Identification of Linkages between EDCs in Personal Care Products and Breast Cancer through Data Integration Combined with Gene Network Analysis
Abstract
1. Introduction
2. Methods
2.1. Data Collection and EDC Selection
2.2. Curating Common Interacting Genes between EDCs and Breast Cancer
2.3. Gene Nework Analysis and Screening Potential EDCs
3. Results
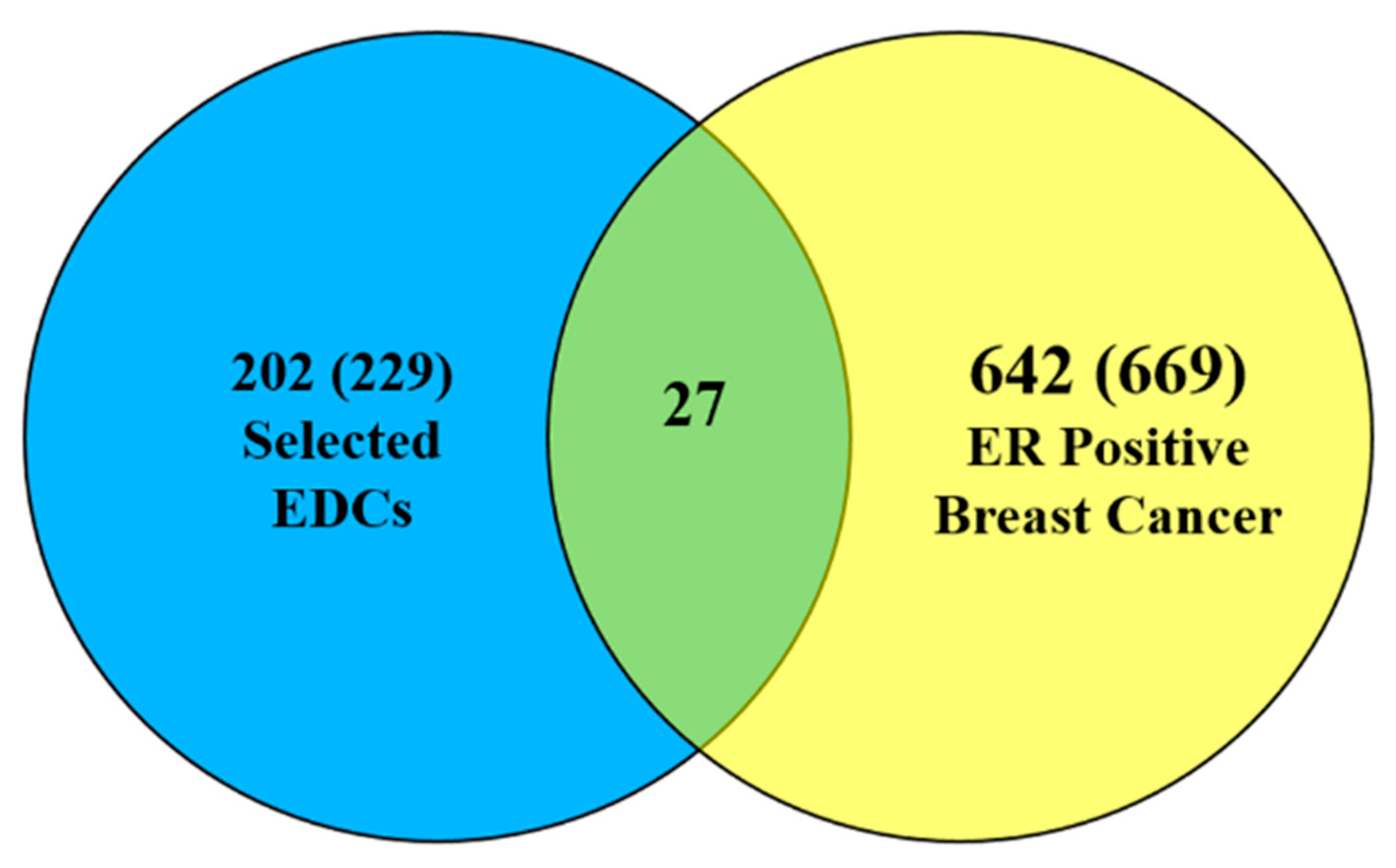
3.1. Data Integration
3.2. Finding EDC-Interacting Genes and Mutated Genes in ER Positive Breast Cancer
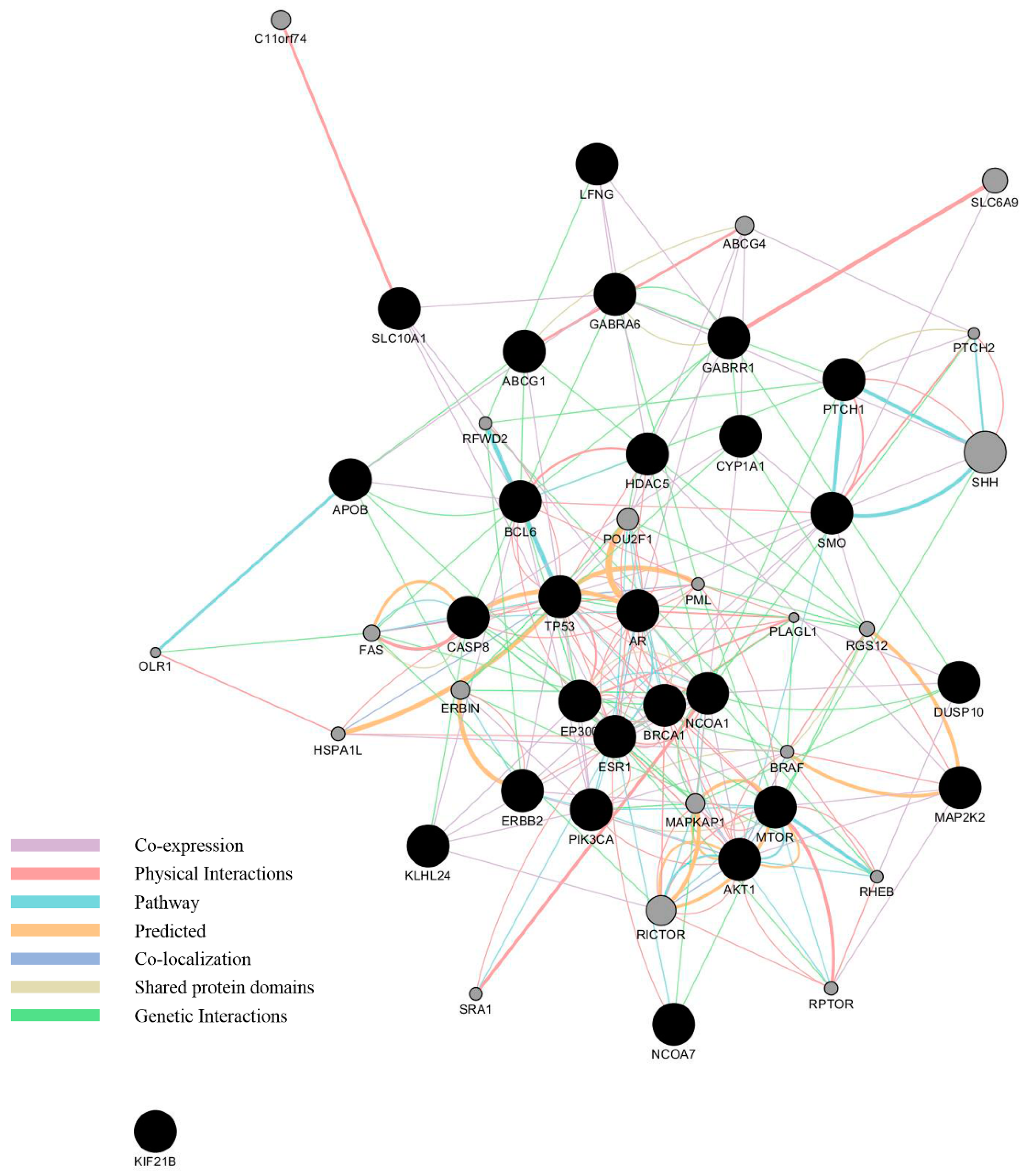
3.3. Network Analysis of Curated Genes and Potential EDCs
4. Discussion
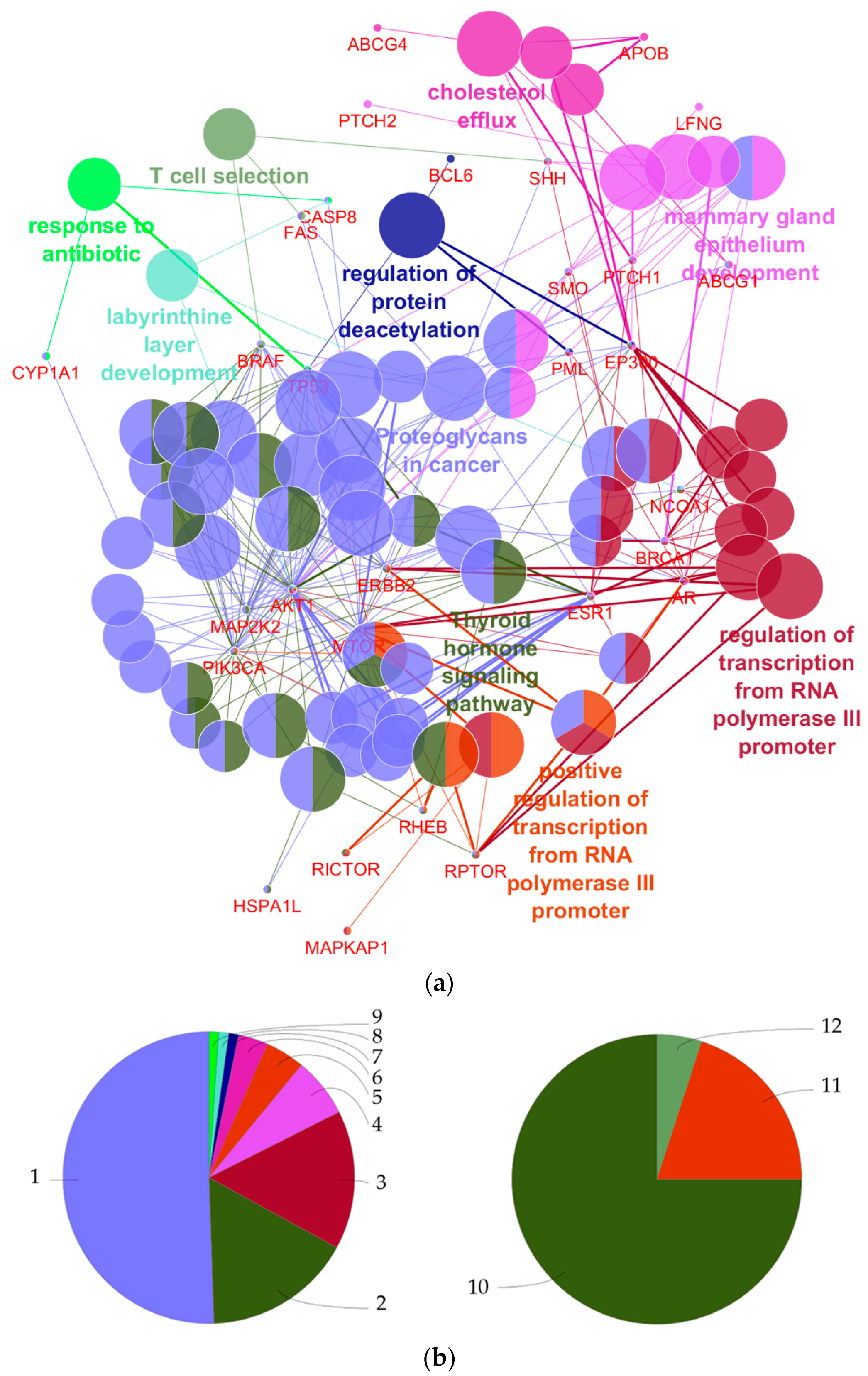
4.1. What Can Be Derived from the Functional Annotations of the 27 Genes with Regard to Breast Cancer Development?
4.2. How Can Other Potential Diseases be Predicted Based on the 20 Predicted Genes?
4.3. Contributions and Limitations of the Study
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bernhardt, E.S.; Rosi, E.J.; Gessner, M.O. Synthetic chemicals as agents of global change. Front. Ecol. Environ. 2017, 15, 84–90. [Google Scholar] [CrossRef]
- Plastics Europe. Plastics-the Facts 2016. In An Analysis of European Plastics Production, Demand and Waste Data; Plastics Europe: Brussels, Belgium, 2014. [Google Scholar]
- Gore, A.C.; Crews, D.; Doan, L.L.; Merrill, M.L.; Patisaul, H.; Zota, A. Introduction to Endocrine Disrupting Chemicals (EDCs). In A Guide for Public Interest Organizations and Policy-Makers; Endocrine Society: Washington, DC, USA, 2014. [Google Scholar]
- Trasande, L.; Zoeller, R.T.; Hass, U.; Kortenkamp, A.; Grandjean, P.; Myers, J.P.; DiGangi, J.; Bellanger, M.; Hauser, R.; Legler, J. Estimating burden and disease costs of exposure to endocrine-disrupting chemicals in the European Union. J. Clin. Endocrinol. Metab. 2015, 100, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Beronius, A.; Vandenberg, L.N. Using systematic reviews for hazard and risk assessment of endocrine disrupting chemicals. Rev. Endocr. Metab. Disord. 2015, 16, 273–287. [Google Scholar] [CrossRef] [PubMed]
- UNEP/WHO. State of the Science of Endocrine Disrupting Chemicals–2012; Bergman, A., Heindel, J.J., Jobling, S., Kidd, K.A., Zoeller, R.T., Eds.; WHO Press: Geneva, Switzerland, 2013; pp. 1–272. [Google Scholar]
- Colborn, T.; Saal, F.S.V.; Soto, A.M. Developmental effects of endocrine-disrupting chemicals in wildlife and humans. Environ. Health Perspect. 1993, 101, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, V.F.; Tal, A.; Arnon, S. Why endocrine disrupting chemicals (EDCs) challenge traditional risk assessment and how to respond. J. Hazard. Mater. 2015, 286, 589–611. [Google Scholar] [CrossRef] [PubMed]
- Schug, T.T.; Johnson, A.F.; Birnbaum, L.S.; Colborn, T.; Guillette, L.J., Jr.; Crews, D.P.; Collins, T.; Soto, A.M.; Vom Saal, F.S.; McLachlan, J.A.; et al. Minireview: Endocrine disruptors: Past lessons and future directions. Mol. Endocrinol. 2016, 30, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Rochester, J.R. Bisphenol a and human health: A review of the literature. Reprod. Toxicol. 2013, 42, 132–155. [Google Scholar] [CrossRef]
- Rubin, B.S. Bisphenol a: An endocrine disruptor with widespread exposure and multiple effects. J. Streroid Biochem. Mol. Biol. 2011, 127, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Degen, G.H.; Bolt, H.M. Endocrine disruptors: Update on xenoestrogens. Int. Arch. Occup. Environ. Health 2000, 73, 433–441. [Google Scholar] [CrossRef]
- Witorsch, R.J.; Thomas, J.A. Personal care products and endocrine disruption: A critical review of the literature. Crit. Rev. Toxicol. 2010, 40, 1–30. [Google Scholar] [CrossRef]
- Roig, B.; Mnif, W.; Hassine, A.I.H.; Zidi, I.; Bayle, S.; Bartegi, A.; Thomas, O. Endocrine disrupting chemicals and human health risk assessment: A critical review. Crit. Rev. Environ. Sci. Technol. 2013, 43, 2297–2351. [Google Scholar] [CrossRef]
- Manickum, T.; John, W. Occurrence, fate and environmental risk assessment of endocrine disrupting compounds at the wastewater treatment works in Pietermaritzburg (South Africa). Sci. Total Environ. 2014, 468, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Kortenkamp, A. Low dose mixture effects of endocrine disrupters and their implications for regulatory thresholds in chemical risk assessment. Curr. Opin. Pharmacol. 2014, 19, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, N.; Silva, E.; Kortenkamp, A. Combining xenoestrogens at levels below individual no-observed-effect concentrations dramatically enhances steroid hormone action. Environ. Health Perspect. 2002, 110, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Patisaul, H.B.; Adewale, H.B. Long-term effects of environmental endocrine disruptors on reproductive physiology and behavior. Front. Behav. Neurosci. 2009, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Harvey, P.W.; Darbre, P. Endocrine disrupters and human health: Could oestrogenic chemicals in body care cosmetics adversely affect breast cancer incidence in women? A review of evidence and call for further research. J. Appl. Toxicol. 2004, 24, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.; Russo, I.H. The role of estrogen in the initiation of breast cancer. J. Steroid Biochem. Mol. Biol. 2006, 102, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Yager, J.D.; Davidson, N.E. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med 2006, 354, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Liehr, J.G. Is estradiol a genotoxic mutagenic carcinogen? Endocr. Rev. 2000, 21, 40–54. [Google Scholar] [PubMed]
- International Agency for Research on Cancer. Monographs on the Evaluation of Carcinogenic Risks to Humans: Hormonal Contraception and Postmenopausal Hormone Therapy; IARC: Lyon, France, 1999; Volume 72. [Google Scholar]
- Lumachi, F.; Brunello, A.; Maruzzo, M.; Basso, U.; Basso, S.M. Treatment of estrogen receptor-positive breast cancer. Curr. Med. Chem. 2013, 20, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Deoraj, A.; Felty, Q.; Roy, D. Environmental estrogen-like endocrine disrupting chemicals and breast cancer. Mol. Cell. Endocrinol. 2016, 457, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jonathan, N.; Steinmetz, R. Xenoestrogens: The emerging story of bisphenol A. Trends Endocrinol. Metab. 1998, 9, 124–128. [Google Scholar] [CrossRef]
- Erkekoglu, P.; Kocer-Gumusel, B. Genotoxicity of phthalates. Toxicol. Mech. Methods 2014, 24, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Palangat, M.; Chen, C.-W.; Thomas, R.D.; Colerangle, J.; Atkinson, A.; Yan, Z.-J. Biochemical and molecular changes at the cellular level in response to exposure to environmental estrogen-like chemicals. J. Toxicol. Environ. Health A 1997, 50, 1–30. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. Gorilla: A tool for discovery and visualization of enriched go terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Su, Z. Easygo: Gene ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genom. 2007, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Zambon, A.C.; Gaj, S.; Ho, I.; Hanspers, K.; Vranizan, K.; Evelo, C.T.; Conklin, B.R.; Pico, A.R.; Salomonis, N. Go-elite: A flexible solution for pathway and ontology over-representation. Bioinformatics 2012, 28, 2209–2210. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.S.; Shah, F.F.M.; Mohamad, M.S.; Moorthy, K.; Deris, S.; Zakaria, Z.; Napis, S. A review on bioinformatics enrichment analysis tools towards functional analysis of high throughput gene set data. Curr. Proteom. 2015, 12, 14–27. [Google Scholar] [CrossRef]
- He, B.; Yin, J.; Gong, S.; Gu, J.; Xiao, J.; Shi, W.; Ding, W.; He, Y. Bioinformatics analysis of key genes and pathways for hepatocellular carcinoma transformed from cirrhosis. Medicine 2017, 96, e6938. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, B.; Yoo, C.; Roy, D. Discovering gene-environment interactions in glioblastoma through a comprehensive data integration bioinformatics method. Neurotoxicology 2013, 35, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Morgan, M.; Yoo, C.; Deoraj, A.; Roy, S.; Yadav, V.K.; Garoub, M.; Assaggaf, H.; Doke, M. Integrated bioinformatics, environmental epidemiologic and genomic approaches to identify environmental and molecular links between endometriosis and breast cancer. Int. J. Mol. Sci. 2015, 16, 25285–25322. [Google Scholar] [CrossRef] [PubMed]
- Vlasblom, J.; Zuberi, K.; Rodriguez, H.; Arnold, R.; Gagarinova, A.; Deineko, V.; Kumar, A.; Leung, E.; Rizzolo, K.; Samanfar, B.; et al. Novel function discovery with genemania: A new integrated resource for gene function prediction in Escherichia coli. Bioinformatics 2015, 31, 306–310. [Google Scholar] [CrossRef] [PubMed]
- TEDX: The Endocrine Disruption Exchange. Available online: https://endocrinedisruption.org/interactive-tools/tedx-list-of-potential-endocrine-disruptors/search-the-tedx-list (accessed on 3 August 2017).
- The Comparative Toxicogenomics Database (CTD). Available online: http://ctdbase.org (accessed on 3 August 2017).
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; King, B.L.; McMorran, R.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. The comparative toxicogenomics database: Update 2017. Nucleic Acids Res. 2017, 45, D972–D978. [Google Scholar] [CrossRef] [PubMed]
- Mattingly, C.J.; Rosenstein, M.C.; Colby, G.T.; Forrest, J.N., Jr.; Boyer, J.L. The comparative toxicogenomics database (CTD): A resource for comparative toxicological studies. J. Exp. Zool. A Comp. Exp. Biol. 2006, 305, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Catalogue of Somatic Mutations in Cancer (COSMIC). Available online: http://cancer.sanger.ac.uk/cosmic (accessed on 3 August 2017).
- Forbes, S.A.; Bindal, N.; Bamford, S.; Cole, C.; Kok, C.Y.; Beare, D.; Jia, M.; Shepherd, R.; Leung, K.; Menzies, A.; et al. Cosmic: Mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011, 39, D945–D950. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 1–24. [Google Scholar]
- Cytoscape. Available online: http://www.cytoscape.org/download.php (accessed on 3 August 2017).
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. Genemania cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef] [PubMed]
- Mostafavi, S.; Ray, D.; Warde-Farley, D.; Grouios, C.; Morris, Q. Genemania: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008, 9, S4. [Google Scholar] [CrossRef] [PubMed]
- Proctor, C.H.; Loomis, C.P. Analysis of Sociometric Data. In Research Methods in Social Relations; Jahoda, M., Deutsch, M., Cook, S.W., Eds.; Dryden Press: Newyork, NY, USA, 1951; pp. 561–586. [Google Scholar]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. Cluego: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Michels, K.B.; Holmberg, L.; Bergkvist, L.; Wolk, A. Coffee, tea, and caffeine consumption and breast cancer incidence in a cohort of swedish women. Ann. Epidemiol. 2002, 12, 21–26. [Google Scholar] [CrossRef]
- Ganmaa, D.; Willett, W.C.; Li, T.Y.; Feskanich, D.; van Dam, R.M.; Lopez-Garcia, E.; Hunter, D.J.; Holmes, M.D. Coffee, tea, caffeine and risk of breast cancer: A 22-year follow-up. Int. J. Cancer 2008, 122, 2071–2076. [Google Scholar] [CrossRef] [PubMed]
- Fagherazzi, G.; Touillaud, M.S.; Boutron-Ruault, M.C.; Clavel-Chapelon, F.; Romieu, I. No association between coffee, tea or caffeine consumption and breast cancer risk in a prospective cohort study. Public Health Nutr. 2011, 14, 1315–1320. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.M.; Habtemariam, S.; Daglia, M.; Nabavi, S.F. Apigenin and breast cancers: From chemistry to medicine. Anticancer Agents Med. Chem. 2015, 15, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.S.; DeNardo, D.G.; Jacquot, Y.; Laios, I.; Vidal, D.S.; Zambrana, C.R.; Leclercq, G.; Brown, P.H. Stimulatory effect of genistein and apigenin on the growth of breast cancer cells correlates with their ability to activate ER alpha. Breast Cancer Res. Treat. 2006, 99, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Konduracka, E.; Krzemieniecki, K.; Gajos, G. Relationship between everyday use cosmetics and female breast cancer. Pol. Arch. Med. Wewn. 2014, 124, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.P.; Xu, S.S.; Tan, T.F.; Lee, S.T.; Cheng, S.H.; Lee, F.W.F.; Xu, S.J.L.; Ho, K.C. Toxicity and estrogenic endocrine disrupting activity of phthalates and their mixtures. Int. J. Environ. Res. Public Health 2014, 11, 3156–3168. [Google Scholar] [CrossRef] [PubMed]
- Engel, L.S.; Hill, D.A.; Hoppin, J.A.; Lubin, J.H.; Lynch, C.F.; Pierce, J.; Samanic, C.; Sandler, D.P.; Blair, A.; Alavanja, M.C. Pesticide use and breast cancer risk among farmers’ wives in the agricultural health study. Am. J. Epidemiol. 2005, 161, 121–135. [Google Scholar] [CrossRef] [PubMed]
- National Research Council Canada. Phthalate Esters in the Aquatic Environment; National Research Council Canada: Ottawa, ON, Canada, 1980.
- U.S. Consumer Product Safety Commission. Statement of Policy: Testing of Component Parts with Respect to Section 108 of the Consumer Product Safety Improvement Act; U.S. Consumer Product Safety Commission: Bethesda, MD, USA, 2009.
- European Commission. Scientific Committee on Consumer Products; European Commission: Brussels, Belgium, 2007. [Google Scholar]
- Lopez-Carrillo, L.; Hernandez-Ramirez, R.U.; Calafat, A.M.; Torres-Sanchez, L.; Galvan-Portillo, M.; Needham, L.L.; Ruiz-Ramos, R.; Cebrian, M.E. Exposure to phthalates and breast cancer risk in northern Mexico. Environ. Health Perspect. 2010, 118, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Ibarluzea, J.; Fernández, M.F.; Santa-Marina, L.; Olea-Serrano, M.F.; Rivas, A.M.; Aurrekoetxea, J.J.; Expósito, J.; Lorenzo, M.; Torné, P.; Villalobos, M.; et al. Breast cancer risk and the combined effect of environmental estrogens. Cancer Causes Control CCC 2004, 15, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Jeselsohn, R.; Yelensky, R.; Buchwalter, G.; Frampton, G.; Meric-Bernstam, F.; Gonzalez-Angulo, A.M.; Ferrer-Lozano, J.; Perez-Fidalgo, J.A.; Cristofanilli, M.; Gómez, H. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor–positive breast cancer. Clin. Cancer Res. 2014, 20, 1757–1767. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Qin, X.-Y.; Yoshida, M.; Fukuda, T.; Nansai, H.; Hayashi, Y.; Nakajima, T.; Sone, H. Peroxisome proliferator-activated receptor α mediates di-(2-ethylhexyl) phthalate transgenerational repression of ovarian ESR1 expression in female mice. Toxicol. Lett. 2014, 228, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.C.; Lee, B.M. DNA methylation of estrogen receptor alpha gene by phthalates. J. Toxicol. Environ. Health A 2005, 68, 1995–2003. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Sanderson, R.D. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J. Cell. Mol. Med. 2011, 15, 1013–1031. [Google Scholar] [CrossRef] [PubMed]
- Schwertfeger, K.L.; Cowman, M.K.; Telmer, P.G.; Turley, E.A.; McCarthy, J.B. Hyaluronan, inflammation, and breast cancer progression. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Stemke-Hale, K.; Gonzalez-Angulo, A.M.; Lluch, A.; Neve, R.M.; Kuo, W.-L.; Davies, M.; Carey, M.; Hu, Z.; Guan, Y.; Sahin, A. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008, 68, 6084–6091. [Google Scholar] [CrossRef] [PubMed]
- Vivanco, I.; Sawyers, C.L. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer 2002, 2, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.C.; Salazar, E.P.; Kane, S.R.; Liu, N. Effects of thyroid hormones on human breast cancer cell proliferation. J. Steroid Biochem. Mol. Biol. 2008, 109, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Pike, M.C.; Spicer, D.V.; Dahmoush, L.; Press, M.F. Estrogens progestogens normal breast cell proliferation and breast cancer risk. Epidemiol. Rev. 1993, 15, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. Tor signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Zhang, J.; Jin, S.; Li, H. Role of PI3K/AKT/MTOR signaling pathway in DBP-induced apoptosis of testicular sertoli cells in vitro. Environ. Toxicol. Pharmacol. 2017, 53, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Dazert, E.; Hall, M.N. Mtor signaling in disease. Curr. Opin. Cell Biol. 2011, 23, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. MTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.; Chen, Q.; Srivastava, R.K. Inhibition of PI3K/AKT and MEK/ERK pathways act synergistically to enhance antiangiogenic effects of EGCG through activation of FOXO transcription factor. J. Mol. Signal. 2008, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The RAS-ERK and PI3K-MTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Samatar, A.A.; Poulikakos, P.I. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat. Rev. Drug Discov. 2014, 13, 928. [Google Scholar] [CrossRef] [PubMed]
- King, P.J.; Guasti, L.; Laufer, E. Hedgehog signalling in endocrine development and disease. J. Endocrinol. 2008, 198, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-J.; Jiang, J.-T.; Ma, L.; Zhang, J.; Hong, Y.; Liao, K.; Liu, Q.; Liu, G.-H. Molecular and toxicologic research in newborn hypospadiac male rats following in utero exposure to di-n-butyl phthalate (DBP). Toxicology 2009, 260, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-P.; Li, E.-H.; Sun, W.-L.; Xu, D.-L.; Liu, Z.-H.; Zhao, W.; Wood, K.; Xia, S.-J.; Jiang, J.-T. Maternal exposure to di-n-butyl phthalate (DBP) induces combined anorectal and urogenital malformations in male rat offspring. Reprod. Toxicol. 2016, 61, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.H.; Baskin, L.S. Endocrine disruptors, genital development, and hypospadias. J. Androl. 2008, 29, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, S.; Matsumaru, D.; Murashima, A.; Omori, A.; Satoh, Y.; Haraguchi, R.; Motoyama, J.; Iguchi, T.; Nakagata, N.; Hui, C.-C. The role of sonic hedgehog-gli2 pathway in the masculinization of external genitalia. Endocrinology 2011, 152, 2894–2903. [Google Scholar] [CrossRef] [PubMed]
- Ormond, G.; Nieuwenhuijsen, M.J.; Nelson, P.; Toledano, M.B.; Iszatt, N.; Geneletti, S.; Elliott, P. Endocrine disruptors in the workplace, hair spray, folate supplementation, and risk of hypospadias: Case–control study. Environ. Health Perspect. 2009, 117, 303. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.F.; Arrebola, J.P.; Jimenez-Diaz, I.; Saenz, J.M.; Molina-Molina, J.M.; Ballesteros, O.; Kortenkamp, A.; Olea, N. Bisphenol a and other phenols in human placenta from children with cryptorchidism or hypospadias. Reprod. Toxicol. 2016, 59, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.F.; Olmos, B.; Granada, A.; Lopez-Espinosa, M.J.; Molina-Molina, J.M.; Fernandez, J.M.; Cruz, M.; Olea-Serrano, F.; Olea, N. Human exposure to endocrine-disrupting chemicals and prenatal risk factors for cryptorchidism and hypospadias: A nested case-control study. Environ. Health Perspect. 2007, 115, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Merid, S.K.; Goranskaya, D.; Alexeyenko, A. Distinguishing between driver and passenger mutations in individual cancer genomes by network enrichment analysis. BMC Bioinform. 2014, 15, 308. [Google Scholar] [CrossRef] [PubMed]

| Name (Cas No.) | Interacting Genes |
|---|---|
| Captan (133-06-2) | 5 genes: GSTP1 | CYP3A4 | ESR1 | NR1I2 | TSC22D1 |
| Diethylhexyl Phthalate (117-81-7) | 115 genes: ABCB1 | ACADM | ACADVL | AHR | AKT1 | AMH | AOX1 | AR | ARRDC3 | BAX | BBC3 | BCL2 | CASP3 | CASP7 | CASP8 | CASP9 | CDKN1A | CDO1 | CELSR2 | CGA | CGB3 | CLDN6 | CSNK1A1 | CTNNB1 | CXCL8 | CYP19A1 | CYP1A1 | CYP1B1 | CYP2C19 | CYP2C9 | CYP3A4 | CYP4A10 | DDIT3 | DHCR24 | DIABLO | DNAJB1 | EP300 | ESR1 | ESR2 | FASN |FLG | FSHB | FSHR | GJA1 | GLI3 | GLRX2 | HDAC4 | HDAC5 | HEXA | HEXB | HMGCR | HSD11B2 | HSPA1B | ID1 | IL17RD | IL4 | KLK3 | LAMP3 | LFNG | LHCGR | LIF | MAPK1 | MAPK3 | MARS | MDM2 | MED1 | MMP2 | MMP9 | MTOR | MYC | NCOA1 | NCOR1 | NGB | NR1H3 | NR1I2 | NR1I3 | NR3C1 | NR4A1 | NR4A2 | NR4A3 | PAPSS1 | PAPSS2 |PIK3CA | PMAIP1 | PPARA | PPARB | PPARD | PPARG | PPARGC1A | PRNP | PTCH1 | PTGS2 | RPS6KB1 | RXRA | RXRB | RXRG | SCARA3 | SCD | SLC7A11 | SMO | SP3 | SQLE | SREBF1 | SREBF2 | STAR | SUOX | TIMP2 | TNF | TP53 | TSPAN6 | TXNRD1 | VCL | VEGFA | VLDLR | ZNF461 |
| Diethyl Phthalate (84-66-2) | 20 genes: AHR | APOA1 | APOB | AR | CASP3 | CXCL8 | CYP19A1 | CYP1B1 | ESR1 | ESR2 | FLG | NR1I2 | NR1I3 | PPARA | PPARB | PPARG | RXRA | RXRB | RXRG | SHBG |
| Lindane (58-89-9) | 134 genes: ABCA1 |ABCB1 | ABCG1 | ABCG2 | ACRC | AGPAT9 | AK4 | ALDH8A1 | AR | ASNS | BAX | BCL2 | BCL6 | BIRC3 | BRCA1 | C15ORF39 | CARS | CAT | CCND1 | CCNG2 | CD69 | CD84 | CDKN1A | CEBPB | CHAC1 | CRIM1 | CX3CR1 | CYP11A1 | CYP11B1 | CYP11B2 | CYP17A1 | CYP19A1 | CYP1A1 | CYP2B6 | CYP2D6 | CYP2E1 | CYP3A4 | CYP3A7 | DAP3 |DNAJB4 | DUSP10 | ERBB2 | ERBB3 | ESR1 | ESR2 | ESRRA | EVI2A | FAM107B | FAM213B | FBXO32 | FRAT1 | GABRA1 | GABRA2 | GABRA4 | GABRA6 | GABRB1 | GABRB2 | GABRB3 | GABRG2 | GABRR1 | GCLC | GCLM | GLRA1 | GLRA2 | GLRA3 | GNRH1 | GPR18 | GPT2 | GSR | GSTM1 | HMGCS1 | HSD3B2 | HSPA1A | HYLS1 | ID1 | IER3 | IFNG | IL5 |ISL2 | JUN | KIF21B | KLHL24 | LIF | MAP2K1 | MAP2K2 | MAPK1 | MAPK3 | MMP9 | MTHFD2 | NANOS1 | NCOA7 | NFE2L3 | NOS2 | NR1I2 | NRF1 | PDCD4 | PELI1 | PGR | PLCL1 | PNRC1 | POMC | PPARGC1A | PPRC1 | RAF1 | RARA | RGS2 | RXRB | SEMA3G | SESN2 | SGK1 | SHBG | SLC10A1 | SLC22A1 | SLC3A2 | SLC7A11 | SQSTM1 | SRC | SRXN1 |STAM2 | STAR | SULT2A1 | TFAM | TFB2M | TFF1 | TM6SF1 | TMCO6 | TMEM177 | TMEM267 | TNF | TP53 | TRIB3 | VEGFA | VLDLR | ZNF628 |
| Degree Centrality | Gene | Official Full Name | Interacting Gene | Networks * |
|---|---|---|---|---|
| 13 | ESR1 | Estrogen receptor 1 | AKT1 | 2 |
| AR | 2, 3, 5, 6 | |||
| BRCA1 | 2, 3 | |||
| CASP8 | 7 | |||
| EP300 | 2, 3 | |||
| ERBB2 | 2 | |||
| HDAC5 | 2 | |||
| NCOA1 | 2, 3 | |||
| NCOA7 | 2, 3 | |||
| PIK3CA | 2, 3 | |||
| SLC10A1 | 1 | |||
| SMO | 1 | |||
| TP53 | 2 | |||
| 12 | TP53 | Tumor protein p53 | AKT1 | 1 |
| AR | 2, 7 | |||
| BCL6 | 2, 3 | |||
| BRCA1 | 2, 3 | |||
| CASP8 | 2 | |||
| EP300 | 2, 7 | |||
| ERBB2 | 1 | |||
| ESR1 | 2 | |||
| HDAC5 | 2 | |||
| MTOR | 2 | |||
| NCOA1 | 2 | |||
| SMO | 1 | |||
| 12 | NCOA1 | Nuclear receptor coactivator 1 | AKT1 | 1, 3 |
| AR | 2, 3 | |||
| BRCA1 | 2 | |||
| CYP1A1 | 1 | |||
| DUSP10 | 1, 7 | |||
| EP300 | 1, 2, 3, 6 | |||
| ESR1 | 2, 3 | |||
| HDAC5 | 7 | |||
| KLHL24 | 1 | |||
| NCOA7 | 7 | |||
| PTCH1 | 7 | |||
| TP53 | 2 | |||
| 11 | AKT1 | AKT serine/threonine kinase 1 | AR | 2 |
| BRCA1 | 2 | |||
| EP300 | 2 | |||
| ERBB2 | 1 | |||
| ESR1 | 2 | |||
| MAP2K2 | 1 | |||
| MTOR | 2, 3, 5, 4 | |||
| NCOA1 | 1, 3 | |||
| PIK3CA | 2, 3 | |||
| SMO | 3 | |||
| TP53 | 1 | |||
| 11 | BCL6 | B-cell CLL/lymphoma 6 | ABCG1 | 7 |
| APOB | 1, 7 | |||
| EP300 | 2, 7 | |||
| GABRR1 | 7 | |||
| HDAC5 | 2, 3 | |||
| KLHL24 | 1 | |||
| PIK3CA | 1 | |||
| PTCH1 | 7 | |||
| SLC10A1 | 1 | |||
| SMO | 2 | |||
| TP53 | 2, 3 |
| Score | Chemical Name (Cas No.) | Interacting Genes |
|---|---|---|
| 40 | Perfluorooctanoic acid (335-67-1) | ABCG1, APOB, CYP1A1, ERBB2, ESR1, TP53, SHH |
| 37 | Stearic acid (57-11-4) | ABCG1, AKT1, AR, ESR1 |
| 35 | Triphenyl phosphate (115-86-6) | AR, ESR1, TP53 |
| 34 | Dibutyl Phthalate (84-74-2) | AKT1, AR, ESR1 |
| 30 | Sodium Fluoride (7681-49-4) | AKT1, CASP8, TP53, FAS |
| GO Term | Ontology Source | p-Value | Annotated Genes |
|---|---|---|---|
| Proteoglycans in cancer | KEGG | 4.0 × 10−14 | AKT1|BRAF|ERBB2|ESR1|FAS|MAP2K2| MTOR|PIK3CA|PTCH1|SHH|SMO|TP53 |
| Thyroid hormone signaling pathway | KEGG | 7.2 × 10−12 | AKT1|EP300|ESR1|MAP2K2|MTOR|NCOA1| PIK3CA|RHEB|TP53 |
| Regulation of transcription from RNA polymerase III promoter | GO-Biological process | 1.6 × 10−8 | AR|BRCA1|ERBB2|MTOR|RPTOR |
| Mammary gland epithelium development | GO-Biological process | 2.5 × 10−8 | AKT1|AR|ESR1|PML|PTCH1|SMO |
| Positive regulation of transcription from RNA polymerase III promoter | GO-Biological process | 2.8 × 10−8 | AR|ERBB2|MTOR|RPTOR |
| Cholesterol efflux | GO-Biological process | 5.0 × 10−8 | ABCG1|ABCG4|APOB|PTCH1|SHH |
| Regulation of protein deacetylation | GO-Biological process | 3.0 × 10−6 | BCL6|EP300|PML|TP53 |
| T cell selection | GO-Biological process | 2.0 × 10−4 | BRAF|FAS|SHH |
| Labyrinthine layer development | GO-Biological process | 2.6 × 10−4 | AKT1|CASP8|NCOA1 |
| Response to antibiotic | GO-Biological process | 3.4 × 10−4 | CASP8|CYP1A1|TP53 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, H.; Kim, J.; Kim, Y. Identification of Linkages between EDCs in Personal Care Products and Breast Cancer through Data Integration Combined with Gene Network Analysis. Int. J. Environ. Res. Public Health 2017, 14, 1158. https://doi.org/10.3390/ijerph14101158
Jeong H, Kim J, Kim Y. Identification of Linkages between EDCs in Personal Care Products and Breast Cancer through Data Integration Combined with Gene Network Analysis. International Journal of Environmental Research and Public Health. 2017; 14(10):1158. https://doi.org/10.3390/ijerph14101158
Chicago/Turabian StyleJeong, Hyeri, Jongwoon Kim, and Youngjun Kim. 2017. "Identification of Linkages between EDCs in Personal Care Products and Breast Cancer through Data Integration Combined with Gene Network Analysis" International Journal of Environmental Research and Public Health 14, no. 10: 1158. https://doi.org/10.3390/ijerph14101158
APA StyleJeong, H., Kim, J., & Kim, Y. (2017). Identification of Linkages between EDCs in Personal Care Products and Breast Cancer through Data Integration Combined with Gene Network Analysis. International Journal of Environmental Research and Public Health, 14(10), 1158. https://doi.org/10.3390/ijerph14101158





