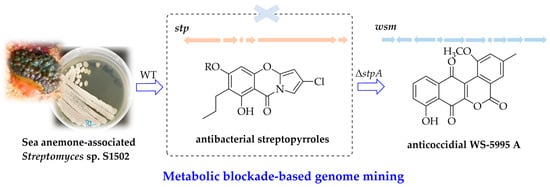
Metabolic Blockade-Based Genome Mining of Sea Anemone-Associated Streptomyces sp. S1502 Identifies Atypical Angucyclines WS-5995 A–E: Isolation, Identification, Biosynthetic Investigation, and Bioactivities
Abstract
:1. Introduction
2. Results
2.1. Identification of Streptopyrroles from Streptomyces sp. S1502 and the Construction of a Streptopyrroles-Null Mutant
2.2. Isolation and Structural Elucidation of Compounds from Streptomyces sp. S1502/Δstp1 Mutant
2.3. Characterization and Heterologous Expression of the wsm Biosynthetic Gene Cluster and Preliminary Investigation into the Genes Involved in Biosynthetic Pathways of Compounds 1–5
2.4. Bioactivities of WS-5995 A–E
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strains, Plasmids, and Culture Conditions
3.3. Genome Sequencing and Bioinformatic Analysis
3.4. Construction of the Δstp1 and Mutants Based on the Δstp1
3.5. Heterologous Expression of the wsm Gene Cluster in S. lividans SBT5
3.6. Small-Scale Fermentations of Streptomyces sp. S1502 and Its Mutants
3.7. Large-Scale Fermentations of Streptomyces sp. S1502 and Δstp1 Mutant
3.8. Cytotoxic Activities of Compounds 1–5
3.9. In Vitro Anticoccidial Activity
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, R.; Zhu, H.; Zhang, H.; Ju, J.; Li, Q.; Fu, S. Six sets of aromatic polyketides differing in size and shape derive from a single biosynthetic gene cluster. J. Nat. Prod. 2023, 86, 1512–1519. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Ding, W.; Sun, C.; Ji, X.; Ling, C.; Zhou, Z.; Chen, Z.; Chen, X.; Ju, J. Julichrome monomers from marine gastropod mollusk-associated Streptomyces and stereochemical revision of julichromes Q(3·5) and Q(3·3). Chem. Biodivers. 2020, 17, e2000057. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.H.; Je, H.W.; Kim, H.; Kang, H.S. Promoter engineering of natural product biosynthetic gene clusters in actinomycetes: Concepts and applications. Nat. Prod. Rep. 2024. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Hug, J.J.; Fu, C.; Bian, X.; Zhang, Y.; Muller, R. Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 2019, 36, 1412–1436. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ding, X.; Du, Y.; Li, Y.; Ren, W.; Lu, Y.; Shi, Y.; Sun, H.; Wang, L.; Li, Y.; et al. Genome-directed discovery of bicyclic cinnamoyl-containing nonribosomal peptides with anticoronaviral activity from Streptomyces griseus. Org. Lett. 2023, 25, 4874–4879. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Bai, J.; Yan, D.; Bao, X.; Li, W.; Liu, B.; Zhang, D.; Qi, X.; Yu, D.; Hu, Y. Genome mining combined metabolic shunting and OSMAC strategy of an endophytic fungus leads to the production of diverse natural products. Acta Pharm. Sin. B 2021, 11, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, C.; Li, Q.; Ma, J.; Ju, J. Metabolic blockade-based genome mining reveals lipochain-linked dihydro-beta-alanine synthetases involved in autucedine biosynthesis. Org. Lett. 2022, 24, 5535–5540. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yi, X.; Zhou, Z.; Yang, J.; Pei, Y.; Shi, S.; Gao, C.; Tian, X.; Ju, J.; Li, Q. Metabolic blockade-based genome mining of Streptomyces cacaoi SCSIO 68063: Isolation and identification of BE-18257 and pentaminomycin analogues. Tetrahedron 2023, 130, 133148. [Google Scholar] [CrossRef]
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef] [PubMed]
- Kharel, M.K.; Zhu, L.; Liu, T.; Rohr, J. Multi-oxygenase complexes of the gilvocarcin and jadomycin biosyntheses. J. Am. Chem. Soc. 2007, 129, 3780–3781. [Google Scholar] [CrossRef]
- Tibrewal, N.; Pahari, P.; Wang, G.; Kharel, M.K.; Morris, C.; Downey, T.; Hou, Y.; Bugni, T.S.; Rohr, J. Baeyer-Villiger C-C bond cleavage reaction in gilvocarcin and jadomycin biosynthesis. J. Am. Chem. Soc. 2012, 134, 18181–18184. [Google Scholar] [CrossRef]
- Huang, C.; Yang, C.; Zhang, W.; Zhang, L.; Zhu, Y.; Zhang, C. Discovery of an unexpected 1,4-oxazepine-linked seco-fluostatin heterodimer by inactivation of the oxidoreductase-encoding gene flsP. J. Nat. Prod. 2021, 84, 2336–2344. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Huang, C.; Fang, C.; Zhang, L.; Chen, S.; Zhang, Q.; Zhang, C.; Zhang, W. Inactivation of flavoenzyme-encoding gene flsO1 in fluostatin biosynthesis leads to diversified angucyclinone derivatives. J. Org. Chem. 2021, 86, 11019–11028. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, L.; Zhang, W.; Huang, C.; Zhu, Y.; Jiang, X.; Liu, W.; Zhao, M.; De, B.C.; Zhang, C. Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation. Nat. Commun. 2022, 13, 5386. [Google Scholar] [CrossRef]
- Tibrewal, N.; Downey, T.E.; Van Lanen, S.G.; Ul Sharif, E.; O’Doherty, G.A.; Rohr, J. Roles of the synergistic reductive O-methyltransferase GilM and of O-methyltransferase GilMT in the gilvocarcin biosynthetic pathway. J. Am. Chem. Soc. 2012, 134, 12402–12405. [Google Scholar] [CrossRef]
- Feng, Z.; Kim, J.H.; Brady, S.F. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA derived type II PKS gene cluster. J. Am. Chem. Soc. 2010, 132, 11902–11903. [Google Scholar] [CrossRef]
- Myronovskyi, M.; Luzhetskyy, A. Native and engineered promoters in natural product discovery. Nat. Prod. Rep. 2016, 33, 1006–1019. [Google Scholar] [CrossRef]
- Yang, C.; Huang, C.; Zhang, W.; Zhu, Y.; Zhang, C. Heterologous expression of fluostatin gene cluster leads to a bioactive heterodimer. Org. Lett. 2015, 17, 5324–5327. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, D.; Xu, M.; Tao, M.; Bai, L.; Deng, Z.; Pfeifer, B.A.; Jiang, M. Reconstitution of kinamycin biosynthesis within the heterologous host Streptomyces albus J1074. J. Nat. Prod. 2018, 81, 72–77. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, C.; Zhang, L. Investigation of the molecular landscape of bacterial aromatic polyketides by global analysis of type II polyketide synthases. Angew. Chem. Int. Ed. 2022, 61, e202202286. [Google Scholar] [CrossRef]
- Trew, S.J.; Wrigley, S.K.; Pairet, L.; Sohal, J.; Shanu-Wilson, P.; Hayes, M.A.; Martin, S.M.; Manohar, R.N.; Chicarelli-Robinson, M.I.; Kau, D.A.; et al. Novel streptopyrroles from Streptomyces rimosus with bacterial protein histidine kinase inhibitory and antimicrobial activities. J. Antibiot. 2000, 53, 1–11. [Google Scholar] [CrossRef]
- Alanjary, M.; Steinke, K.; Ziemert, N. AutoMLST: An automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019, 47, W276–W282. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Ding, X.B.; Brimble, M.A.; Furkert, D.P. Nitropyrrole natural products: Isolation, biosynthesis and total synthesis. Org. Biomol. Chem. 2016, 14, 5390–5401. [Google Scholar] [CrossRef]
- Jaremko, M.J.; Lee, D.J.; Opella, S.J.; Burkart, M.D. Structure and substrate sequestration in the pyoluteorin type II peptidyl carrier protein PltL. J. Am. Chem. Soc. 2015, 137, 11546–11549. [Google Scholar] [CrossRef]
- Purdy, T.N.; Kim, M.C.; Cullum, R.; Fenical, W.; Moore, B.S. Discovery and biosynthesis of tetrachlorizine reveals enzymatic benzylic dehydrogenation via an ortho-quinone methide. J. Am. Chem. Soc. 2021, 143, 3682–3686. [Google Scholar] [CrossRef]
- Qiao, Y.; Yan, J.; Jia, J.; Xue, J.; Qu, X.; Hu, Y.; Deng, Z.; Bi, H.; Zhu, D. Characterization of the biosynthetic gene vluster for the antibiotic armeniaspirols in Streptomyces armeniacus. J. Nat. Prod. 2019, 82, 318–323. [Google Scholar] [CrossRef]
- Lehr, N.A.; Adomas, A.; Asiegbu, F.O.; Hampp, R.; Tarkka, M.T. WS-5995 B, an antifungal agent inducing differential gene expression in the conifer pathogen Heterobasidion annosum but not in Heterobasidion abietinum. Appl. Microbiol. Biotechnol. 2009, 85, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Ikushima, H.; Okamoto, M.; Tanaka, H.; Ohe, O.; Kohsaka, M.; Aoki, H.; Imanaka, H. New anticoccidial antibiotics, WS-5995 A and B. I. isolation and characterization. J. Antibiot. 1980, 33, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Bai, T.; Yu, Y.; Xu, Z.; Tao, M. Construction of Streptomyces lividans SBT5 as an efficient heterologous expression host. J. Huazhong Agric. Univ. 2014, 33, 1–6. [Google Scholar]
- Yang, Z.; Liu, C.; Wang, Y.; Chen, Y.; Li, Q.; Zhang, Y.; Chen, Q.; Ju, J.; Ma, J. MGCEP 1.0: A genetic-engineered marine-derived chassis cell for a scaled heterologous expression platform of microbial bioactive metabolites. ACS Synth. Biol. 2022, 11, 3772–3784. [Google Scholar] [CrossRef] [PubMed]
- van den Belt, M.; Gilchrist, C.; Booth, T.J.; Chooi, Y.H.; Medema, M.H.; Alanjary, M. CAGECAT: The Comparative gene cluster analysis toolbox for rapid search and visualisation of homologous gene clusters. BMC Bioinform. 2023, 24, 181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, J.; Alam, M.; Wang, F.; Shang, Z.; Chen, Y.; Sun, C.; Lu, Z.; Gao, Y.; Zhang, T.; et al. Eficient mutasynthesis of “non-natural” antitubercular ilamycins with low cytotoxicity. ACS Synth. Biol. 2024, 13, 930–941. [Google Scholar] [CrossRef]
- Luo, M.; Wang, Y.; Frisch, D.; Joobeur, T.; Wing, R.; Dean, R. Melon bacterial artificial chromosome (BAC) library construction using improved methods and identification of clones linked to the locus conferring resistance to melon Fusarium wilt (Fom-2). Genome 2001, 44, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.A.; Kim, R.R.; Lawton, E.S.; Tran, J.; Lewis, S.K.; Deol, A.S.; Van Arnam, E.B. Bacterial associates of a desert specialist fungus-growing ant antagonize competitors with a nocamycin analog. ACS Chem. Biol. 2022, 17, 1824–1830. [Google Scholar] [CrossRef]
- Ji, X.; Dong, Y.; Ling, C.; Zhou, Z.; Li, Q.; Ju, J. Elucidation of the tailoring steps in julichrome biosynthesis by marine gastropod mollusk-associated Streptomyces sampsonii SCSIO 054. Org. Lett. 2020, 22, 6927–6931. [Google Scholar] [CrossRef]





| Position | Compound | ||
|---|---|---|---|
| WS-5995 E (1) | |||
| δC Type | δH Mult. (J in Hz) | HMBC | |
| 1 | 182.9, C | - | - |
| 2 | 117.9, C | - | - |
| 3 | 131.4, C | - | - |
| 3-OH | - | - | - |
| 4 | 185.0, C | - | - |
| 5 | 113.6, C | - | - |
| 6 | 161.4, C | - | - |
| 6-OH | - | 11.21, s | 113.6 (5); 123.2 (7); 137.7 (8); 161.4 (6) |
| 7 | 123.2, CH | 7.23 (d, J = 8.4 Hz) | 113.6 (5); 120.1 (9) |
| 8 | 137.7, CH | 7.64 (dd, J = 8.4, 7.9 Hz) | 133.2 (7); 161.4 (10) |
| 9 | 120.1, CH | 7.69 (d, J = 7.9 Hz) | 113.6 (5); 123.2 (7); 182.9 (1) |
| 10 | 133.2, C | - | - |
| 11 | 122.4, C | - | - |
| 12 | 157.3, C | - | - |
| 13 | 116.1, CH | 6.99, br. s | 21.9 (18); 117.9 (2); 123.5 (15); 157.3 (12) |
| 14 | 140.4, C | - | - |
| 15 | 123.5, CH | 7.53, br. s | 21.9 (18); 116.1 (13); 166.5 (19) |
| 16 | 133.2, C | - | - |
| 17 | 56.3, CH3 | 3.77, s | 157.3 (12) |
| 18 | 21.9, CH3 | 2.44, s | 123.5 (15); 116.1 (13); 140.4 (14) |
| 19 | 166.5, C | - | - |
| 20 | 61.1, CH2 | 4.17, (q, J = 7.1 Hz) | 14.2 (21); 166.5 (19) |
| 21 | 14.2, CH3 | 1.19 (t, J = 7.1 Hz) | 61.1 (20) |
| Compounds | IC50 (μM) for 72 h | |||||
|---|---|---|---|---|---|---|
| MCF-7 | BT-549 | RBE | A549 | NCM-460 | Huvec | |
| 1 | >50 | >50 | >50 | >50 | >50 | >50 |
| 2 | >50 | >50 | 21.39 | >50 | 40.84 | 31.23 |
| 3 | 19.60 | 5.97 | 16.79 | >50 | 18.47 | 7.67 |
| 4 | >50 | >50 | >50 | >50 | >50 | >50 |
| 5 | >50 | >50 | >50 | >50 | >50 | >50 |
| Adriamycin | 2.96 | 6.42 | 3.62 | 4.62 | 3.24 | 3.87 |
| Cisplatin | 4.49 | 14.21 | 4.26 | 9.34 | 19.03 | 5.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhou, L.; Pan, X.; Liao, Z.; Qi, N.; Sun, M.; Zhang, H.; Ju, J.; Ma, J. Metabolic Blockade-Based Genome Mining of Sea Anemone-Associated Streptomyces sp. S1502 Identifies Atypical Angucyclines WS-5995 A–E: Isolation, Identification, Biosynthetic Investigation, and Bioactivities. Mar. Drugs 2024, 22, 195. https://doi.org/10.3390/md22050195
Wang Y, Zhou L, Pan X, Liao Z, Qi N, Sun M, Zhang H, Ju J, Ma J. Metabolic Blockade-Based Genome Mining of Sea Anemone-Associated Streptomyces sp. S1502 Identifies Atypical Angucyclines WS-5995 A–E: Isolation, Identification, Biosynthetic Investigation, and Bioactivities. Marine Drugs. 2024; 22(5):195. https://doi.org/10.3390/md22050195
Chicago/Turabian StyleWang, Yuyang, Le Zhou, Xiaoting Pan, Zhangjun Liao, Nanshan Qi, Mingfei Sun, Hua Zhang, Jianhua Ju, and Junying Ma. 2024. "Metabolic Blockade-Based Genome Mining of Sea Anemone-Associated Streptomyces sp. S1502 Identifies Atypical Angucyclines WS-5995 A–E: Isolation, Identification, Biosynthetic Investigation, and Bioactivities" Marine Drugs 22, no. 5: 195. https://doi.org/10.3390/md22050195