Community Structure Detection for Directed Networks through Modularity Optimisation
Abstract
:1. Introduction
2. Iterative Mathematical Programming Model for Modularity Optimisation on Directed Networks
| Sets | |
| node | |
| m | module |
| directed edge pointing from node n to e | |
| Parameters | |
| weight of edge point from node n to e | |
| sum of weights over all edges points to node n; incoming edge weight | |
| sum of weights over all edges points from node n; outgoing edge weight | |
| L | total amount of weights over all edges in the given network |
| Binary Variables | |
| 1 if node n belongs to module m; 0 otherwise | |
| Free Variables | |
| sum of for all nodes that belong to module m () | |
| sum of for all nodes that belong to module m () | |
| sum of edge weights in module m | |
| a positive intermediate variable. if both nodes n to e belong to module m; 0 otherwise | |
| represent the product of and , used as an intermediate variable for the MIP model | |
| represent the product of and , used as an intermediate variable for the MIP model | |
2.1. First Model—MINLP
2.2. Second Model—MIP
2.3. Full Algorithm
| Algorithm 1: Our proposed algorithm DiMod for detecting modules in directed networks. |
 |
3. Results
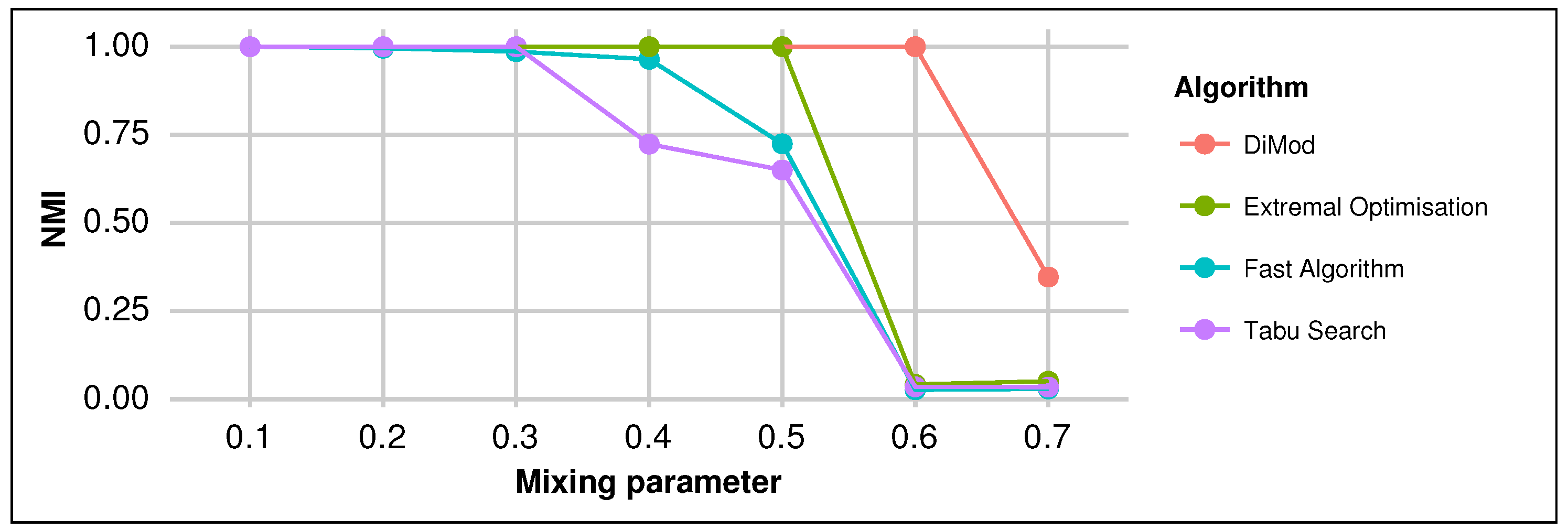
3.1. Synthetic Networks
3.2. Real Networks
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MINLP | Mixed Integer Non-Linear Programming |
| MIP | Mixed Integer Linear Programming |
References
- Boccaletti, S.; Latora, V.; Moreno, Y.; Chavez, M.; Hwang, D. Complex networks: Structure and dynamics. Phys. Rep. 2006, 424, 175–308. [Google Scholar] [CrossRef]
- Fortunato, S. Community detection in graphs. Phys. Rep. 2010, 486, 75–174. [Google Scholar] [CrossRef]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [PubMed]
- Brandes, U.; Delling, D.; Gaertler, M.; Gorke, R.; Hoefer, M.; Nikoloski, Z.; Wagner, D. On Modularity Clustering. IEEE Trans. Knowl. Data Eng. 2008, 20, 172–188. [Google Scholar] [CrossRef]
- Lancichinetti, A.; Fortunato, S. Limits of modularity maximization in community detection. Phys. Rev. E 2011, 84, 066122. [Google Scholar] [CrossRef] [PubMed]
- Good, B.H.; De Montjoye, Y.A.; Clauset, A. Performance of modularity maximization in practical contexts. Phys. Rev. E 2010, 81, 046106. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, S.; Hric, D. Community Detection in Networks: A User Guide. Phys. Rep. 2016. [Google Scholar] [CrossRef]
- Danon, L.; Díaz-Guilera, A.; Duch, J.; Arenas, A. Comparing community structure identification. J. Stat. Mech. Theory Exp. 2005, 2005, P09008. [Google Scholar] [CrossRef]
- Medus, A.; Acuña, G.; Dorso, C. Detection of community structures in networks via global optimization. Phys. A Stat. Mech. Appl. 2005, 358, 593–604. [Google Scholar] [CrossRef]
- Newman, M.E.J. Fast algorithm for detecting community structure in networks. Phys. Rev. E 2004, 69, 066133. [Google Scholar] [CrossRef] [PubMed]
- Duch, J.; Arenas, A. Community detection in complex networks using extremal optimization. Phys. Rev. E 2005, 72, 027104. [Google Scholar] [CrossRef] [PubMed]
- Aloise, D.; Cafieri, S.; Caporossi, G.; Hansen, P.; Perron, S.; Liberti, L. Column generation algorithms for exact modularity maximization in networks. Phys. Rev. E 2010, 82, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Bennett, L.; Papageorgiou, L.G.; Tsoka, S. Module detection in complex networks using integer optimisation. Algorithms Mol. Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Bennett, L.; Liu, S.; Papageorgiou, L.G.; Tsoka, S. Detection of Disjoint and Overlapping Modules in Weighted Complex Networks. Adv. Complex Syst. 2012, 15. [Google Scholar] [CrossRef]
- Cafieri, S.; Hansen, P.; Liberti, L. Improving heuristics for network modularity maximization using an exact algorithm. Discret. Appl. Math. 2014, 163, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Bennett, L.; Kittas, A.; Muirhead, G.; Papageorgiou, L.G.; Tsoka, S. Detection of Composite Communities in Multiplex Biological Networks. Sci. Rep. 2015, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.C.; Bennett, L.; Papageorgiou, L.G.; Tsoka, S. A mathematical programming approach for sequential clustering of dynamic networks. Eur. Phys. J. B 2016. [Google Scholar] [CrossRef]
- Dourisboure, Y.; Geraci, F.; Pellegrini, M. Extraction and Classification of Dense Communities in the Web. In Proceedings of the 16th International Conference on World Wide Web, Banff, AB, Canada, 8–12 May 2007; pp. 461–470.
- Liu, Y.; Moser, J.; Aviyente, S. Network community structure detection for directional neural networks inferred from multichannel multisubject EEG data. IEEE Trans. Biomed. Eng. 2014, 61, 1919–1930. [Google Scholar] [PubMed]
- Guimerà, R.; Nunes Amaral, L.A. Functional cartography of complex metabolic networks. Nature 2005, 433, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Lu, H.; Nardini, C. Extracting weights from edge directions to find communities in directed networks. J. Stat. Mech. Theory Exp. 2010, 2010, P06003. [Google Scholar] [CrossRef]
- Satuluri, V.; Parthasarathy, S. Symmetrizations for Clustering Directed Graphs. In Proceedings of the 14th International Conference on Extending Database Technology, Uppsala, Sweden, 21–24 March 2011; pp. 343–354.
- Zheng, Q.; Skillicorn, D.B. Spectral embedding of directed networks. Soc. Netw. Anal. Min. 2016, 6, 1–15. [Google Scholar] [CrossRef]
- Ning, X.; Liu, Z.; Zhang, S. Local community extraction in directed networks. Phys. A Stat. Mech. Appl. 2016, 452, 258–265. [Google Scholar] [CrossRef]
- Malliaros, F.D.; Vazirgiannis, M. Clustering and community detection in directed networks: A survey. Phys. Rep. 2013, 533, 95–142. [Google Scholar] [CrossRef]
- Leicht, E.A.; Newman, M.E.J. Community structure in directed networks. Phys. Rev. Lett. 2008, 100, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S. Radatools—Communities Detection in Complex Networks and Other tools. Available online: http://deim.urv.cat/~sergio.gomez/radatools.php (accessed on 20 July 2016).
- Lancichinetti, A.; Fortunato, S. Benchmarks for testing community detection algorithms on directed and weighted graphs with overlapping communities. Phys. Rev. E 2009, 80, 016118. [Google Scholar] [CrossRef] [PubMed]
- Arenas, A.; Fernández, A.; Gómez, S. Analysis of the structure of complex networks at different resolution levels. New J. Phys. 2008, 10, 53039. [Google Scholar] [CrossRef]
- Rosenthal, R.E. GAMS—A User’s Guide; GAMS Development Corporation: Washington, DC, USA, 2016. [Google Scholar]
- Bussieck, M.R.; Drud, A. SBB: A New Solver for Mixed Integer Nonlinear Programming. Available online: http://ww.atlatec.-port.gams.com/presentations/present_sbb.pdf (accessed on 26 October 2016).
- IBM CPLEX Optimizer—United States. Available online: https://www-01.ibm.com/software/commerce/optimization/cplex-optimizer/ (accessed on 27 September 2016).
- Esquivel, A.V.; Rosvall, M. Comparing network covers using mutual information. arXiv, 2012; arXiv:1202.0425. [Google Scholar]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of “small-world” networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Kittas, A.; Bennett, L.; Hermjakob, H.; Tsoka, S. Organizational principles of the Reactome human BioPAX model using graph theory methods. J. Complex Netw. 2016. [Google Scholar] [CrossRef]
- Batagelj, V. Pajek Data: Roget’s Thesaurus, 1879. Available online: http://vlado.fmf.uni-lj.si/pub/networks/data/dic/roget/Roget.htm (accessed on 20 July 2016).
- Leskovec, J.; Krevl, A. SNAP Datasets: Stanford Large Network Dataset Collection. Available online: http://snap.stanford.edu/data (accessed on 26 October 2016).
- Xu, G.; Tsoka, S.; Papageorgiou, L.G. Finding community structures in complex networks using mixed integer optimisation. Eur. Phys. J. B 2007, 60, 231–239. [Google Scholar] [CrossRef]
- Bennett, L.; Kittas, A.; Liu, S.; Papageorgiou, L.G.; Tsoka, S. Community Structure Detection for Overlapping Modules through Mathematical Programming in Protein Interaction Networks. PLoS ONE 2014, 9, e112821. [Google Scholar] [CrossRef] [PubMed]
| Nodes | Edges | Type of Network | |
|---|---|---|---|
| Mycobacterium tuberculosis | 194 | 849 | unweighted |
| Caenorhabditis elegans | 297 | 2345 | weighted |
| Roget’s thesaurus | 994 | 5058 | unweighted |
| Plasmodium falciparum | 1390 | 6497 | unweighted |
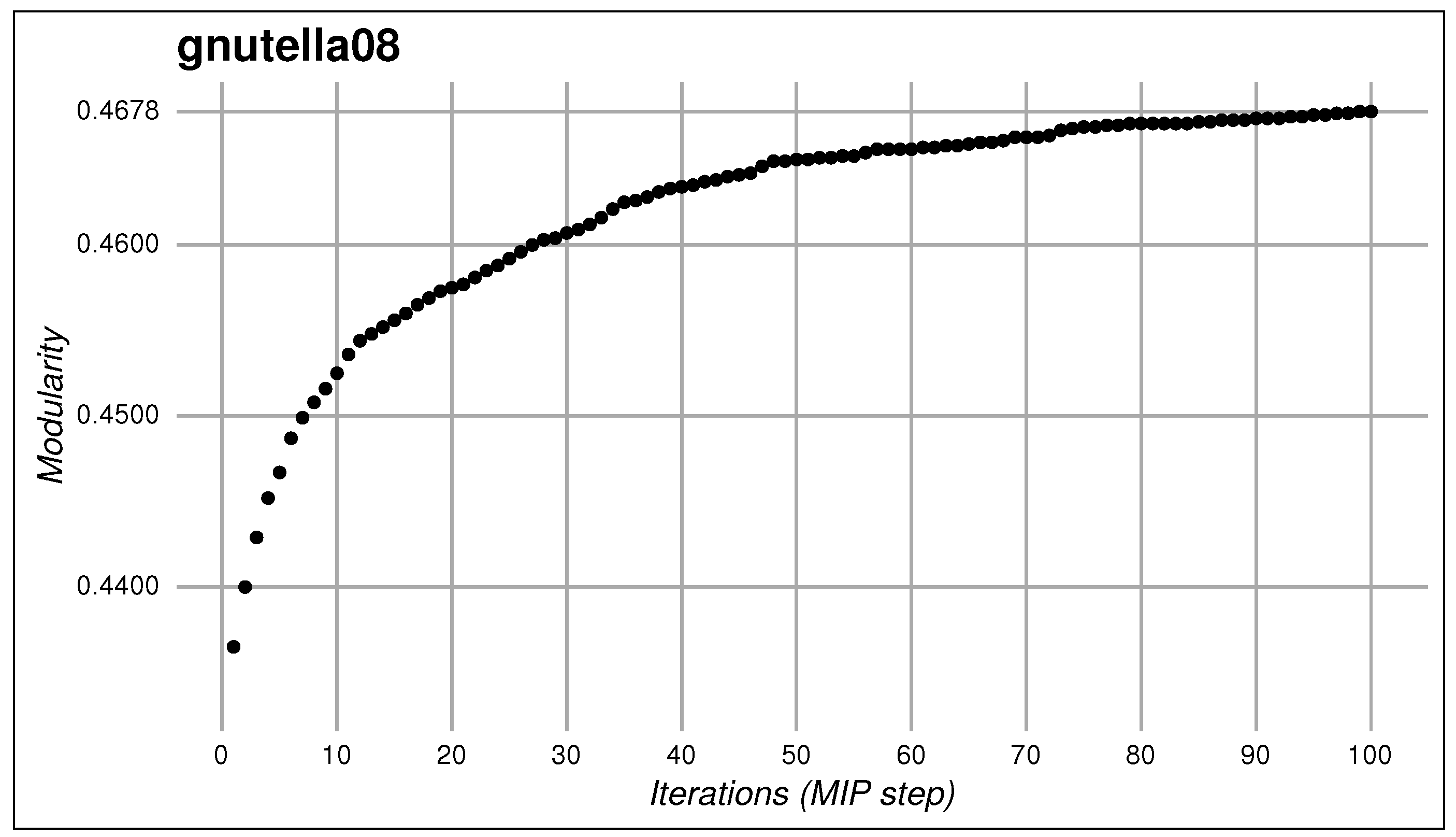
| gnutella08 | 6301 | 20,777 | unweighted |
| Myc. tub. | C. elegans | Roget | P. falc. | gnutella08 | |
|---|---|---|---|---|---|
| Initial modularity | 0.4636 | 0.4877 | 0.5063 | 0.6978 | 0.4333 |
| Final modularity | 0.5073 | 0.5076 | 0.5860 | 0.7238 | 0.4678 |
| Improvement | 9.43% | 4.08% | 15.75% | 3.72% | 7.97% |
| Number of modules | 9 | 5 | 13 | 20 | 24 |
| Myc. tub. | C. elegans | Roget | P. falc. | gnutella08 | |
|---|---|---|---|---|---|
| Extremal | 0.4802 | 0.4731 | 0.5582 | 0.6685 | 0.2475 |
| Fast algorithm | 0.4567 | 0.5058 | 0.5002 | 0.6846 | 0.4624 |
| Tabu search | 0.4635 | 0.4438 | 0.5021 | 0.6496 | 0.2281 |
| DiMod | 0.5073 | 0.5076 | 0.5860 | 0.7238 | 0.4678 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Silva, J.C.; Papageorgiou, L.G.; Tsoka, S. Community Structure Detection for Directed Networks through Modularity Optimisation. Algorithms 2016, 9, 73. https://doi.org/10.3390/a9040073
Yang L, Silva JC, Papageorgiou LG, Tsoka S. Community Structure Detection for Directed Networks through Modularity Optimisation. Algorithms. 2016; 9(4):73. https://doi.org/10.3390/a9040073
Chicago/Turabian StyleYang, Lingjian, Jonathan C. Silva, Lazaros G. Papageorgiou, and Sophia Tsoka. 2016. "Community Structure Detection for Directed Networks through Modularity Optimisation" Algorithms 9, no. 4: 73. https://doi.org/10.3390/a9040073






