Isolation and Characterization of Native Microalgae from the Peruvian Amazon with Potential for Biodiesel Production
Abstract
:1. Introduction
2. Experimental Section
2.1. Sample Collection and Isolation of Microalgae
2.2. Morphological Identification of Microalgae
2.3. Genomic DNA Isolation
2.4. PCR Amplification and Sequencing of ITS2-rDNA Region
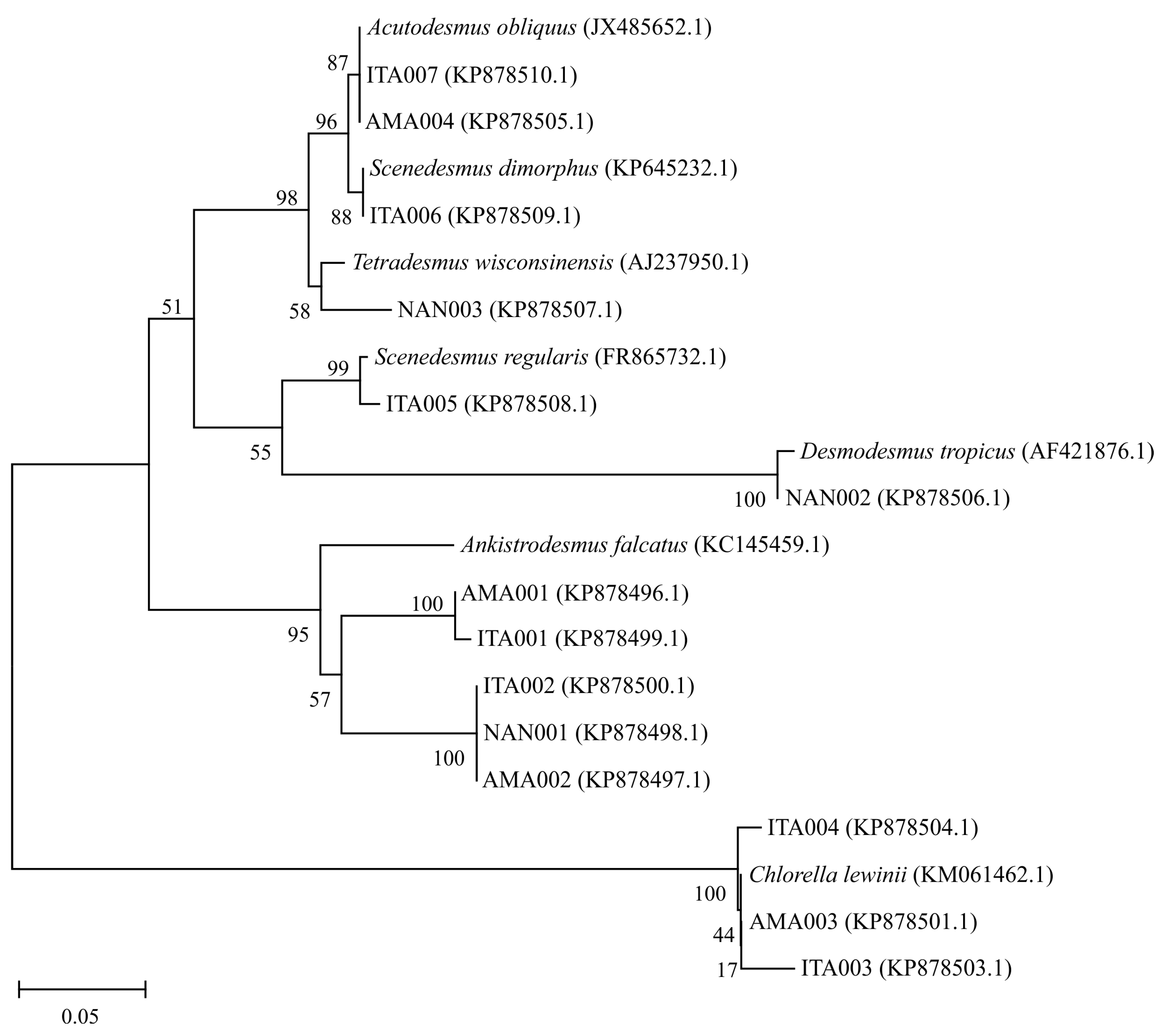
2.5. Phylogenetic Analysis
2.6. Microalgae Cultivation, Biomass and Total Lipids Measurements
2.7. Strains Culture under Nitrogen-Sufficient and Nitrogen-Deficient Medium
2.8. Determination of Total Protein, Total Carbohydrate, Ash Content, Esterification and Fatty Acids Profiling
2.9. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Identification of the Microalgae Strains
3.2. Microalgae Cultivation, Biomass and Total Lipids Measurements
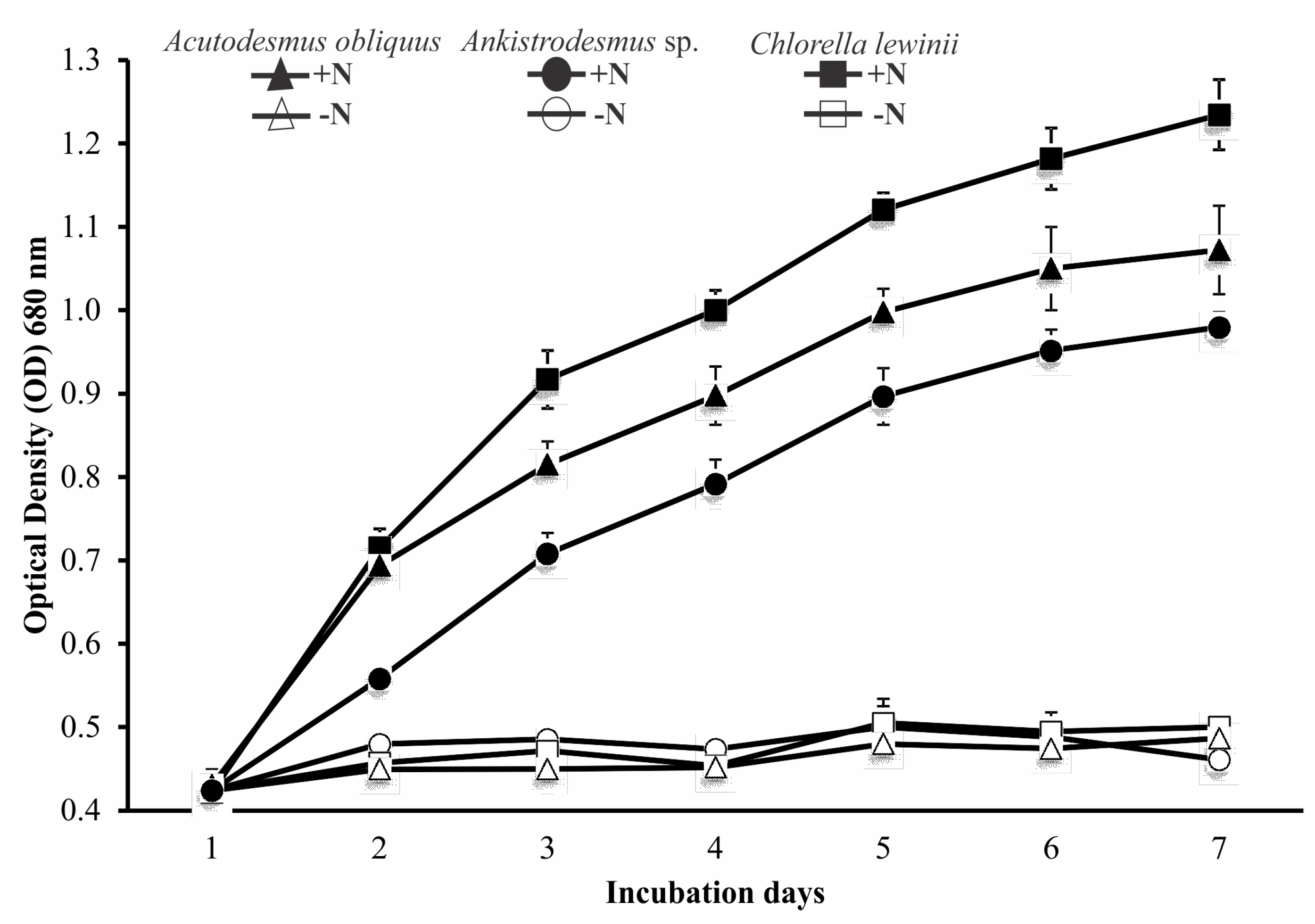
3.3. Strains Culture under Nitrogen-Sufficient and Nitrogen-Deficient Medium
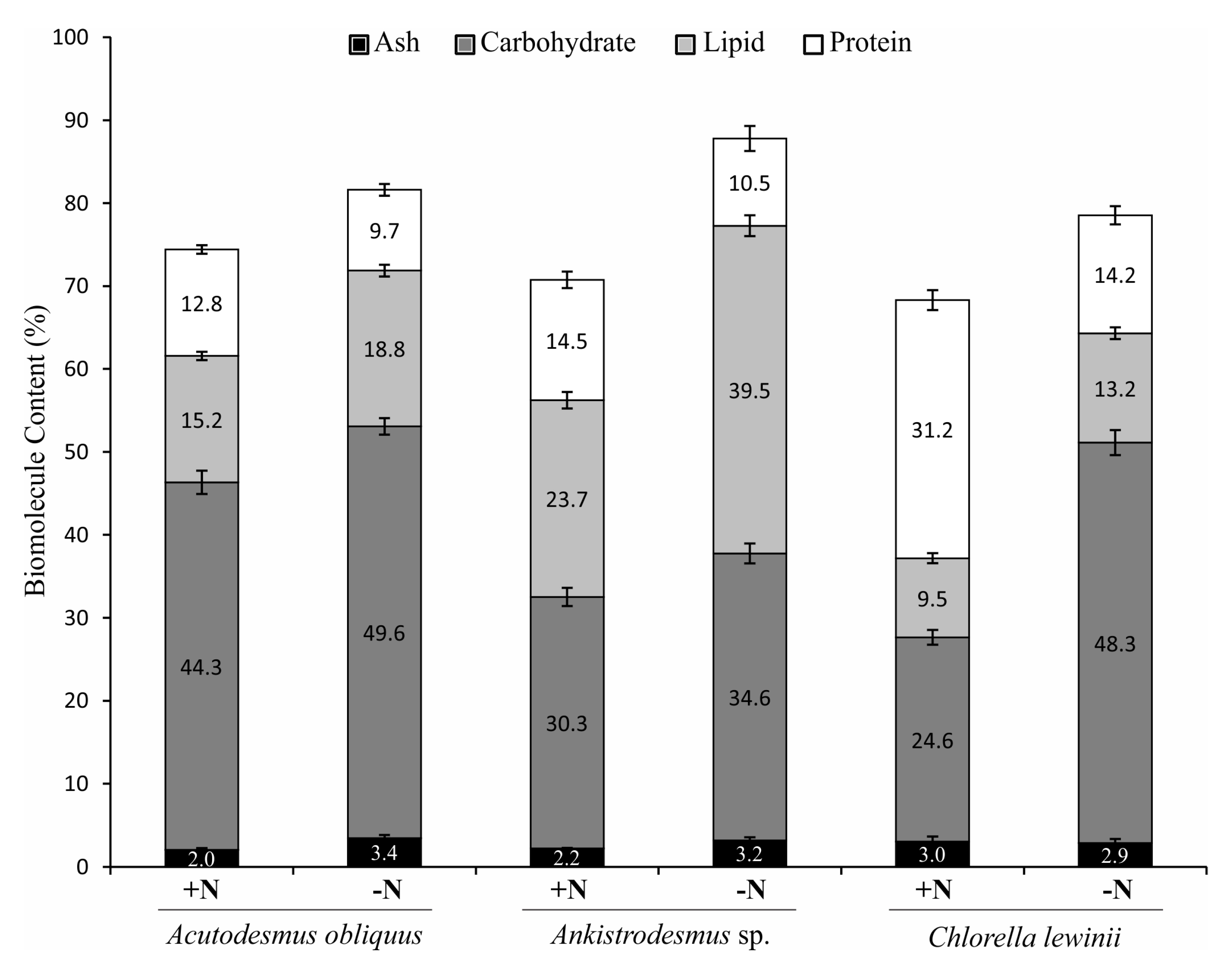
3.4. Total Protein, Total Lipid and Total Carbohydrate Content
3.5. Fatty Acids Profiling
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- BP Global BP Statisical Review of World Energy 2014. Available online: http://www.bp.com/content/dam/bp/pdf/Energy-economics/statistical-review-2014/BP-statistical-review-of-world-energy-2014-full-report.pdf (accessed on 20 June 2014).
- Pachauri, R.K.; Reisinger, A. Cambio Climático 2007: Informe de Síntesis. Contribución de los Grupos de Trabajo I, II y III al Cuarto Informe de Evaluación del Grupo Intergubernamental de Expertos Sobre el Cambio Climático; IPCC: Geneva, Switzerland, 2007; p. 104. [Google Scholar]
- Perú. Comisión Permanente del Congreso de la República. Ley de Promoción del Mercado de Biocombustibles. Available online: http://www2.congreso.gob.pe/sicr/cendocbib/con4_uibd.nsf/9E95620CC059138105257C9E005AB2F9/$FILE/28054.pdf (accessed on 12 February 2017).
- Perú. Comisión Permanente del Congreso de la República. Reglamento de la Ley de Promoción del Mercado de Biocombustibles. Available online: http://www2.osinerg.gob.pe/MarcoLegal/docrev/DS-013-2005-EM-CONCORDADO.pdf (accessed on 12 February 2017).
- Arévalo, L.F.; Nalvarte, J.; Torres, J.; Ramírez, Y. Impactos Socio-Económicos de la Producción de Biocombustibles en la Amazonía Peruana; Primera; SNV-WWF: Lima, Perú, 2009. [Google Scholar]
- Sharma, K.K.; Schuhmann, H.; Schenk, P.M. High Lipid Induction in Microalgae for Biodiesel Production. Energies 2012, 5, 1532–1553. [Google Scholar] [CrossRef]
- Schenk, P.M.; Thomas-Hall, S.R.; Stephens, E.; Marx, U.C.; Mussgnug, J.H.; Posten, C.; Kruse, O.; Hankamer, B. Second Generation Biofuels: High-Efficiency Microalgae for Biodiesel Production. BioEnergy Res. 2008, 1, 20–43. [Google Scholar] [CrossRef]
- Foley, J.A.; Asner, G.P.; Costa, M.H.; Coe, M.T.; de Fries, R.; Gibbs, H.K.; Howard, E.A.; Olson, S.; Patz, J.; Ramankutty, N.; et al. Amazonia revealed: Forest degradation and loss of ecosystem goods and services in the Amazon Basin. Front. Ecol. Environ. 2007, 5, 25–32. [Google Scholar] [CrossRef]
- Duong, V.T.; Li, Y.; Nowak, E.; Schenk, P.M. Microalgae Isolation and Selection for Prospective Biodiesel Production. Energies 2012, 5, 1835–1849. [Google Scholar] [CrossRef]
- Georgianna, D.R.; Mayfield, S.P. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 2012, 488, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Wijffels, R.H.; Barbosa, M.J. An Outlook on Microalgal Biofuels. Science 2010, 329, 796–799. [Google Scholar] [CrossRef] [PubMed]
- Richmond, A. Handbook of Microalgal Culture: Biotechnology and Applied Phycology; Blackwell Sci. Ltd.: Hudson, NJ, USA, 2004. [Google Scholar]
- Menetrez, M.Y. An overview of algae biofuel production and potential environmental impact. Environ. Sci. Technol. 2012, 46, 7073–7085. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.H.; Sharratt, P.N.; Das, P.; Balasubramanian, R.K.; Naraharisetti, P.K.; Shaik, S. Life cycle energy and CO2 analysis of microalgae-to-biodiesel: Preliminary results and comparisons. Bioresour. Technol. 2011, 102, 5800–5807. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.P. The Influence of the Mineral Composition of the Medium on the Growth of Planktonic Algae: Part I. Methods and Culture Media. J. Ecol. 1942, 30, 284–325. [Google Scholar] [CrossRef]
- Carlos, E.M.B.; Mariãngela, M. Gêneros de Algas de Águas Continentais do Brasil—Chave Para Identificação e Descrições, 2nd ed.; Rima: Sao Carlos, Brasil, 2006. [Google Scholar]
- Greenspan, P.; Mayer, E.P.; Fowler, S.D. Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985, 100, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Kaur, S.; Sarkar, M.; Srivastava, R.B.; Gogoi, H.K.; Kalita, M.C. Fatty acid profiling and molecular characterization of some freshwater microalgae from India with potential for biodiesel production. New Biotechnol. 2012, 29, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resource Coordinators. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015, 43, D6–D17. [Google Scholar]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef] [PubMed]
- BioEdit Sequence Alignment Editor for Windows 95/98/NT/XP/Vista/7. Available online: http://www.mbio.ncsu.edu/bioedit/bioedit.html (accessed on 2 December 2015).
- Sievers, F.; Higgins, D.G. Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. (Clifton NJ) 2014, 1079, 105–116. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhao, P.; He, C.; Li, J.; Tang, X.; Zhou, J.; Huang, Z. Isolation of a novel strain of Monoraphidium sp. and characterization of its potential application as biodiesel feedstock. Bioresour. Technol. 2012, 121, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Hartree, E.F. Determination of protein: A modification of the lowry method that gives a linear photometric response. Anal. Biochem. 1972, 48, 422–427. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- AOAC International. AOAC Official Methods of Analysis of the Association of Official Analytical Chemists (AOAC), 15th ed.; AOAC International: Washington, DC, USA, 1990; Volume 1. [Google Scholar]
- Ichihara, K.; Fukubayashi, Y. Preparation of fatty acid methyl esters for gas-liquid chromatography. J. Lipid Res. 2010, 51, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L.; Owende, P. Biofuels from microalgae—A review of technologies for production, processing, and extractions of biofuels and co-products. Renew. Sustain. Energ Rev. 2010, 14, 557–577. [Google Scholar] [CrossRef]
- Raja, R.; Hemaiswarya, S.; Kumar, N.A.; Sridhar, S.; Rengasamy, R. A perspective on the biotechnological potential of microalgae. Crit. Rev. Microbiol. 2008, 34, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Mata, T.M.; Martins, A.A.; Caetano, N.S. Microalgae for biodiesel production and other applications: A review. Renew. Sustain. Energy Rev. 2010, 14, 217–232. [Google Scholar] [CrossRef]
- De Wilt, A.; Butkovskyi, A.; Tuantet, K.; Leal, L.H.; Fernandes, T.V.; Langenhoff, A.; Zeeman, G. Micropollutant removal in an algal treatment system fed with source separated wastewater streams. J. Hazard. Mater. 2015, 304, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lin, G.; Podola, B.; Melkonian, M. Continuous removal of zinc from wastewater and mine dump leachate by a microalgal biofilm PSBR. J. Hazard. Mater. 2015, 297, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Sioli, H. The Amazon and its main affluents: Hydrography, morphology of the river courses, and river types. In Monographiae Biologicae; Sioli, H., Ed.; Springer: Dordrecht, The Netherlands, 1984; pp. 127–165. [Google Scholar]
- Moniz, M.B.J.; Kaczmarska, I. Barcoding of Diatoms: Nuclear Encoded ITS Revisited. Protist 2010, 161, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 2009, 50, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.C.; Coleman, A.W. The Internal Transcribed Spacer 2 Exhibits a Common Secondary Structure in Green Algae and Flowering Plants. J. Mol. Evol. 1997, 44, 258–271. [Google Scholar] [CrossRef] [PubMed]
- Abou-Shanab, R.A.I.; Matter, I.A.; Kim, S.-N.; Oh, Y.-K.; Choi, J.; Jeon, B.-H. Characterization and identification of lipid-producing microalgae species isolated from a freshwater lake. Biomass Bioenergy 2011, 35, 3079–3085. [Google Scholar] [CrossRef]
- Do Nascimento, M.; Ortiz-Marquez, J.C.F.; Sanchez-Rizza, L.; Echarte, M.M.; Curatti, L. Bioprospecting for fast growing and biomass characterization of oleaginous microalgae from South-Eastern Buenos Aires, Argentina. Bioresour. Technol. 2012, 125, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Araujo, G.S.; Matos, L.J.B.L.; Gonçalves, L.R.B.; Fernandes, F.A.N.; Farias, W.R.L. Bioprospecting for oil producing microalgal strains: Evaluation of oil and biomass production for ten microalgal strains. Bioresour. Technol. 2011, 102, 5248–5250. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Li, Y.; Min, M.; Hu, B.; Chen, P.; Ruan, R. Local bioprospecting for high-lipid producing microalgal strains to be grown on concentrated municipal wastewater for biofuel production. Bioresour. Technol. 2011, 102, 6909–6919. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, E.A.; Farahat, L.A.; Abdel Aziz, Z.K.; Fatthallah, N.A.; Salah El Din, R.A. Evaluation of the potential for some isolated microalgae to produce biodiesel. Egypt. J. Pet. 2015, 24, 97–101. [Google Scholar] [CrossRef]
- Ho, S.; Lai, Y.-Y.; Chiang, C.-Y.; Chen, C.-N.N.; Chang, J.-S. Selection of elite microalgae for biodiesel production in tropical conditions using a standardized platform. Bioresour. Technol. 2013, 147, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cui, J.; Liu, Q.; Ding, Y.; Liu, J. Screening and phylogenetic analysis of lipid-rich microalgae. Algal Res. 2015, 11, 381–386. [Google Scholar] [CrossRef]
- Rodolfi, L.; Chini Zittelli, G.; Bassi, N.; Padovani, G.; Biondi, N.; Bonini, G.; Tredici, M.R. Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 2009, 102, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Miao, X.; Li, R.; Zhong, J. In situ biodiesel production from fast-growing and high oil content Chlorella pyrenoidosa in rice straw hydrolysate. J. Biomed. Biotechnol. 2011, 2011, 141207. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.T.Y.; Sivaloganathan, B.; Obbard, J.P. Screening of marine microalgae for biodiesel feedstock. Biomass Bioenergy 2011, 35, 2534–2544. [Google Scholar] [CrossRef]
- Abou-Shanab, R.A.I.; Hwang, J.-H.; Cho, Y.; Min, B.; Jeon, B.-H. Characterization of microalgal species isolated from fresh water bodies as a potential source for biodiesel production. Appl. Energy 2011, 88, 3300–3306. [Google Scholar] [CrossRef]
- Chaichalerm, S.; Pokethitiyook, P.; Yuan, W.; Meetam, M.; Sritong, K.; Pugkaew, W.; Kungvansaichol, K.; Kruatrachue, M.; Damrongphol, P. Culture of microalgal strains isolated from natural habitats in Thailand in various enriched media. Appl. Energy 2012, 89, 296–302. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, P.; Yang, X.; Hao, Z.; Zhang, L.; Luo, N.; Shi, J. Isolation and identification by 18S rDNA sequence of high lipid potential microalgal species for fuel production in Hainan Dao. Biomass Bioenergy 2014, 66, 197–203. [Google Scholar] [CrossRef]
- Chiu, S.-Y.; Kao, C.-Y.; Tsai, M.-T.; Ong, S.-C.; Chen, C.-H.; Lin, C.-S. Lipid accumulation and CO2 utilization of Nannochloropsis oculata in response to CO2 aeration. Bioresour. Technol. 2009, 100, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Huerlimann, R.; de Nys, R.; Heimann, K. Growth, lipid content, productivity, and fatty acid composition of tropical microalgae for scale-up production. Biotechnol. Bioeng. 2010, 107, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Sarkany, N.; Cui, Y. Biomass and lipid productivities of Chlorella vulgaris under autotrophic, heterotrophic and mixotrophic growth conditions. Biotechnol. Lett. 2009, 31, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Hempel, N.; Petrick, I.; Behrendt, F. Biomass productivity and productivity of fatty acids and amino acids of microalgae strains as key characteristics of suitability for biodiesel production. J. Appl. Phycol. 2012, 24, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Yeesang, C.; Cheirsilp, B. Effect of nitrogen, salt, and iron content in the growth medium and light intensity on lipid production by microalgae isolated from freshwater sources in Thailand. Bioresour. Technol. 2011, 102, 3034–3040. [Google Scholar] [CrossRef] [PubMed]
- Msanne, J.; Xu, D.; Konda, A.R.; Casas-Mollano, J.A.; Awada, T.; Cahoon, E.B.; Cerutti, H. Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry 2012, 75, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Lin, J. Effects of nitrogen source and concentration on biomass and oil production of a Scenedesmus rubescens like microalga. Bioresour. Technol. 2011, 102, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, J.; Dunahay, T.; Benemann, J.; Roessler, P. A Look Back at the U.S. Department of Energy’s Aquatic Species Program—Biodiesel from Algae; U.S. Department of Energy’s Office of Fuels Development: Golden, CO, USA, 1998; p. 328.
- Vaulot, D.; Olson, R.J.; Merkel, S.; Chisholm, S.W. Cell-cycle response to nutrient starvation in two phytoplankton species, Thalassiosira weissflogii and Hymenomonas carterae. Mar. Biol. 1987, 95, 625–630. [Google Scholar] [CrossRef]
- Olson, R.J.; Chisholm, S.W. Effects of light and nitrogen limitation on the cell cycle of the dinoflagellate Amphidinium carteri. J. Plankton Res. 1986, 8, 785–793. [Google Scholar] [CrossRef]
- Li, Y.; Fei, X.; Deng, X. Novel molecular insights into nitrogen starvation-induced triacylglycerols accumulation revealed by differential gene expression analysis in green algae Micractinium pusillum. Biomass Bioenergy 2012, 42, 199–211. [Google Scholar] [CrossRef]
- Abdelaziz, A.E.M.; Leite, G.B.; Belhaj, M.A.; Hallenbeck, P.C. Screening microalgae native to Quebec for wastewater treatment and biodiesel production. Bioresour. Technol. 2014, 157, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Valdez-Ojeda, R.; González-Muñoz, M.; Us-Vázquez, R.; Narváez-Zapata, J.; Chavarria-Hernandez, J.C.; López-Adrián, S.; Barahona-Pérez, F.; Toledano-Thompson, T.; Garduño-Solórzano, G.; Escobedo-Gracia Medrano, R.M. Characterization of five fresh water microalgae with potential for biodiesel production. Algal Res. 2015, 7, 33–44. [Google Scholar] [CrossRef]
- Shrivastav, A.; Mishra, S.K.; Suh, W.I.; Farooq, W.; Moon, M.; Kim, T.-H.; Kumar, K.; Choi, G.-G.; Park, M.S.; Yang, J.-W. Characterization of newly isolated oleaginous microalga Monoraphidium sp. for lipid production under different conditions. Algal Res. 2015, 12, 289–294. [Google Scholar] [CrossRef]
- Yen, H.-W.; Hu, I.-C.; Chen, C.-Y.; Ho, S.-H.; Lee, D.-J.; Chang, J.-S. Microalgae-based biorefinery—From biofuels to natural products. Bioresour. Technol. 2013, 135, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Y.; Cheng, J.J.; Mos, M.; Daroch, M. Biological potential of microalgae in China for biorefinery-based production of biofuels and high value compounds. New Biotechnol. 2015, 32, 588–596. [Google Scholar] [CrossRef] [PubMed]
- El-Sheekh, M.; Abomohra, A.E.-F.; Hanelt, D. Optimization of biomass and fatty acid productivity of Scenedesmus obliquus as a promising microalga for biodiesel production. World J. Microbiol. Biotechnol. 2013, 29, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Guldhe, A.; Kumari, S.; Rawat, I.; Bux, F. Investigation of combined effect of nitrogen, phosphorus and iron on lipid productivity of microalgae Ankistrodesmus falcatus KJ671624 using response surface methodology. Biochem. Eng. J. 2015, 94, 22–29. [Google Scholar] [CrossRef]
- Kilham, S.; Kreeger, D.; Goulden, C.; Lynn, S. Effects of nutrient limitation on biochemical constituents of Ankistrodesmus falcatus. Freshw. Biol. 1997, 38, 591–596. [Google Scholar] [CrossRef]
- Breuer, G.; Lamers, P.P.; Martens, D.E.; Draaisma, R.B.; Wijffels, R.H. The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour. Technol. 2012, 124, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Breuer, G.; de Jaeger, L.; Artus, V.G.; Martens, D.E.; Springer, J.; Draaisma, R.B.; Eggink, G.; Wijffels, R.H.; Lamers, P.P. Superior triacylglycerol (TAG) accumulation in starchless mutants of Scenedesmus obliquus: (II) evaluation of TAG yield and productivity in controlled photobioreactors. Biotechnol. Biofuels 2014, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.; Prakash-Rai, M.; Sharma, R. Effect of Nitrogen on Growth and Lipid Content of Chlorella pyrenoidosa. Am. J. Biochem. Biotechnol. 2011, 7, 126–131. [Google Scholar] [CrossRef]
- Piligaev, A.V.; Sorokina, K.N.; Bryanskaya, A.V.; Peltek, S.E.; Kolchanov, N.A.; Parmon, V.N. Isolation of prospective microalgal strains with high saturated fatty acid content for biofuel production. Algal Res. 2015, 12, 368–376. [Google Scholar] [CrossRef]
- Weiss, S.B.; Kennedy, E.P. The enzymatic synthesis of triglycerides. J. Am. Chem. Soc. 1956, 78, 3550. [Google Scholar] [CrossRef]
- Miller, R.; Wu, G.; Deshpande, R.R.; Vieler, A.; Gärtner, K.; Li, X.; Moellering, E.R.; Zäuner, S.; Cornish, A.J.; Liu, B.; et al. Changes in Transcript Abundance in Chlamydomonas reinhardtii following Nitrogen Deprivation Predict Diversion of Metabolism. Plant Physiol. 2010, 154, 1737–1752. [Google Scholar] [CrossRef] [PubMed]
- Rismani-Yazdi, H.; Haznedaroglu, B.Z.; Bibby, K.; Peccia, J. Transcriptome sequencing and annotation of the microalgae Dunaliella tertiolecta: Pathway description and gene discovery for production of next-generation biofuels. BMC Genom. 2011, 12, 148. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.J.; Fernández, C.M.; Casas, A.; Rodríguez, L.; Pérez, A. Influence of fatty acid composition of raw materials on biodiesel properties. Bioresour. Technol. 2009, 100, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Knothe, G. Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Process. Technol. 2005, 86, 1059–1070. [Google Scholar] [CrossRef]
- Knothe, G. “Designer” Biodiesel: Optimizing Fatty Ester Composition to Improve Fuel Properties. Energy Fuels 2008, 22, 1358–1364. [Google Scholar] [CrossRef]
- Smith, P.C.; Ngothai, Y.; Dzuy Nguyen, Q.; O’Neill, B.K. Improving the low-temperature properties of biodiesel: Methods and consequences. Renew. Energy 2010, 35, 1145–1151. [Google Scholar] [CrossRef]
- Dunn, R.O. Effect of antioxidants on the oxidative stability of methyl soyate (biodiesel). Fuel Process. Technol. 2005, 86, 1071–1085. [Google Scholar] [CrossRef]
- Knothe, G. Improving biodiesel fuel properties by modifying fatty ester composition. Energy Environ. Sci. 2009, 2, 759–766. [Google Scholar] [CrossRef]
| Source of Collection | Strain | Genera/Species | GenBank Accession Number | Biomass Productivity (mg·L−1·d−1) | Lipid Productivity (mg·L−1·d−1) | Lipid Content (% dw) |
|---|---|---|---|---|---|---|
| Amazon River (03°41′0.6′′ S, 73°14′8.9′′ W) | AMA001 | Ankistrodesmus sp. | KP878496.1 | 12.2 ± 0.3 | 3.7 ± 0.1 | 30.7 ± 1.5 |
| AMA002 | Ankistrodesmus sp. | KP878497.1 | 3.6 ± 0.4 | 1.0 ± 0.2 | 27.7 ± 2.1 | |
| AMA003 | Chlorella lewinii | KP878501.1 | 22.3 ± 0.7 | 5.1 ± 0.5 | 22.7 ± 1.5 | |
| AMA004 | Acutodesmus abliquus | KP878505.1 | 31.6 ± 0.4 | 6.4 ± 0.7 | 20.3 ± 2.1 | |
| Itaya River (03°43′1.4′′ S, 73°14′17.8′′ W) | ITA001 | Ankistrodesmus sp. | KP878499.1 | 5.9 ± 0.2 | 4.0 ± 0.3 | 43.7 ± 1.7 |
| ITA002 | Ankistrodesmus sp. | KP878500.1 | 13.6 ± 0.4 | 5.7 ± 0.5 | 30.6 ± 2.0 | |
| ITA003 | Chlorella lewinii | KP878503.1 | 12.9 ± 0.3 | 4.0 ± 0.5 | 22.4 ± 2.7 | |
| ITA004 | Chlorella lewinii | KP878504.1 | 22.1 ± 0.7 | 3.1 ± 0.4 | 21.5 ± 2.3 | |
| ITA005 | Scenedesmus regularis | KP878508.1 | 9.3 ± 0.4 | 5.6 ± 0.4 | 24.2 ± 1.4 | |
| ITA006 | Scenedesmus dimorphus | KP878509.1 | 18.6 ± 0.6 | 3.2 ± 0.3 | 26.0 ± 1.7 | |
| ITA007 | Acutodesmus abliquus | KP878510.1 | 17.9 ± 0.4 | 2.2 ± 0.3 | 15.6 ± 1.9 | |
| Nanay River (03°42′0.2′′ S, 73°15′32′′ W) | NAN001 | Ankistrodesmus sp. | KP878498.1 | 14.3 ± 0.6 | 2.8 ± 0.2 | 46.7 ± 1.5 |
| NAN002 | Desmodesmus tropicus | KP878506.1 | 12.2 ± 0.4 | 2.3 ± 0.2 | 17.5 ± 1.1 | |
| NAN003 | Tetradesmus sp. | KP878507.1 | 13.9 ± 0.4 | 3.0 ± 0.3 | 13.5 ± 1.2 |
| Fatty Acids | Acutodesmus obliquus | Ankistrodesmus sp. | Chlorella lewinii | |||
|---|---|---|---|---|---|---|
| +N | −N | +N | −N | +N | −N | |
| 14:0 | - | - | 0.52 ± 0.02 | 0.88 ± 0.02 | - | 0.58 ± 0.01 |
| 16:0 | 27.24 ± 0.07 | 28.67 ± 0.17 | 18.68 ± 0.19 | 22.61 ± 0.06 | 19.64 ± 0.21 | 21.23 ± 0.10 |
| 18:0 | 1.84 ± 0.05 | 2.55 ± 0.51 | 1.01 ± 0.02 | 1.81 ± 0.02 | 3.71 ± 0.26 | 1.53 ± 0.15 |
| 16:1n-7 | 0.23 ± 0.00 | 0.25 ± 0.01 | 0.71 ± 0.03 | 0.55 ± 0.01 | 0.85 ± 0.43 | 0.38 ± 0.00 |
| 18:1n-7 | 1.31 ± 0.02 | 1.31 ± 0.00 | 1.41 ± 0.05 | 0.94 ± 0.03 | 1.46 ± 0.06 | 1.23 ± 0.02 |
| 18:1n-9 | 32.13 ± 0.21 | 36.08 ± 0.21 | 36.99 ± 0.04 | 40.47 ± 0.38 | 13.71 ± 0.15 | 23.26 ± 0.41 |
| 18:2n-6 | 17.32 ± 0.04 | 16.51 ± 0.16 | 7.47 ± 0.18 | 6.56 ± 0.07 | 20.34 ± 0.39 | 19.57 ± 0.01 |
| 18:3n-3 | 11.30 ± 0.17 | 8.54 ± 0.31 | 14.86 ± 0.04 | 11.73 ± 0.12 | 18.15 ± 0.22 | 13.71 ± 0.16 |
| 18:3n-6 | 1.01 ± 0.01 | 0.78 ± 0.02 | 0.55 ± 0.00 | 0.54 ± 0.02 | 1.24 ± 0.06 | 1.00 ± 0.20 |
| 18:4n-3 | 2.50 ± 0.05 | 1.77 ± 0.08 | 6.31 ± 0.02 | 6.01 ± 0.05 | 4.91 ± 0.25 | 4.03 ± 0.20 |
| 20:4n-6 | - | - | - | 0.84 ± 0.08 | - | - |
| 22:5n-3 | - | - | - | 0.62 ± 0.02 | - | - |
| Unidentified | 5.12 ± 0.03 | 3.55 ± 0.06 | 11.49 ± 0.04 | 6.45 ± 0.03 | 15.99 ± 0.34 | 13.48 ± 0.06 |
| SFA | 29.07 | 31.22 | 20.22 | 25.31 | 23.35 | 23.34 |
| UFA | 70.93 | 68.78 | 79.78 | 74.69 | 76.65 | 76.66 |
| MUFA | 33.67 | 37.64 | 39.11 | 41.95 | 16.02 | 24.87 |
| PUFA | 32.13 | 27.59 | 29.18 | 26.29 | 44.64 | 38.31 |
| SFA + MUFA | 62.74 | 68.86 | 59.33 | 67.26 | 39.36 | 48.21 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cobos, M.; Paredes, J.D.; Maddox, J.D.; Vargas-Arana, G.; Flores, L.; Aguilar, C.P.; Marapara, J.L.; Castro, J.C. Isolation and Characterization of Native Microalgae from the Peruvian Amazon with Potential for Biodiesel Production. Energies 2017, 10, 224. https://doi.org/10.3390/en10020224
Cobos M, Paredes JD, Maddox JD, Vargas-Arana G, Flores L, Aguilar CP, Marapara JL, Castro JC. Isolation and Characterization of Native Microalgae from the Peruvian Amazon with Potential for Biodiesel Production. Energies. 2017; 10(2):224. https://doi.org/10.3390/en10020224
Chicago/Turabian StyleCobos, Marianela, Jae D. Paredes, J. Dylan Maddox, Gabriel Vargas-Arana, Leenin Flores, Carla P. Aguilar, Jorge L. Marapara, and Juan C. Castro. 2017. "Isolation and Characterization of Native Microalgae from the Peruvian Amazon with Potential for Biodiesel Production" Energies 10, no. 2: 224. https://doi.org/10.3390/en10020224
APA StyleCobos, M., Paredes, J. D., Maddox, J. D., Vargas-Arana, G., Flores, L., Aguilar, C. P., Marapara, J. L., & Castro, J. C. (2017). Isolation and Characterization of Native Microalgae from the Peruvian Amazon with Potential for Biodiesel Production. Energies, 10(2), 224. https://doi.org/10.3390/en10020224






