL) |
| M. luteus (MIC 8 µg/mL) | ||
| A. niger (IC50 4 µg/mL) | ||
| T. mentagrophytes (IC50 4 µg/mL) | ||
| C. albicans (IC50 0.25 µg/mL) | ||
| C. neoformans (IC50 2 µg/mL) | ||
| Ianthelliformisamine A (120) | Suberea ianthelliformis | P. aeruginosa (IC50 7 µM) |
| Ianthelliformisamine C (122) | Suberea ianthelliformis | P. aeruginosa (IC50 9 µM) |
| S. aureus (IC50 4µM) | ||
| Polyketides | ||
| N-acetyl-N-demethylmayamycin (123) | Streptomyces sp. 182SMLY | MRSA (MIC 20 µM) |
| Lindgomycin (124) | Lindgomycetaceae strain | B. subtilis (MIC 2.2 µM) |
| X. campestris (MIC 17.8 µM) | ||
| S. epidermidis (MIC 4.6 µM) | ||
| S. aureus (MIC 2.7 µM) | ||
| S. aureus (MRSA) (MIC 5.1 µM) | ||
| C. albicans (MIC 5.7 µM) | ||
| S. tritici (MIC 5.1 µM) | ||
| P. acnes (MIC 4.7 µM) | ||
| Ascosetin (125) | Lindgomycetaceae strain | B. subtilis (MIC 3.4 µM) |
| X. campestris (MIC 14.8 µM) | ||
| S. epidermidis (MIC 6.3 µM) | ||
| S. aureus (MIC 2.9 µM) | ||
| S. aureus (MRSA) (MIC 3.2 µM) | ||
| C. albicans (MIC 8 µM) | ||
| S. tritici (MIC 10 µM) | ||
| P. acnes (MIC 2.8 µM) | ||
| Manadodioxan E (127) | Plakortis bergquistae | E. coli (Zone of inhibition 16 mm at 10 µg/disk) |
| B. cereus (Zone of inhibition 9 mm at 10 µg/disk) | ||
| 2-(7-(2-Ethylbutyl)-2,3,4,4a,6,7-hexahydro-2-oxopyrano-[3,2b]-pyran-3-yl)-ethyl benzoate (128) | B. subtilis MTCC 10407 | V. parahemolyticus (Zone of inhibition 11 mm at 10 µg/disk) |
| V. vulnificus (Zone of inhibition 13 mm at 10 µg/disk) | ||
| A. hydrophilla (Zone of inhibition 18 mm at 10 µg/disk) | ||
| 2-((4Z)-2-ethyl-octahydro-6-oxo-3-((E)-pent-3-enylidene)-pyrano-[3,2b]-pyran-7-yl)-ethyl benzoate (129) | B. subtilis MTCC 10407 | V. parahemolyticus (Zone of inhibition 11 mm at 10 µg/disk/mL) |
| V. vulnificus (Zone of inhibition 14 mm at 10 µg/disk) | ||
| A. hydrophilla (Zone of inhibition 15 mm at 10 µg/disk) | ||
| 3-(methoxycarbonyl)-4-(5-(2-ethylbutyl)-5,6-dihydro-3-methyl-2H-pyran-2-yl)-butyl benzoate (130) | Sargassum myriocystum | V. parahemolyticus (Zone of inhibition 7 mm at 10 µg/disk) |
| V. vulnificus (Zone of inhibition 7 mm at 10 µg/disk) | ||
| A. hydrophilla (Zone of inhibition 8 mm at 10 µg/disk) | ||
| 2-(8-butyl-3-ethyl-3,4,4a,5,6,8ahexahydro-2H-chromen-6-yl)-ethyl benzoate (131) | Sargassum myriocystum | V. parahemolyticus (Zone of inhibition 9 mm at 10 µg/disk) |
| V. vulnificus (Zone of inhibition 8 mm at 10 µg/disk | ||
| A. hydrophilla (Zone of inhibition 7 mm at 10 µg/disk) | ||
| Fradimycin A (132) | Streptomyces fradiae | S. aureus (MIC 6 µg/mL) |
| Fradimycin B (133) | Streptomyces fradiae | S. aureus (MIC 2 µg/mL) |
| Glycosylated Macrolactin A1 (134) | Streptomyces sp. | B. subtilis (MIC 0.027 µg/mL) |
| E. coli (MIC 0.022 µg/mL) | ||
| P. aeruginosa (MIC 0.055 µg/mL) | ||
| S. aureus (MIC 0.055 µg/mL) | ||
| S. cerevisiae (MIC 0.022 µg/mL) | ||
| Glycosylated Macrolactin B1 (135) | Streptomyces sp. | B. subtilis (MIC 0.055 µg/mL) |
| E. coli (MIC 0.022 µg/mL) | ||
| P. aeruginosa (MIC 0.055 µg/mL) | ||
| S. aureus (MIC 0.055 µg/mL) | ||
| S. cerevisiae (MIC 0.022 µg/mL) | ||
| Macrolactin X (136) | Bacillus sp. | B. subtilis (MIC 16 µg/mL) |
| E. coli (MIC 16 µg/mL) | ||
| S. cerevisiae (MIC 64 µg/mL) | ||
| Macrolactin Y (137) | Bacillus sp. | B. subtilis (MIC 16 µg/mL) |
| E. coli (MIC 32 µg/mL) | ||
| S. cerevisiae (MIC 64 µg/mL) | ||
| Macrolactin Z (138) | Bacillus sp. | B. subtilis (MIC 16 µg/mL) |
| E. coli (MIC 32 µg/mL) | ||
| S. cerevisiae (MIC 64 µg/mL) | ||
| Macrolactinic acid (139) | Bacillus sp. | B. subtilis (MIC 64 µg/mL) |
| E. coli (MIC 32 µg/mL) | ||
| S. cerevisiae (MIC 128 µg/mL) | ||
| Heronamycin A (140) | Streptomyces sp. | B. subtilis ATCC 6051 (MIC 18 µM) |
| B. subtilis ATCC (MIC 14 µM) | ||
| Herbimycin A (141) | Streptomyces sp. | B. subtilis ATCC 6051 (MIC 5 µM) |
| B. subtilis ATCC (MIC 5 µM) | ||
| C. albicans ATCC 90028 (MIC 7 µM) | ||
| 3-(Octahydro-9-isopropyl-2H-benzo[h]chromen-4-yl)-2-methylpropyl benzoate (142) | Bacillus amyloliquefaciens | V. parahemolyticus (Zone of inhibition 14 mm at 25 µg/disk) |
| V. vulnificus (Zone of inhibition 16 mm at 25 µg/disk) | ||
| A. hydrophilla (Zone of inhibition 18 mm at 25 µg/disk) | ||
| Methyl 8-(2-(benzoyloxy)ethyl)-hexahydro-4-((E)-pent-2-enyl)-2H-chromene-6-carboxylate (143) | Bacillus amyloliquefaciens | V. parahemolyticus (Zone of inhibition 12 mm at 25 µg/disk) |
| V. vulnificus (Zone of inhibition 14 mm at 25 µg/disk) | ||
| A. hydrophilla (Zone of inhibition 16 mm at 25 µg/disk) | ||
| 3-O-(α-d-ribofuranosyl)questin (144) | Eurotium cristatum | E. coli (MIC 32 µg/mL) |
| Eurorubrin (145) | Eurotium cristatum | E. coli (MIC 64 µg/mL) |
| Isocoumarins | ||
| Penicisimpins A (146) | Penicillium simplicissimum MA-332 | A. hydrophilia (MIC > 64µg/mL) |
| E. coli (MIC 4 µg/mL) | ||
| M. luteus (MIC 8 µg/mL) | ||
| P. aeruginosa (MIC 4 µg/mL) | ||
| V. alginolyticus (MIC 8µg/mL) | ||
| V. harveyi (MIC 4 µg/mL) | ||
| V. parahaemolyticus (MIC 4 µg/mL) | ||
| C. gloeosprioides (MIC 4 µg/mL) | ||
| Penicisimpins B (147) | Penicillium simplicissimum MA-332 | A. hydrophilia (MIC 32 µg/mL) |
| E. coli (MIC 32 µg/mL) | ||
| M. luteus (MIC 64 µg/mL) | ||
| P. aeruginosa (MIC 32 µg/mL) | ||
| V. alginolyticus (MIC 32 µg/mL) | ||
| V. harveyi (MIC 16 µg/mL) | ||
| V. parahaemolyticus (MIC 32 µg/mL) | ||
| C. gloeosprioides (MIC 16 µg/mL) | ||
| Penicisimpins C (148) | Penicillium simplicissimum MA-332 | A. hydrophilia (MIC 16 µg/mL) |
| E. coli (MIC 8 µg/mL) | ||
| M. luteus (MIC 16 µg/mL) | ||
| P. aeruginosa (MIC 8 µg/mL) | ||
| V. alginolyticus (MIC 16 µg/mL) | ||
| V. harveyi (MIC 8 µg/mL) | ||
| V. parahaemolyticus (MIC 8 µg/mL) | ||
| C. gloeosprioides (MIC 8 µg/mL | ||
| Citreoisocoumarin (149) | Penicillium vinaceum | S. aureus (Zone of inhibition 19 mm at 100 µg/disk) |
| Nucleosides | ||
| Rocheicoside A (150) | Streptomyces rochei 06CM016 | E. coli O157:H7 (MIC 16 µg/mL) |
| MRSA DSM (MIC 8 µg/mL) | ||
| C. albicans DSM (MIC 4 µg/mL) | ||
| Plicacetin (151) | Streptomyces rochei 06CM016 | E. coli O157:H7 (MIC 8 µg/mL) |
| MRSA DSM (MIC 4 µg/mL) | ||
| C. albicans DSM (MIC 8 µg/mL) | ||
| Norplicacetin (152) | Streptomyces rochei 06CM016 | E. coli O157:H7 (MIC 16 µg/mL) |
| MRSA DSM (MIC 8 µg/mL) | ||
| C. albicans DSM (MIC 4 µg/mL) | ||
| p-amino benzamido uracil (153) | Streptomyces rochei 06CM016 | E. coli O157:H7 (MIC 16 µg/mL) |
| MRSA DSM (MIC 16 µg/mL) | ||
| C. albicans DSM (MIC 8 µg/mL) | ||
| Cytosamine (154) | Streptomyces rochei 06CM016 | E. coli O157:H7 (MIC 16 µg/mL) |
| MRSA DSM (MIC 16 µg/mL) | ||
| C. albicans DSM (MIC 8 µg/mL) | ||
| Miscellaneous Compounds | ||
| Terretrione A (151) | Penicillium vinaceum | C. albicans (Zone of inhibition 27 mm at 100 µg/disk) |
| Terretrione C (152) | Penicillium sp. CYE-87 | C. albicans (MIC 32 µg/mL) |
| Terretrione D (153) | Penicillium sp. CYE-87 | C. albicans (MIC 32 µg/mL) |
| α-Cyclopiazonic acid (154) | Penicillium vinaceum | E. coli (Zone of inhibition 20 mm at 100 µg/disk) |
| 5-methoxydihydrosterigmato-cystin (155) | Aspergillus versicolor | S. aureus (MIC 13 µg/mL) |
| B. subtilis (MIC 3 µg/mL) | ||
| Caerulomycin A (156) | Actinoalloateichus cyanogriseus | C. albicans (MIC 0.8–1.6 µg/mL) |
| C. albicans CO9 (MIC 0.8–1.6 µg/mL) | ||
| C. glabrata HO5FlucR (MIC 0.4–0.8 µg/mL) | ||
| C. krusei GO3FlucR (MIC 0.8–1.6 µg/mL) | ||
| Trichodin A (157) | Trichoderma sp. | B. subtilis (IC50 27 µM) |
| S. epidermidis (IC50 24 µM) | ||
| C. albicans (IC50 25 µM) | ||
| Pyridoxatin (158) | Trichoderma sp. | B. subtilis (IC50 4 µM) |
| S. epidermidis (IC50 4 µM) | ||
| S aureus (MRSA), (IC50 4 µM) | ||
| C. albicans (IC50 26 µM) | ||
| T. rubrum (IC50 4 µM) | ||
| 1-(10-Aminodecyl) Pyridinium (159) | Amycolatopsis alba | B. subtilis NCIM 2439 (MIC > 70 µg/mL) |
| B. pumilus NCIM 2327 (MIC > 90 µg/mL) | ||
| S. aureus NCIM 5021 (MIC > 160 µg/mL) | ||
| A. formicans NCIM 2319 (MIC > 150 µg/mL) | ||
| E. coli NCIM 2067 (MIC > 90 µg/mL) | ||
| Porric acid D (160) | Alternaria sp. | S. aureus (MIC 100 µg/mL) |
| Altenusin (161) | Alternaria sp. | S. aureus (MIC 25 µg/mL) |
| p-Hydroxybenzoic acid (162) | Pseudoalteromonas flavipulchra | V. anguillarum (MIC 1.25 mg/mL) |
| trans-Cinnamic acid (163) | Pseudoalteromonas flavipulchra | V. anguillarum (MIC 1.25 mg/mL) |
| N-hydroxybenzoisoxazolone (164) | Pseudoalteromonas flavipulchra | V. anguillarum (MIC 0.25 mg/mL) |
| Bacilosarcin B (165) | Bacillus subtilis | S. aureus (MIC 4 µM) |
| amicoumacin A (167) | Bacillus subtilis | S. aureus (MIC 19 µM) |
| B. subtillis (MIC 19 µM) | ||
| Microluside A (169) | Micrococcus sp. EG45 | E. faecalis JH212 (MIC 10 µM) |
| S. aureus NCTC 8325 (MIC 13 µM) | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choudhary, A.; Naughton, L.M.; Montánchez, I.; Dobson, A.D.W.; Rai, D.K. Current Status and Future Prospects of Marine Natural Products (MNPs) as Antimicrobials. Mar. Drugs 2017, 15, 272. https://doi.org/10.3390/md15090272
Choudhary A, Naughton LM, Montánchez I, Dobson ADW, Rai DK. Current Status and Future Prospects of Marine Natural Products (MNPs) as Antimicrobials. Marine Drugs. 2017; 15(9):272. https://doi.org/10.3390/md15090272
Chicago/Turabian StyleChoudhary, Alka, Lynn M. Naughton, Itxaso Montánchez, Alan D. W. Dobson, and Dilip K. Rai. 2017. "Current Status and Future Prospects of Marine Natural Products (MNPs) as Antimicrobials" Marine Drugs 15, no. 9: 272. https://doi.org/10.3390/md15090272