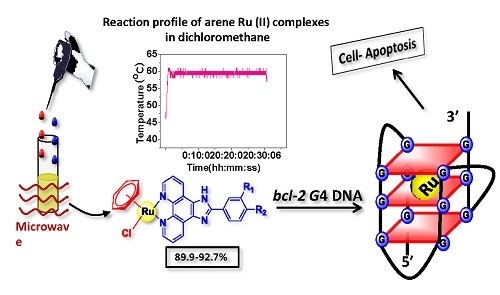
Microwave-Assisted Synthesis of Arene Ru(II) Complexes Induce Tumor Cell Apoptosis Through Selectively Binding and Stabilizing bcl-2 G-Quadruplex DNA
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Instruments
2.3. Synthesis and Characterization
2.3.1. Synthesis of 1a, 2a, 3a, and 4a
2.3.2. Microwave-Assisted Synthesis of Complex [(η6-C6H6)Ru(p-ClPIP)Cl]Cl 1b
2.3.3. Microwave-Assisted Synthesis of Complex [(η6-C6H6)Ru(m-ClPIP)Cl]Cl 2b
2.3.4. Microwave-Assisted Synthesis of Complex [(η6-C6H6)Ru(p-NPIP)Cl]Cl 3b
2.3.5. Microwave-Assisted Synthesis of Complex [(η6-C6H6)Ru(m-NPIP)Cl]Cl 4b
2.4. Biochemical Mechanism
2.4.1. Cell Lines and Cell Culture
2.4.2. MTT Assay
2.4.3. Flow Cytometric Analysis
2.4.4. Bcl-2 G-Quadruplex DNA-Binding Studies
Electronic Spectra
EB-Quenching Experiments
Circular Dichroism
CD Melting
3. Results and Discussion
3.1. Microwave-Assisted Synthesis of Arene Ru(II) Complexes
3.2. Inhibit the Growth of A549 Cells through Inducing Cell Apoptosis
3.3. Bindingand Stabilizing bcl-2 G-Quadruplex DNA
3.3.1. Electronic Spectra
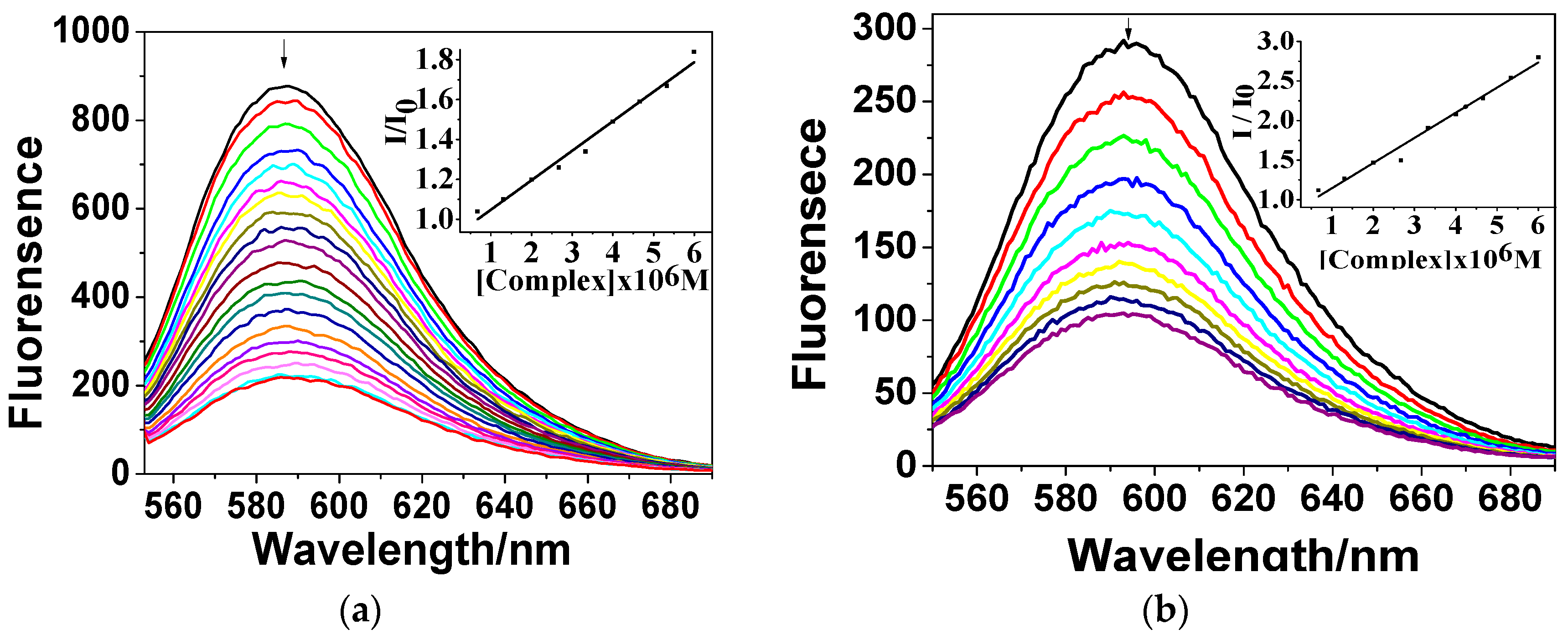
3.3.2. Fluorescence Quenching
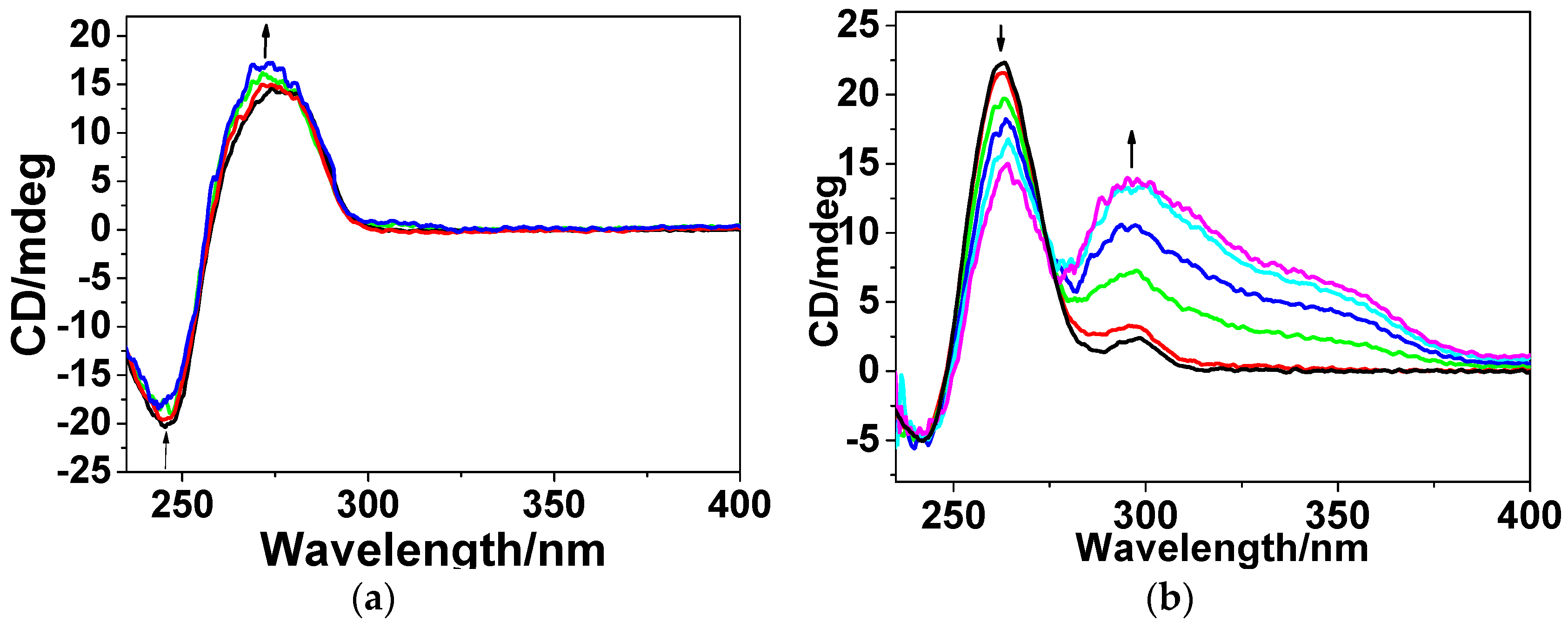
3.3.3. Circular Dichroism
3.3.4. Melting Point Experiments
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Scolaro, C.; Bergamo, A.; Brescacin, L.; Delfino, R.; Cocchietto, M.; Laurenczy, G.; Geldbach, T.J.; Sava, G.; Dyson, P.J. In vitro and in vivo evaluation of ruthenium(II)-arene PTA complexes. J. Med. Chem. 2005, 48, 4161–4171. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-K.; Sadler, P.J. Metal complexes as DNA intercalators. Acc. Chem. Res. 2011, 44, 349–359. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liao, S.Y.; Tan, C.P.; Ye, R.R.; Xu, Y.W.; Zhao, M.; Ji, L.N.; Mao, Z.W. Ruthenium-arene-β-carboline complexes as potent inhibitors of cyclin-dependent Kinase 1: Synthesis, characterization and anticancer mechanism studies. Chem. Eur. J. 2013, 19, 12152–12160. [Google Scholar] [CrossRef] [PubMed]
- Vajpayee, V.; Lee, S.; Kim, S.H.; Kang, S.C.; Cook, T.R.; Kim, H.; Kim, D.W.; Verma, S.S.; Lah, M.S.; Kim, I.S.; et al. Self-assembled metalla-rectangles bearing azodipyridyl ligands: Synthesis, characterization and antitumor activity. Dalton Trans. 2013, 42, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Qian, Q.; Li, P.; Lei, X.; Xiao, Q.; Huang, S.; Huang, C.; Cui, J. Synthesis, characterization, and anticancer activity of a series of ketone-N4-substituted thiosemicarbazones and their Ruthenium(II) arene complexes. Inorg. Chem. 2013, 52, 12440–12449. [Google Scholar] [CrossRef] [PubMed]
- Adriana, G.; Ovidiu, B.; Loredana, B.; Thomas, C.; Ioana, B.N.; Brun, T. Synthesis, anticancer activity, and genome profiling of thiazolo aene Ruthenium complexes. J. Med. Chem. 2015, 58, 8475–8490. [Google Scholar]
- Pettinari, R.; Pettinari, C.; Marchetti, F.; Skelton, B.W.; White, A.H.; Bonfili, L.; Cuccioloni, M.; Mozzicafreddo, M.; Cecarini, V.; Angeletti, M.; et al. Arene-ruthenium(II) acypyrazolonato complexes: Apoptosis-promoting effects on human cancer cells. J. Med. Chem. 2014, 57, 4532–4542. [Google Scholar] [CrossRef] [PubMed]
- Caruso, F.; Monti, E.; Matthews, J.; Rossi, M.; Gariboldi, M.B.; Pettinari, C.; Pettinari, R.; Marchetti, F. Synthesis, characterization, and antitumor activity of water-soluble (arene)Ruthenium(II) derivatives of 1,3-Dimethyl-4-acylpyrazolon-5-ato ligands. First example of Ru(arene)(ligand) antitumor species involving simultaneous Ru–N7(guanine) bonding and ligand intercalation to DNA. Inorg. Chem. 2014, 53, 3668–3677. [Google Scholar] [PubMed]
- Olga, N.; Jana, K.; Vendula, B.; Ctirad, H.; Marie, V.; Haimei, C.; Sadler, P.J.; Viktor, B. Conformation of DNA modified by monofunctional Ru(II) arene complexes: Recognition by DNA binding proteins and repair. Relationship to cytotoxicity. Chem. Biol. 2005, 12, 121–129. [Google Scholar]
- Patrycja, N.S.; van Beijnum, J.R.; Angela, C.; Nazarov, A.A.; Georges, W.; Hubert, V.D.B.; Dyson, P.J.; Griffioen, A.W. Organometallic ruthenium(II) arene compounds with antiangiogenic activity. J. Med. Chem. 2011, 54, 3895–3902. [Google Scholar]
- Bergamo, A.; Masi, A.; Peacock, A.F.A.; Habtemariam, A.; Sadler, P.J.; Sava, G. In vivo tumour and metastasis reduction and in vitro effects on invasion assays of the ruthenium RM175 and osmium AFAP51 organometallics in the mammary cancer model. Inorg. Biochem. 2010, 104, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; He, J.; Mei, W.J.; Zhang, Z.; Wu, X.H.; Sun, F.Y. Arene ruthenium(II) complex, a potent inhibitor against proliferation, migration and invasion of breast cancer cells, reduces stress fibers, focal adhesions and invadopodia. Metallomics 2014, 6, 2204–2212. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Fan, C.D.; Chen, T.F.; Liu, C.R.; Mei, W.J.; Chen, S.D.; Wang, B.G.; Chen, Y.Y.; Zheng, W.J. Microwave-assisted synthesis of arene ruthenium(II) complexes that induce S-phase arrest in cancer cells by DNA damage-mediated p53 phosphorylation. Eur. J. Med. Chem. 2013, 63, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zheng, K.D.; Liao, S.; Ding, Y.; Li, Y.Q.; Mei, W.J. Arene Ruthenium (II) complexes as low-toxicity inhibitor against the proliferation, migration, and invasion of MDA-MB-231 cells through binding and stabilizing c-myc G-quadruplex DNA. Organometallics 2016, 35, 317–326. [Google Scholar] [CrossRef]
- Winkhaus, G.; Singer, H. Ruthen(II)-komplexe mit zweizähnigem cycloheptatrien und benzol. J. Organomet. Chem. 1967, 7, 487–491. [Google Scholar] [CrossRef]
- Renfrew, A.K.; Egger, A.E.; Scopelliti, R.; Hartinger, C.G.; Dyson, P.J. Synthesis and characterisation of the water soluble bis-phosphine complex [Ru(η6-cymene)(PPh2(o-C6H4O)-κ2-P,O)(pta)]+ and an investigation of its cytotoxic effects. Comptes Rendus Chim. 2010, 13, 1144–1150. [Google Scholar] [CrossRef]
- Giguere, R.J.; Bray, T.L.; Duncan, S.M.; Majetich, G. Application of Commercial Microwave Ovens to Organic Synthesis. Tetrahedron Lett. 1986, 27, 4945–4948. [Google Scholar] [CrossRef]
- Kitchen, H.J.; Vallance, S.R.; Kennedy, J.L.; Tapia-Ruiz, N.; Carassiti, L.; Harrison, A.; Whittaker, A.G.; Drysdale, T.D.; Kingman, S.W.; Gregory, D.H. Modern microwave methods in solid-state inorganic materials chemistry: From fundamentals to manufacturing. Chem. Rev. 2014, 114, 1170–1206. [Google Scholar] [CrossRef] [PubMed]
- Antonio, D.L.H.; Angel, D.O.; Andres, M. Microwaves in organic synthesis. Thermal and non-thermal microwave effects. Chem. Soc. Rev. 2005, 34, 164–178. [Google Scholar]
- Iannelli, M.; Ritter, H. Microwave-assisted direct synthesis and polymerization of chiral acrylamide. Macromol. Chem. Phys. 2005, 206, 349–353. [Google Scholar] [CrossRef]
- Keith, O.P.; Maksymilian, K.; Anna, Z.; Dorota, G. Microwave-assisted cobinamide synthesis. J. Org. Chem. 2014, 79, 7752–7757. [Google Scholar]
- Dallinger, D.; Kappe, C.O. Microwave-assisted synthesis in water as solvent. J. Cheminform. 2007, 38, 2563–2591. [Google Scholar] [CrossRef]
- Zhang, Z.; Mei, W.J.; Wu, X.H.; Wang, X.C.; Wang, B.G.; Chen, S.D. Synthesis and characterization of chiral ruthenium(II) complexes Λ/Δ-[Ru(bpy)2(H2iip)](ClO4)2 as stabilizers of c-myc G-quadruplex DNA. J. Coord. Chem. 2015, 68, 1465–1475. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, T.F.; Zhang, Z.; Liao, S.Y.; Wu, X.H.; Wu, J.; Mei, W.; Chen, Y.H.; Wu, W.L.; Zeng, L.L.; et al. Microwave-assisted synthesis of arene ruthenium(II) complexes [(η6-RC6H5)Ru(m-MOPIP)Cl]Cl (R = -H and -CH3) as groove binder to c-myc G4 DNA. Dalton Trans. 2014, 43, 9216–9225. [Google Scholar] [CrossRef] [PubMed]
- Bolink, H.J.; Luca, C.; Eugenio, C.; Michael, G.T.; Enrique, O.; Costa, R.D.; Viruela, P.M.; Nazeeruddin, M.K. Stable single-layer light-emitting electrochemical cell using 4,7-diphenyl-1,10-phenanthroline-bis(2-phenylpyridine)iridium(III) hexafluorophosphate. J. Am. Chem. Soc. 2006, 128, 14786–14787. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsson, M.; Becker, H.C.; Hammarström, L.; Bonnefous, C.; Chamchoumis, C.; Thummel, R.P. Six-Membered Ring Chelate Complexes of Ru(II): Structural and Photophysical Effects. Inorg. Chem. 2007, 46, 10354–10364. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C.; Kitada, S.; Takayama, S.; Miyashita, T. Regulation of chemoresistance by the bcl-2 oncoprotein in non-Hodgkin’s lymphoma and lymphocytic leukemia cell lines. Ann. Oncol. 1994, 5, 61–65. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Daekyu, S.; Hurley, L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. [Google Scholar] [CrossRef] [PubMed]
- Onyshchenko, M.I.; Gaynutdinov, T.I.; Englund, E.A.; Appella, D.H.; Neumann, R.D.; Panyutin, I.G. Stabilization of G-quadruplex in the BCL2 promoter region in double-stranded DNA by invading short PNAs. Nucleic Acid. Res. 2009, 37, 7570–7580. [Google Scholar] [CrossRef] [PubMed]
- Stephen, N.; Gary, P. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002, 1, 383–393. [Google Scholar]
- Reed, J.E.; Anna Arola, A.; Stephen, N.; Ramón, V. Stabilization of G-quadruplex DNA and inhibition of telomerase activity by square-planar nickel(II) complexes. J. Am. Chem. Soc. 2006, 128, 5992–5993. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.; Williamson, M.P.; Thomas, J.A. Differentiating quadruplexes: Binding preferences of a luminescent dinuclear ruthenium(II) complex with four-stranded DNA structures. Org. Biomol. Chem. 2010, 8, 2617–2621. [Google Scholar] [CrossRef] [PubMed]
- Amrita, G.; Priyadip, D.; Gill, M.R.; Prasenjit, K.; Walker, M.G.; Thomas, J.A.; Amitava, D. Photoactive Ru(II)-polypyridyl complexes that display sequence selectivity and high-affinity binding to duplex DNA through groove binding. Chemistry 2011, 17, 2089–2098. [Google Scholar]
- Chan, D.S.; Yang, H.; Kwan, M.H.; Cheng, Z.; Lee, P.; Bai, L.P.; Jiang, Z.H.; Wong, C.Y.; Fong, W.F.; Leung, C.H.; et al. Structure-based optimization of FDA-approved drug methylene blue as a c-myc G-quadruplex DNA stabilizer. Biochimie 2011, 93, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Rajendiran, V.; Murali, M.; Suresh, E.; Sinha, S.; Somasundaram, K.; Palaniandavar, M. Mixed ligand ruthenium(II) complexes of bis(pyrid-2-yl)-/bis(benzimidazol-2-yl)-dithioether and diimines: Study of non-covalent DNA binding and cytotoxicity. Dalton Trans. 2008, 1, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Therrien, B. Functionalised η6-arene ruthenium complexes. Coord. Chem. Rev. 2009, 253, 493–519. [Google Scholar] [CrossRef]
- Chen, T.; Wong, Y. Selenocystine induces reactive oxygen species-mediated apoptosis in human cancer cells. Biomed. Pharmacother. 2008, 63, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wong, Y.S. Selenocystine induces S-phase arrest and apoptosis in human breast adenocarcinoma MCF-7 cells by modulating ERK and Akt phosphorylation. J. Agric. Food Chem. 2008, 56, 10574–10581. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.T.; Rodriguez, M.; Bard, A.J. Voltammetric studies of the interaction of metal chelates with DNA. II. Tris-chelated complexes of cobalt(III) and iron(II) with 1,10-phenanthroline and 2,2′-bipyridine. J. Am. Chem. Soc. 1989, 111, 8901–8911. [Google Scholar] [CrossRef]
- Mei, W.J.; Liu, J.; Chao, H.; Ji, L.N.; Li, A.X.; Liu, J.Z. DNA-binding and cleavage studies of a novel porphyrin ruthenium mixed complex [MPyTPP—Ru(pip)2Cl]+. Trans. Met. Chem. 2003, 28, 852–857. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Brabec, V.; Kleinwächter, V.; Butour, J.L.; Johnson, N.P. Biophysical studies of the modification of DNA by antitumour platinum coordination complexes. Biophys. Chem. 1990, 35, 129–141. [Google Scholar] [CrossRef]
- Ju, C.C.; Zhang, A.G.; Yuan, C.L.; Zhao, X.L.; Wang, K.Z. The interesting DNA-binding properties of three novel dinuclear Ru(II) complexes with varied lengths of flexible bridges. J. Inorg. Biochem. 2011, 105, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Beckford, F.A.; Shaloski, M., Jr.; Leblanc, G.; Thessing, J.; Lewis-Alleyne, L.C.; Holder, A.A.; Li, L.; Seeram, N.P. Microwave synthesis of mixed ligand diimine-thiosemicarbazone complexes of ruthenium(II): Biophysical reactivity and cytotoxicity. Dalton Trans. 2009, 48, 10757–10764. [Google Scholar] [CrossRef] [PubMed]
- Pasternack, R.F.; Caccam, M.; Keogh, B.; Stephenson, T.A.; Williams, A.P.; Gibbs, E.J. Long-range fluorescence quenching of ethidium ion by cationic porphyrins in the presence of dna. J. Am. Chem. Soc. 1991, 113, 6835–6840. [Google Scholar] [CrossRef]
- Yu, H.J.; Zhao, Y.; Mo, W.J.; Hao, Z.F.; Yu, L. Ru-indoloquinoline complex as a selective and effective human telomeric G-quadruplex binder. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 132, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Tarushi, A.; Psomas, G.; Raptopoulou, C.P.; Kessissoglou, D.P. Zinc complexes of the antibacterial drug oxolinic acid: Structure and DNA-binding properties. J. Inorg. Biochem. 2009, 103, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Roxanne, K.; Pablo, E.; Johans, F.; Chantal, A.; Nicolas, M.; Sleiman, H.F. A platinum supramolecular square as an effective G-quadruplex binder and telomerase inhibitor. J. Am. Chem. Soc. 2008, 130, 10040–10041. [Google Scholar]
- Anne, D.C.; Elsa, D.; Jean-Louis, M.; Marie-Paule, T.F.; David, M. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007, 129, 1856–1857. [Google Scholar]
- Peng, D.; Tan, J.H.; Chen, S.B.; Ou, T.M.; Gu, L.Q.; Huang, Z.S. Bisaryldiketene derivatives: A new class of selective ligands for c-myc G-quadruplex DNA. Bioorg. Med. Chem. 2010, 18, 8235–8242. [Google Scholar] [CrossRef] [PubMed]


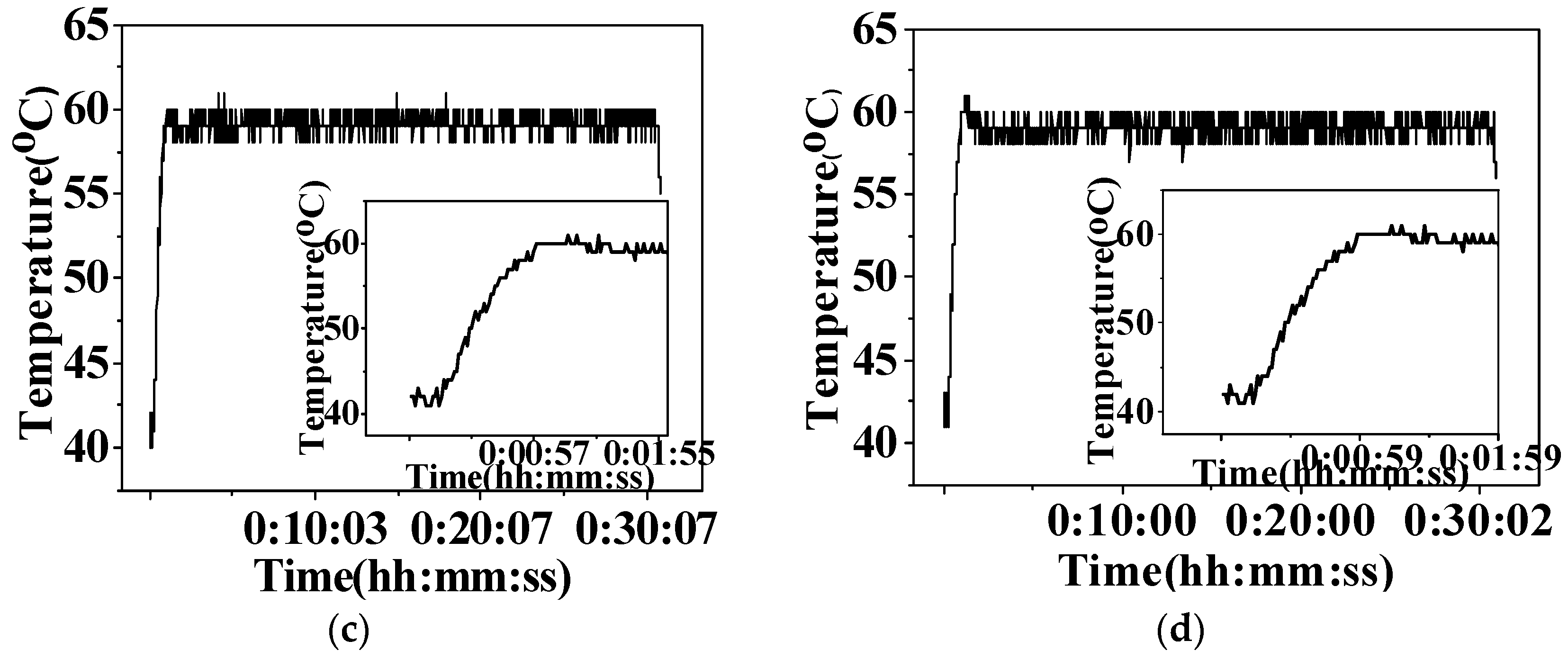



| Complexes | Microwave-Assisted | Conventional | ||||
|---|---|---|---|---|---|---|
| Temperature/°C | Time/h | Yield/% | Temperature/°C | Time/h | Yield/% | |
| 1b | 60 | 0.5 | 90.7 | 60 | 4 | 76.2 |
| 2b | 60 | 0.5 | 92.7 | 60 | 4 | 81.2 |
| 3b | 60 | 0.5 | 89.9 | 60 | 4 | 70.6 |
| 4b | 60 | 0.5 | 91.7 | 60 | 4 | 76.0 |
| Complexes | IC50/μmol | ||
|---|---|---|---|
| A549 | SMCC7721 | SW620 | |
| 1b | 16.59 | >100 | >100 |
| 2b | 26.55 | >100 | >100 |
| 3b | >100 | >100 | >100 |
| 4b | 91.88 | >100 | >100 |
| [Ru(arene)Cl2]2Cl2 | 73.19 | >100 | >100 |
| Cis-platin | 16.54 | 2.92 | 5.74 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wu, Q.; Wang, X.; Xie, Q.; Tang, Y.; Lan, Y.; Zhang, S.; Mei, W. Microwave-Assisted Synthesis of Arene Ru(II) Complexes Induce Tumor Cell Apoptosis Through Selectively Binding and Stabilizing bcl-2 G-Quadruplex DNA. Materials 2016, 9, 386. https://doi.org/10.3390/ma9050386
Chen Y, Wu Q, Wang X, Xie Q, Tang Y, Lan Y, Zhang S, Mei W. Microwave-Assisted Synthesis of Arene Ru(II) Complexes Induce Tumor Cell Apoptosis Through Selectively Binding and Stabilizing bcl-2 G-Quadruplex DNA. Materials. 2016; 9(5):386. https://doi.org/10.3390/ma9050386
Chicago/Turabian StyleChen, Yanhua, Qiong Wu, Xicheng Wang, Qiang Xie, Yunyun Tang, Yutao Lan, Shuangyan Zhang, and Wenjie Mei. 2016. "Microwave-Assisted Synthesis of Arene Ru(II) Complexes Induce Tumor Cell Apoptosis Through Selectively Binding and Stabilizing bcl-2 G-Quadruplex DNA" Materials 9, no. 5: 386. https://doi.org/10.3390/ma9050386