The Construction of a High-Density Genetic Map for the Interspecific Cross of Castanea mollissima × C. henryi and the Identification of QTLs for Leaf Traits
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Materials and DNA Extraction
2.2. Sequencing Library Construction and Illumina Sequencing
2.3. Quality Assessment
2.4. SNP Identification and Genotyping
2.5. Genetic Map Construction
2.6. Phenotypic Data Analysis
2.7. Analysis of QTLs
2.8. Statistical Analysis
3. Results
3.1. Genotyping by Sequencing
3.2. Linkage Map Construction
3.3. Phenotypic Analysis
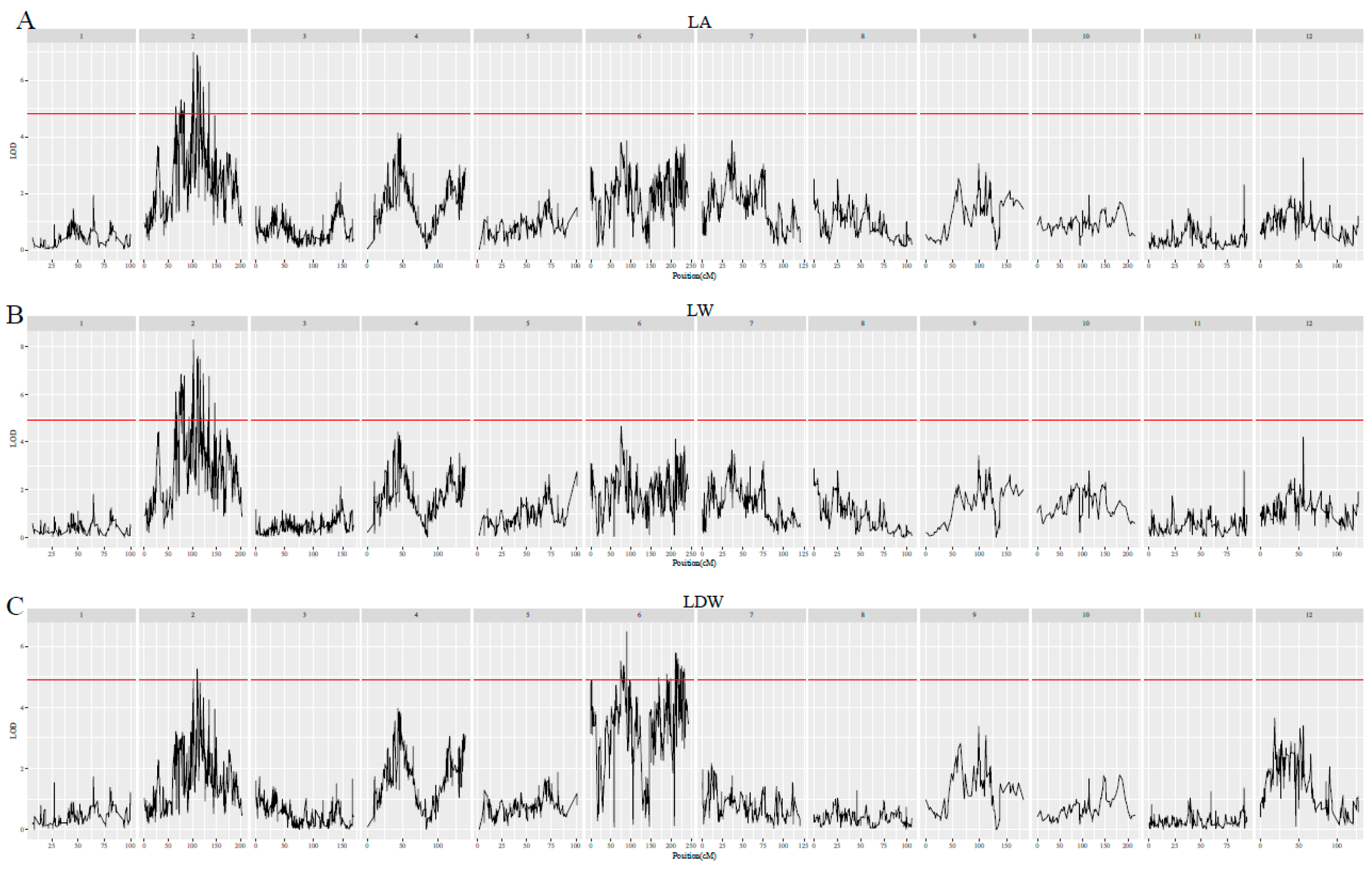
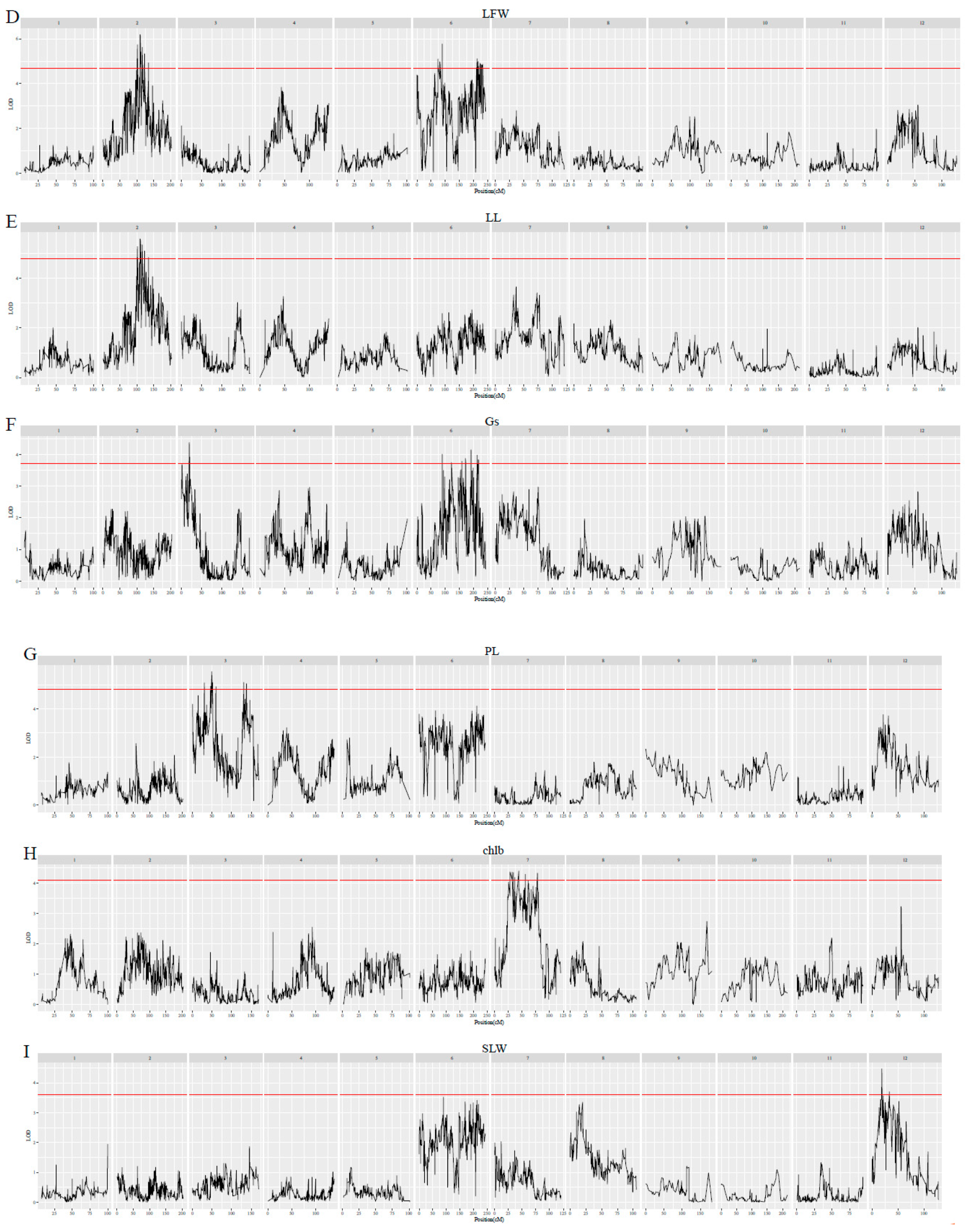
3.4. QTLs for Leaf Traits
4. Discussion
4.1. Construction of a High-Density Genetic Map of Chestnut
4.2. Analysis of QTLs for Leaf Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, S.; Wang, L.T.; Fu, Y.J.; Jiang, J.C. Bioactive constituents, nutritional benefits and woody food applications of Castanea mollissima: A comprehensive review. Food Chem. 2022, 393, 133380. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Lorenzo, S.; Fernández-López, J.; Moreno-González, J. Variability and Grouping of Northwestern Spanish Chestnut Cultivars. II. Isoenzyme Traits. J. Amer. Soc. Hort. Sci. 1996, 121, 190–197. [Google Scholar] [CrossRef]
- Kim, C.Y.; Kim, J.W.; Kim, J.H.; Jeong, J.S.; Ko, J.W.; Kim, T.W. Inner Shell of the Chestnut (Castanea crenatta) Suppresses Inflammatory Responses in Ovalbumin-Induced Allergic Asthma Mouse Model. Nutrients 2022, 14, 2067. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.; Cádiz-Gurrea, M.d.l.L.; Silva, A.M.; Macedo, C.; Rodrigues, F.; Costa, P. Development and Optimization of a Topical Formulation with Castanea sativa Shells Extract Based on the Concept “Quality by Design”. Sustainability 2022, 14, 129. [Google Scholar] [CrossRef]
- Cristina, C.; Lillian, B.; João, B.; Marina, S.; Ricardo, C.C.; Albino, B.; Maria, O.; Isabel, C.F.R.F. Castanea sativa male flower extracts as an alternative additive in the Portuguese pastry delicacy “pastel de nata”. Food Funct. 2020, 11, 1. [Google Scholar]
- Conedera, M.; Krebs, P.; Gehring, E.; Wunder, J.; Hülsmann, L.; Abegg, M.; Maringer, J. How future-proof is Sweet chestnut (Castanea sativa) in a global change context? Forest Ecol. Manag. 2021, 494, 119320. [Google Scholar] [CrossRef]
- Paletto, A.; Focacci, M.; De Meo, I. Farmers’ opinions on chestnut (Castanea sativa Mill.) supply chain development strategies: A case study in Central Italy. Forest Syst. 2018, 27, eSC02. [Google Scholar] [CrossRef]
- Barakat, A.; Staton, M.; Cheng, C.H.; Park, J.; Yassin, N.B.M.; Ficklin, S.; Yeh, C.C.; Hebard, F.; Baier, K.; Powell, W.; et al. Chestnut resistance to the blight disease: Insights from transcriptome analysis. BMC Plant Biol. 2012, 12, 38. [Google Scholar] [CrossRef]
- Sevarika, M.; Stacconi, V.R.; Romani, R. Fine Morphology of Antennal and Ovipositor Sensory Structures of the Gall Chestnut Wasp, Dryocosmus kuriphilus. Insects 2021, 12, 231. [Google Scholar] [CrossRef]
- Bounous, G. Perspectives and future of the chestnut industry in Europe and all over the world. Acta Hortic. 2014, 1043, 19–22. [Google Scholar] [CrossRef]
- Jiang, X.B.; Fang, Z.; Lai, J.S.; Wu, Q.; Wu, J.; Gong, B.C.; Wang, Y.P. Genetic Diversity and Population Structure of Chinese Chestnut (Castanea mollissima Blume) Cultivars Revealed by GBS Resequencing. Plants 2022, 11, 3524. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.Y.; Wei, W.; Liu, Y.; Wang, G.P.; Zhang, Q.; Xing, Y.; Zhang, S.H.; Liu, Z.H.; Cao, Q.Q.; Qin, L. Construction of a SNP-Based High-Density Genetic Map Using Genotyping by Sequencing (GBS) and QTL Analysis of Nut Traits in Chinese Chestnut (Castanea mollissima Blume). Front. Plant Sci. 2018, 9, 816. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Deng, H.; Liang, G.; Ye, X.; Qin, Y.; Huang, L. Construction of a High-Density Genetic Map for Pitaya Using the Whole Genome Resequencing Approach. Horticulturae 2021, 7, 534. [Google Scholar] [CrossRef]
- Lv, D.; Zhang, C.; Yv, R.; Yao, J.; Wu, J.; Song, X.; Jian, J.; Song, P.; Zhang, Z.; Han, D.; et al. Utilization of a Wheat50K SNP microarray-derived high-density genetic map for QTL mapping of plant height and grain traits in wheat. Plants 2021, 10, 1167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rashid, M.A.R.; Li, X.; Yao, C.; Lu, L.; Bai, J.; Li, Y.; Xu, N.; Yang, Q.; Zhang, L. Collection and evaluation of genetic diversity and population structure of potato landraces and varieties in China. Front. Plant Sci. 2019, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.A.R.; Zhao, Y.; Zhang, H.; Li, J.; Li, Z. Nucleotide diversity, natural variation, and evolution of Flexible culm-1 and Strong culm-2 lodging resistance genes in rice. Genome 2016, 59, 473–483. [Google Scholar] [CrossRef]
- Talukder, Z.I.; Gong, L.; Hulke, B.S.; Pegadaraju, V.; Song, Q.; Schultz, Q.; Qi, L.L. A high-density SNP Map of sunflower derived from RAD-sequencing facilitating fine-mapping of the rust resistance gene R12. PLoS ONE 2014, 9, e98628. [Google Scholar] [CrossRef]
- Rifat, T.; Khan, K.; Islam, M.S. Genetic diversity in dragon fruit (Hylocereus sp.) germplasms revealed by RAPD marker. J. Anim. Plant Sci. 2019, 29, 809–818. [Google Scholar]
- Jiang, S.; Luo, J.; Wang, X.Q.; An, H.S.; Zhang, J.Y.; Li, S.G. QTL mapping and transcriptome analysis to identify genes associated with green/russet peel in Pyrus pyrifolia. Sci. Hortic-Amsterdam 2022, 293, 110714. [Google Scholar] [CrossRef]
- Soto, J.C.; Ortiz, J.F.; Perlaza-Jimenez, L.; Vasquez, A.X.; Lopez-Lavalle, L.A.; Mathew, B.; Léon, J.; Bernal, A.J.; Ballvora, A.; López, C.E. A genetic map of cassava (Manihot esculenta Crantz) with integrated physical mapping of immunity-related genes. BMC Genomics 2015, 16, 190. [Google Scholar] [CrossRef]
- Casasoli, M.; Mattioni, C.; Cherubini, M.; Villani, F. A genetic linkage map of European chestnut (Castanea sativa Mill.) based on RAPD, ISSR and isozyme markers. Theor. Appl. Genet. 2001, 102, 1190–1199. [Google Scholar] [CrossRef]
- Casasoli, M.; Pot, D.; Plomion, C.; Monteverdi, M.C.; Barreneche, T.; Lauteri, M.; Villani, F. Identification of QTLs affecting adaptive traits in Castanea sativa Mill. Plant Cell Environ. 2004, 27, 1088–1101. [Google Scholar] [CrossRef]
- Qi, Z.M.; Sun, Y.N.; Wang, J.L.; Zhang, D.W.; Liu, C.Y.; Hu, G.H.; Chen, Q.S. Meta-Analysisof100-Seed Weight QTLs in Soybean. Agr. Sci. China 2011, 10, 327–334. [Google Scholar] [CrossRef]
- Zhu, P.Y.; Meng, Y.J.; Zhang, K.J.; Wang, X.Y.; Liang, K.; Wang, T.T.; Xu, J.; Qin, X.D.; Wu, Z.; Cheng, C.Y.; et al. Mapping of fruit apex shape related QTLs across multi-genetic backgrounds in cucumber (Cucumis sativus L.). Horticl. Plant J. 2022, 8, 328–340. [Google Scholar] [CrossRef]
- Amin, G.M.A.; Kong, K.; Sharmin, R.A.; Kong, J.J.; Bhat, J.A.; Zhao, T.J. Characterization and apid gene-mapping of leaf lesionmimicphenotypeof spl-1mutantinsoybean (Glycine max(L.)Merr.). Int. J. Mol. Sci. 2019, 20, 2193. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- van Ooijen, J.W. JoinMap®4; Software for the calculation of genetic linkage maps in experimental populations; Kyazma B.V.: Wageningen, The Netherlands, 2006. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugenics 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Wang, Y.P.; Gao, T.T.; Zhang, Z.J.; Yuan, X.; Chen, Q.; Zheng, J.Z.; Chen, S.Y.; Ma, F.W.; Li, C. Overexpression of the tyrosine decarboxylase gene MdTyDC confers salt tolerance in apple. Environ. Exp. Bot. 2020, 180, 104244. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Feng, Y.L. Relationship between leaf photosynthetic capacity and specific leaf weight, nitrogen content and allocation of two ficus species under different light intensities. J. Plant Physiol. Mol. Biol. 2004, 30, 269–276. [Google Scholar]
- Ooijen, J.W.v. MapQTL®6; Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species; Kyazma B.V.: Wageningen, The Netherlands, 2009. [Google Scholar]
- Tisne, S.; Reymond, M.; Vile, D.; Fabre, J.; Dauzat, M.; Koornneef, M.; Granier, C. Combined genetic and modeling approaches reveal that epidermal cell area and number in leaves are controlled by leaf and plant developmental processes in Arabidopsis. Plant Physiol. 2008, 148, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Guo, E.C.; Wang, G.P.; Kong, D.J.; Liu, Q.X.; Zhang, X.Z.; and Han, J.C. The genetic linkage map of Chinese chestnut based on RAPD markers. Mol. Plant Breed 2008, 6, 207–212. [Google Scholar]
- Nishio, S.; Takada, N.; Yamamoto, T.; Terakami, S.; Hayashi, T.; Sawamura, Y.; Saito, T. Mapping and pedigree analysis of the gene that controls the easy peel pellicle trait in Japanese chestnut (Castanea crenata Sieb. et Zucc.). Tree Genet. Genomes 2013, 9, 723–730. [Google Scholar] [CrossRef]
- Kubisiak, T.L.; Nelson, C.D.; Staton, M.E.; Zhebentyayeva, T.; Smith, C.; Olukolu, B.A.; Fang, G.C.; Hebrard, F.V.; Anagnostakis, S.; Wheeler, N.; et al. A transcriptome-based genetic map of Chinese chestnut (Castanea mollissima) and identification of regions of segmental homology with peach (Prunus persica). Tree Genet. Genomes 2013, 9, 557–571. [Google Scholar] [CrossRef]
- Kubisiak, T.L.; Hebard, F.V.; Nelson, C.D.; Zhang, J.; Bernatzky, R.; Huang, H.; Anagnostakis, S.L.; Doudrick, R.L. Molecular mapping of resistance to blight in an interspecific cross in the genus Castanea. Phytopathology 1997, 87, 751–759. [Google Scholar] [CrossRef]
- Donnelly, K.; Cavers, S.; Cottrell, J.E.; Ennos, R.A. Genetic variation for needle traits in Scots pine (Pinus sylvestris L.). Tree Genet. Genomes 2016, 12, 40. [Google Scholar] [CrossRef]
- Bozkurt, A.E.; Coskuncelebi, K.; Terzioglu, S. Population variability of scots pine (Pinus sylvestris L.) in Turkey according to the needle morphology. Hrvatsko Sumarsko Drustvo. 2021, 145, 347–354. [Google Scholar]
- Popovic, V.; Nikolic, B.; Lucic, A.; Rakonjac, L.; Jovanovic, D.S.; Miljkovic, D. Morpho-anatomical trait variability of the Norway spruce (Picea abies (L.) Karst.) needles in natural populations along elevational diversity gradient. Trees-Struct. Funct. 2022, 36, 1131–1147. [Google Scholar] [CrossRef]
- Flores, A.; Climent, J.; Pando, V.; Lopez-Upton, J.; Alia, R. Intraspecific Variation in Pines from the Trans-Mexican Volcanic Belt Grown under Two Watering Regimes: Implications for Management of Genetic Resources. Forests 2018, 9, 71. [Google Scholar] [CrossRef]
- Jankowski, A.; Wyka, T.P.; Zytkowiak, R.; Nihlgard, B.; Reich, P.B.; Oleksyn, J. Cold adaptation drives variability in needle structure and anatomy in Pinus sylvestris L. along a 1900 km temperate-boreal transect. Funct. Ecol. 2017, 31, 2212–2223. [Google Scholar] [CrossRef]
- Du, M.; Xiong, M.; Chang, Y.; Liu, Z.; Wang, R.; Lin, X.; Zhou, Z.; Lu, M.; Liu, C.; Liu, E. Mining candidate genes and favorable haplotypes for flag leaf shape in Rice (Oryza sativa L.) based on a genome-wide association study. Agronomy 2022, 12, 1814. [Google Scholar] [CrossRef]
- Bu, S.; Zhan, P.; Huang, L.; Tang, J.; Chen, L.; Zhu, H.; Liu, Z.; Meng, L.; Liu, G.; Wang, S. Identification, interaction, expression, and function of QTLs on leaf numbers with single-segment substitution lines in Rice. Agronomy 2022, 12, 2968. [Google Scholar] [CrossRef]
- Yi, Q.; López-Malvar, A.; Álvarez-Iglesias, L.; Romay, M.C.; Revilla, P. Genome-wide association analysis identified newly natural variation for photosynthesis-related traits in a large maize panel. Agronomy 2023, 13, 801. [Google Scholar] [CrossRef]
- Chen, H.; Chen, S.; Wang, X.; Liu, X.; Wang, X.; Zhu, R.; Mo, W.; Wang, R.; Zhang, S. Different environmental and phylogenetic controls over the altitudinal variation in leaf N and P Resorption traits between woody and herbaceous plants. Forests 2023, 14, 5. [Google Scholar] [CrossRef]
- Cifuentes, L.; Moreno, F. Trait coordination at leaf level explains the resistance to excess light stress in shade-tolerant tropical tree species. Tree Physiol. 2022, 42, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Siddique, M.I.; Han, K.; Lee, J.; Lee, E.S.; Lee, Y.R.; Lee, H.E.; Lee, S.Y.; Kim, D.S. QTL Analysis for Chlorophyll content in Strawberry (Fragaria × ananassa Duch.) leaves. Agriculture 2021, 11, 1163. [Google Scholar] [CrossRef]
- Li, D.M.; Zhu, G.F. High-density genetic linkage map construction and QTLs identification associated with four leaf-related traits in Lady’s Slipper Orchids (Paphiopedilum concolor × Paphiopedilum hirsutissimum). Horticulturae 2022, 8, 842. [Google Scholar] [CrossRef]
- Sohi, H.S.; Gill, M.I.S.; Chhuneja, P.; Arora, N.K.; Maan, S.S.; Singh, J. Construction of genetic linkage map and mapping QTL specific to leaf anthocyanin colouration in mapping population ‘Allahabad Safeda’ × ‘Purple Guava (Local)’ of Guava (Psidium guajava L.). Plants 2022, 11, 2014. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Y.; Wang, Z.; Ye, M.; Wu, R. Functional mapping of genes modulating plant shade avoidance using leaf traits. Plants 2023, 12, 608. [Google Scholar] [CrossRef]
- Schoonmaker, A.N.; Hulse-Kemp, A.M.; Youngblood, R.C.; Rahmat, Z.; Atif Iqbal, M.; Rahman, M.; Kochan, K.J.; Scheffler, B.E.; Scheffler, J.A. Detecting cotton leaf curl virus resistance quantitative trait loci in Gossypium hirsutum and iCottonQTL a new R/Shiny app to streamline genetic mapping. Plants 2023, 12, 1153. [Google Scholar] [CrossRef]


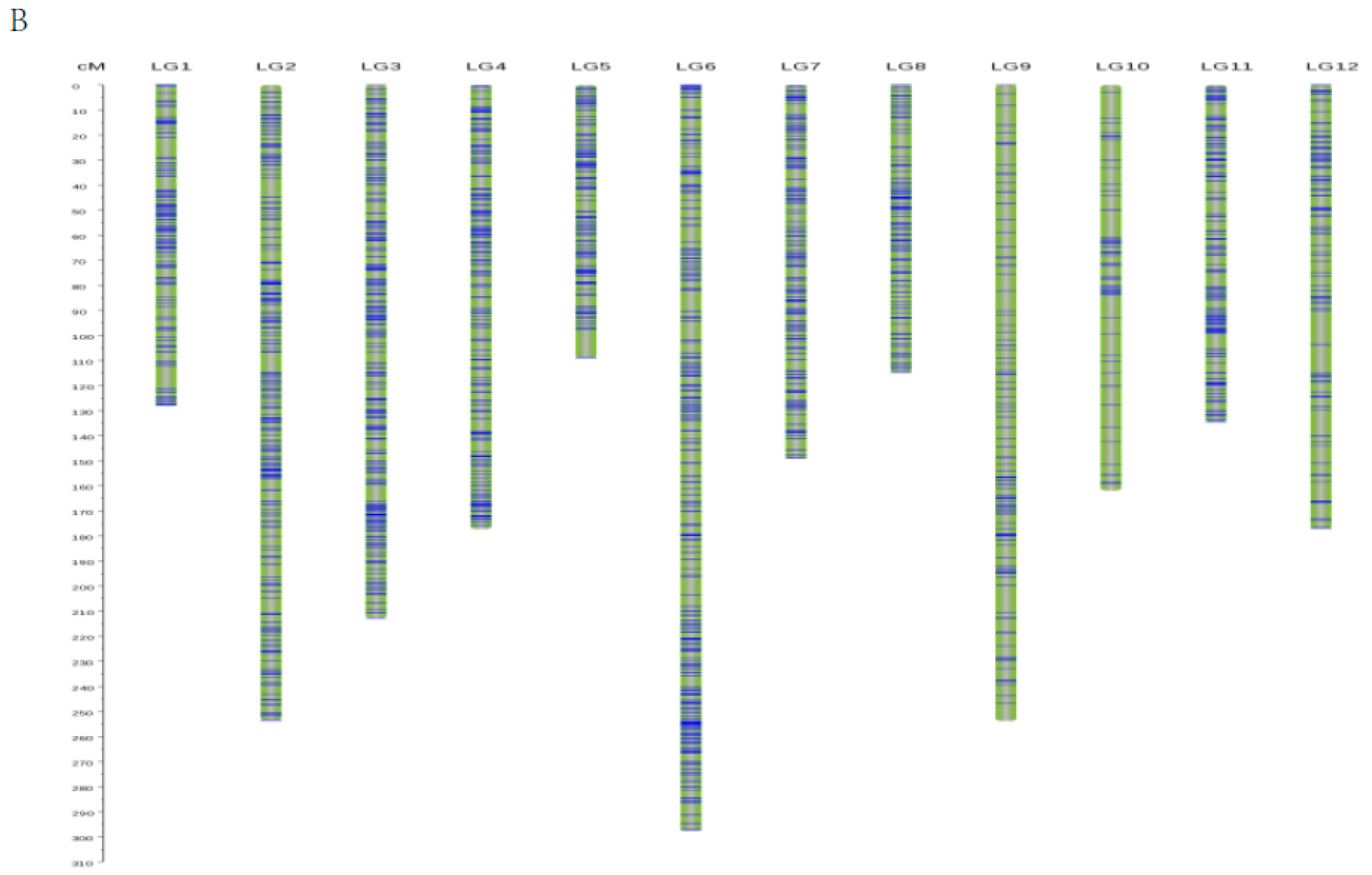
| Linkage Group | Average Distance between Two Markers (cM) | Number of Intervals-D (cM) | |||
|---|---|---|---|---|---|
| <5 | 5~10 | 10~20 | >20 | ||
| Lg01 | 0.31 | 315 | 2 | 0 | 0 |
| Lg02 | 0.42 | 487 | 0 | 0 | 0 |
| Lg03 | 0.29 | 584 | 0 | 0 | 0 |
| Lg04 | 0.28 | 485 | 0 | 0 | 0 |
| Lg05 | 0.3 | 338 | 1 | 0 | 0 |
| Lg06 | 0.36 | 676 | 3 | 0 | 0 |
| Lg07 | 0.33 | 367 | 0 | 0 | 0 |
| Lg08 | 0.34 | 310 | 0 | 0 | 0 |
| Lg09 | 1.15 | 164 | 7 | 0 | 0 |
| Lg10 | 1.01 | 206 | 8 | 0 | 0 |
| Lg11 | 0.27 | 343 | 0 | 0 | 0 |
| Lg12 | 0.45 | 279 | 3 | 0 | 0 |
| Total | 0.4 | 4554 | 24 | 0 | 0 |
| Trait/Trait | LL | LW | LSI | LA | PL | LFW | LDW | LMC | SLW | Pn | Gs | Ci | Tr | WUE | Chl·a | Chl·b | Chl·a + b | Chl·a/b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LL | 1.000 | |||||||||||||||||
| LW | 0.728 ** | 1.000 | ||||||||||||||||
| LSI | 0.410 ** | −0.322 ** | 1.000 | |||||||||||||||
| LA | 0.899 ** | 0.940 ** | −0.009 | 1.000 | ||||||||||||||
| PL | 0.640 ** | 0.442 ** | 0.289 ** | 0.573 ** | 1.000 | |||||||||||||
| LFW | 0.814 ** | 0.832 ** | 0.024 | 0.904 ** | 0.657 ** | 1.000 | ||||||||||||
| LDW | 0.799 ** | 0.808 ** | 0.032 | 0.882 ** | 0.689 ** | 0.959 ** | 1.000 | |||||||||||
| LMC | 0.284 ** | 0.288 ** | 0.028 | 0.308 ** | 0.019 | 0.285 ** | 0.070 | 1.000 | ||||||||||
| SLW | 0.170 * | 0.113 | 0.093 | 0.165 * | 0.484 ** | 0.476 ** | 0.601 ** | −0.405 ** | 1.000 | |||||||||
| Pn | 0.342 ** | 0.325 ** | 0.041 | 0.352 ** | 0.372 ** | 0.431 ** | 0.429 ** | 0.065 | 0.308 ** | 1.000 | ||||||||
| Gs | 0.316 ** | 0.289 ** | 0.052 | 0.325 ** | 0.361 ** | 0.419 ** | 0.400 ** | 0.068 | 0.279 ** | 0.825 ** | 1.000 | |||||||
| Ci | −0.062 | −0.043 | −0.023 | −0.059 | −0.086 | −0.022 | −0.059 | 0.124 | −0.034 | 0.188 * | 0.521 ** | 1.000 | ||||||
| Tr | 0.369 ** | 0.421 ** | −0.050 | 0.431 ** | 0.263 ** | 0.474 ** | 0.427 ** | 0.202 ** | 0.175 * | 0.616 ** | 0.723 ** | 0.391 ** | 1.000 | |||||
| WUE | −0.105 | −0.179 * | 0.096 | −0.166 * | 0.049 | −0.122 | −0.067 | −0.184 * | 0.137 | 0.301 ** | 0.004 | −0.254 ** | −0.532 ** | 1.000 | ||||
| Chl·a | 0.359 ** | 0.316 ** | 0.078 | 0.358 ** | 0.297 ** | 0.291 ** | 0.296 ** | 0.052 | 0.017 | 0.327 ** | 0.283 ** | 0.029 | 0.226 ** | 0.049 | 1.000 | |||
| Chl·b | 0.349 ** | 0.305 ** | 0.080 | 0.341 ** | 0.275 ** | 0.276 ** | 0.276 ** | 0.064 | 0.001 | 0.312 ** | 0.278 ** | 0.064 | 0.190 * | 0.069 | 0.965 ** | 1.000 | ||
| Chl·a + b | 0.360 ** | 0.321 ** | 0.074 | 0.360 ** | 0.295 ** | 0.291 ** | 0.295 ** | 0.062 | 0.009 | 0.330 ** | 0.283 ** | 0.037 | 0.223 ** | 0.052 | 0.997 ** | 0.979 ** | 1.000 | |
| Chl·a/b | −0.002 | −0.017 | 0.013 | 0.011 | 0.052 | 0.007 | 0.032 | −0.088 | 0.060 | 0.023 | −0.014 | −0.108 | 0.113 | −0.072 | 0.010 | −0.240 ** | −0.050 | 1.000 |
| Traits | lg | QTLs | Position (cM) | Marker | LOD | PVE (%) |
|---|---|---|---|---|---|---|
| chlb | 7 | qchlb-7-1 | 43.967 | lm1378 | 4.4 | 10.5 |
| 7 | qchlb-7-2 | 27.848 | hk1061 | 4.37 | 10.4 | |
| 7 | qchlb-7-3 | 31.841 | hk727 | 4.36 | 10.4 | |
| 7 | qchlb-7-5 | 34.207 | np2184 | 4.35 | 10.4 | |
| 7 | qchlb-7-6 | 77.799 | lm2630 | 4.33 | 10.3 | |
| 7 | qchlb-7-7 | 78.076 | np2214 | 4.32 | 10.3 | |
| 7 | qchlb-7-8 | 55.739 | lm2145 | 4.29 | 10.2 | |
| 7 | qchlb-7-10 | 34.207 | np3304 | 4.28 | 10.2 | |
| 7 | qchlb-7-11 | 27.848 | lm2 | 4.26 | 10.2 | |
| 7 | qchlb-7-12 | 36.824 | hk725 | 4.19 | 10 | |
| Gs | 3 | qGs-3-1 | 19.122 | np10917 | 4.37 | 10.4 |
| 3 | qGs-3-2 | 19.131 | np1854 | 4.33 | 10.3 | |
| 3 | qGs-3-3 | 19.131 | np1855 | 4.29 | 10.2 | |
| LA | 2 | qLA-2-1 | 101.906 | hk1025 | 6.99 | 16.1 |
| 2 | qLA-2-2 | 109.674 | hk189 | 6.89 | 15.9 | |
| 2 | qLA-2-3 | 110.502 | hk1261 | 6.75 | 15.6 | |
| 2 | qLA-2-4 | 110.034 | lm574 | 6.68 | 15.5 | |
| 2 | qLA-2-5 | 101.563 | lm2215 | 6.63 | 15.4 | |
| 2 | qLA-2-6 | 111.088 | lm2450 | 6.57 | 15.2 | |
| 2 | qLA-2-7 | 115.47 | hk1267 | 6.5 | 15.1 | |
| 2 | qLA-2-8 | 115.501 | lm2373 | 6.47 | 15 | |
| 2 | qLA-2-9 | 102.18 | hk1203 | 6.39 | 14.9 | |
| 2 | qLA-2-10 | 115.501 | lm2293 | 6.36 | 14.8 | |
| 2 | qLA-2-11 | 111.333 | np10396 | 6.33 | 14.7 | |
| 2 | qLA-2-12 | 102.318 | lm1661 | 6.28 | 14.6 | |
| 2 | qLA-2-13 | 115.439 | lm3035 | 6.11 | 14.2 | |
| 2 | qLA-2-14 | 101.906 | hk1030 | 6.09 | 14.2 | |
| 2 | qLA-2-15 | 111.811 | hk1266 | 6.07 | 14.2 | |
| LDW | 6 | qLDW-6-1 | 89.042 | hk1386 | 6.47 | 15 |
| 6 | qLDW-6-2 | 210.151 | hk155 | 5.78 | 13.5 | |
| 6 | qLDW-6-3 | 212.351 | lm1676 | 5.77 | 13.5 | |
| 6 | qLDW-6-4 | 89.042 | lm2681 | 5.62 | 13.2 | |
| 6 | qLDW-6-5 | 216.417 | lm2824 | 5.59 | 13.1 | |
| 6 | qLDW-6-6 | 212.351 | lm438 | 5.58 | 13.1 | |
| 6 | qLDW-6-8 | 213.178 | hk1433 | 5.54 | 13 | |
| 6 | qLDW-6-9 | 74.256 | np4288 | 5.52 | 13 | |
| 6 | qLDW-6-10 | 210.151 | lm1018 | 5.5 | 12.9 | |
| LFW | 2 | qLFW-2-1 | 109.674 | hk189 | 6.19 | 14.4 |
| 2 | qLFW-2-2 | 110.502 | hk1261 | 5.81 | 13.6 | |
| 2 | qLFW-2-3 | 110.034 | lm574 | 5.77 | 13.5 | |
| 2 | qLFW-2-4 | 89.042 | hk1386 | 5.77 | 13.5 | |
| 2 | qLFW-2-5 | 101.906 | hk1025 | 5.74 | 13.4 | |
| 2 | qLFW-2-6 | 101.563 | lm2215 | 5.67 | 13.3 | |
| 2 | qLFW-2-7 | 115.47 | hk1267 | 5.62 | 13.2 | |
| 2 | qLFW-2-8 | 115.501 | lm2373 | 5.61 | 13.2 | |
| LL | 2 | qLL-2-1 | 109.674 | hk189 | 5.59 | 13.1 |
| 2 | qLL-2-2 | 110.502 | hk1261 | 5.51 | 12.9 | |
| 2 | qLL-2-3 | 110.034 | lm574 | 5.35 | 12.6 | |
| 2 | qLL-2-4 | 115.501 | lm2373 | 5.35 | 12.6 | |
| 2 | qLL-2-5 | 115.47 | hk1267 | 5.33 | 12.6 | |
| 2 | qLL-2-6 | 101.906 | hk1025 | 5.28 | 12.4 | |
| 2 | qLL-2-7 | 101.563 | lm2215 | 5.27 | 12.4 | |
| 2 | qLL-2-8 | 111.088 | lm2450 | 5.26 | 12.4 | |
| LW | 2 | qLW-2-1 | 101.906 | hk1025 | 8.28 | 18.8 |
| 2 | qLW-2-2 | 102.18 | hk1203 | 7.81 | 17.8 | |
| 2 | qLW-2-3 | 111.088 | lm2450 | 7.59 | 17.4 | |
| 2 | qLW-2-4 | 101.563 | lm2215 | 7.57 | 17.4 | |
| 2 | qLW-2-5 | 109.674 | hk189 | 7.47 | 17.1 | |
| 2 | qLW-2-6 | 115.47 | hk1267 | 7.46 | 17.1 | |
| 2 | qLW-2-7 | 110.502 | hk1261 | 7.45 | 17.1 | |
| 2 | qLW-2-8 | 102.318 | lm1661 | 7.4 | 17 | |
| 2 | qLW-2-9 | 111.333 | np10396 | 7.38 | 17 | |
| 2 | qLW-2-10 | 115.501 | lm2373 | 7.35 | 16.9 | |
| 2 | qLW-2-11 | 115.501 | lm2293 | 7.31 | 16.8 | |
| 2 | qLW-2-12 | 110.034 | lm574 | 7.28 | 16.7 | |
| 2 | qLW-2-13 | 101.906 | hk1030 | 6.99 | 16.1 | |
| PL | 3 | qPL-3-1 | 49.107 | lm897 | 5.55 | 13 |
| 3 | qPL-3-2 | 48.833 | hk898 | 5.54 | 13 | |
| 3 | qPL-3-3 | 49.107 | hk472 | 5.4 | 12.7 | |
| 3 | qPL-3-4 | 130.784 | hk763 | 5.1 | 12 | |
| SLW | 12 | qSLW-12-1 | 18.321 | hk1366 | 4.47 | 10.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Wang, Y.; Lai, J.; Wu, J.; Wu, C.; Hu, W.; Wu, X.; Gong, B. The Construction of a High-Density Genetic Map for the Interspecific Cross of Castanea mollissima × C. henryi and the Identification of QTLs for Leaf Traits. Forests 2023, 14, 1684. https://doi.org/10.3390/f14081684
Jiang X, Wang Y, Lai J, Wu J, Wu C, Hu W, Wu X, Gong B. The Construction of a High-Density Genetic Map for the Interspecific Cross of Castanea mollissima × C. henryi and the Identification of QTLs for Leaf Traits. Forests. 2023; 14(8):1684. https://doi.org/10.3390/f14081684
Chicago/Turabian StyleJiang, Xibing, Yanpeng Wang, Junsheng Lai, Jian Wu, Conglian Wu, Weiyun Hu, Xiaolong Wu, and Bangchu Gong. 2023. "The Construction of a High-Density Genetic Map for the Interspecific Cross of Castanea mollissima × C. henryi and the Identification of QTLs for Leaf Traits" Forests 14, no. 8: 1684. https://doi.org/10.3390/f14081684





