Full-Waveform Airborne Laser Scanning in Vegetation Studies—A Review of Point Cloud and Waveform Features for Tree Species Classification
Abstract
:1. Introduction
- Which point cloud and waveform features have been used to classify trees into species classes?
- How accurate are the classification strategies based on the derived features, and what are their limitations?
- Which point cloud and waveform features have emerged as indicative features for a specific tree species?
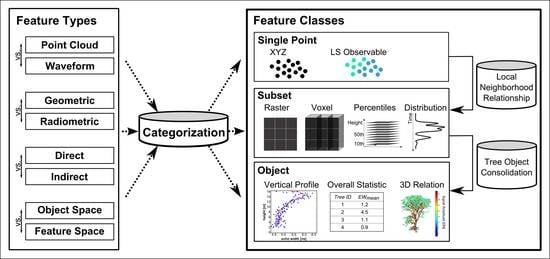
2. Derived and Applied Features for Tree Species Classification
2.1. Full-Waveform Data and Single Tree Classification
2.2. Derived Features for Tree Species Classification
2.3. Applied Feature Sets for Tree Species Classification
3. Discussion
3.1. Influencing and Limiting Factors on Feature Characteristic and Tree Classification
3.1.1. Factors related to Vegetation Structure
3.1.2. Technical Factors related to Data Acquisition and Processing
3.2. Tree Species Specific Feature Characteristics
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
| Abbreviation | |
|---|---|
| ALS | Airborne Laser Scanning |
| CHM | Canopy Height Model |
| DBH | Diameter at Breast Height |
| DSM | Digital Surface Model |
| DTM | Digital Terrain Model |
| FWF | Full-WaveForm |
| LAI | Leaf Area Index |
| MS | Manual tree Segmentation |
| nDSM | Normalized Digital Surface Model |
| PC | Principal Component |
| VTMR | Variance-To-Mean-Ratio |
| WS | Watershed Segmentation |
| Abbreviation | Unit | Definition |
|---|---|---|
| Astats,obj | (DN) | The statistics of amplitude A, e.g., mean A of tree object (Amean,obj). |
| Asum,obj | (DN) | Sum of all waveform’s amplitude peaks. |
| CRlt | (m/m)(%) | Ratio of crown length and tree height. |
| CRlw | (m/m) | Ratio of crown length and width. |
| CRvol | (m³) | Crown volume. |
| CRvol,derivatives | (m³/m)(%) | Crown volume derivatices, e.g., CRvol in relation to crown length (CRvol,l), width (CRvol,w) or to tree height (CRvol,t). |
| EAW | (DN) | The product of echo amplitude and width, e.g., mean EAW of tree object (EAWmean,obj). |
| ER3D/2D | Echo ratio. The number of points in 3D in a fixed search distance is related to the number of points in 2D found in the same distance in 2D. | |
| ERsingle/multiple,obj | Ratio of the number of single echoes to the number of multiple echoes. | |
| EWstats,obj | (ns) | The statistics of echo-width, e.g., mean EW of tree object (EWmean,obj). |
| EWstats,h-layer | The statistics of echo width of a height layer, e.g., mean EW of the upper 2 m (EWmean,u2m). | |
| FS | (°) | Front slope angle from waveform beginning to first peak. |
| Hm,energy | (m)(%) | Height, at which a specific amount of energy is reached, e.g., 50% of the returned energy. |
| Hmin | (m)(%) | Height threshold as the minimum height. |
| HTi | Haralick’s texture features calculated from 3D grey level co-occurrence matrix based on number of points per voxel in different directions in the 3D space. | |
| Pdens,bin,norm | (%) | The number of echoes normalized by the total number of echoes of the tree object at given height layer. |
| Pdens,L-func | The L-function features of echo number. | |
| Pdens,L-func_Npeak | Number of echoes, that are determined by the number of local minimums per height layer of a L-function. | |
| Pgrid,VTMR | Variance-to-mean-ratio of number of echoes of gridded height layer. | |
| PSa, PSb | Function parameter of the parabolic surface fitted to the tree crown. | |
| PSheight | (m)(%) | Vertical length of the parabolic surface fitted to the tree crown. |
| PSradius | (m) | Radius of the parabolic surface fitted to the tree crown. |
| PTIN,Edge | (m) | Variance of edge lengths from Delaunay triangulated points per height layer and their frequency distribution. |
| RWE | (DN) | Total returned waveform energy. |
| TNo | Total number of echoes. | |
| TNoobj,Hmin | Number of points above a defined height threshold. | |
| TNorast,stats,filter | The number of echoes within a defined height layer based on raster-based calculations. For example TNorast,mean,single as the average of the number of single echoes of all raster cells at a defined height. | |
| TNovoxel,column | The number of echoes per voxel is related to the number of echoes of all subjacent voxels. | |
| TNowave,stats | The statistics of the number of echoes of all waveforms. | |
| VA | Vertical profile of amplitude values. | |
| VEW | Vertical profile of echo width values. | |
| VH | Vertical profile of number of echoes. | |
| Vσ | Vertical profile of backscatter cross-section values. | |
| V*_derivative | Derivative of the vertical profile of a feature, e.g., skewness of VEW (VEW_skewness). | |
| Zobj,ellip | (m) | Vertical length of ellipsoid fitted to tree crown. |
| ΔDtrunk- dist,horiz | (m) | The mean horizontal distance of an echo to the previously detected tree trunk. |
| ΔRij | (m) | Distance between two waveform echoes i and j calculated by difference in range, e.g., the distance between the first and the last echo in meter (ΔR1st/last). |
| ΔTij | (ns) | Distance between two waveform echoes i and j calculated by time difference, e.g., the distance between the first and the last echo in nanoseconds (ΔT1st/last). |
| ΔWRij | (m)(ns) | Distance measure of a waveform, e.g., the distance between waveform centroid and ground (ΔWRcentroid,ground), waveform beginning to ground (ΔWRbeginning,ground) and beginning and first peak (ΔWRbeginning,1stpeak). |
| γstats,obj | (m2/m2) | The statistics of backscatter coefficient, e.g., mean γ of tree object. |
| σstats,obj | (m2) | The statistics of backscatter cross-section, e.g., mean σ of tree object. |
| Dominant Species and Study Area (Number of Trees, if Given) | Tree Detection and Classification Method | Applied Feature | Overall Accuracy (c: Class, s: Species Level) | Reference |
|---|---|---|---|---|
| Masson pine (Pinus massoniana (Lamb.), 85), Chinese fir (Cunninghamia lanceolate (Lamb.), 82), Slash pines (Pinus elliottii Engelm., 71), Sawtooth oak (Querus acutissima Carruth., 73), Chinese sweet gum (Liquidambar formosana Hance, 74), Chinese holly (Ilex chinensis Sims., 52) [Yushan Forest, China, leaf-on] | Local Maxima Algorithm on CHM; Random Forest with Mean Decrease Accuracy [2/3 for model training] | ΔWRcentroid,ground, ΔWRbeginning,ground, ΔWRbeginning,1stpeak, TNowave,stats, FS, RWE | c: 86.2% | [15] |
| s: 68.6% | ||||
| Red pine (Pinus koraiensis), Koyama spruce (Picea koraiensis), Dahurian larch (Larix gmelinii), fir (Abies nephrolepis), white birch (Betula platyphylla Suk.), ribbed birch (Betula Costata), Manchurian linden (Tilia Mandschurica), elm (Ulmus Laciniata), Mongolian oak (Quercus Mongolica); (in total: 1847 trees) [Forest, China, leaf-on] | Watershed Segmentation on CHM; Support Vector Machine | Range corrected Astats,obj, EAWstats,obj, EWstats,obj, TNostats,ob | c: 85.7% | [24] |
| s: 55.1% | ||||
| Scots pine (Pinus sylvestris), Norway spruce (Picea abies), sessile oak (Quercus petraea), red oak (Quercus rubra), European beech (Fagus sylvatica) [Forest, SW Germany, leaf-on] | Linear Discriminant Analysis with 18-fold cross-validation | Astats,obj, EAWstats,obj, EWstats,obj, TNostats,ob | c: 91.7% | [25] |
| s: 80.4% | ||||
| Spruce (Picea abies, 42), European larch (Larix decidua, 23), red beech (Fagus sylvatica, 76) [Forest, Austria, leaf-off] | Edge-based segmentation on nDSM; FuzzyRule Set & Decision Tree [1/2 for model training] | ER3D/2D, TNoHmin, EWmean, EWstddev, σmean, σstdev, Hmin > 50th height | c: 83% | [26] |
| s: 75% | ||||
| Scots pine (Pinus sylvestris L.), Norway spruce (Picea abies (L.) Karst.), birches (Betula pendula Roth and Betula pubescens Ehrh.); (in total: 9930 trees) [Forest, Hyytiälä, Finland, leaf-on] | Watershed segmentation on CHM & delineation by cylinders; Quadratic Discriminant Analysis and leave-one-out cross validation | Astats, FWHMstats, RWE, Hm,50%energy, pulse length | 84%–91% | [27] |
| Red beech (Fagus sylvatica, 11), oaks (Quercus robur and Quercus petraea, 10), hornbeam, larch (Carpinus betulus, 4) [Vienna Woods, Austria, leaf-off] | Edge-based segmentation on nDSM; Descriptive feature analysis | EWmean, EWCV, σmean, σcv | (exploratory analysis) | [47] |
| Norway spruce (Picea abies, 256), European beech (Fagus sylvatica, 397), Sycamore maple (Acer pseudoplatanus, 20) [Mountainous, mixed forest, SE Germany] | Watershed segmentation on CHM; k-Means clustering | leaf-on: ΔDtrunk dist, horiz., EAWobj,mean, EW mean_single, EW mean_first leaf-off: EWsingle, EWfirst, ERsingle/multiple | c: | [52] |
| leaf-on: 85.4% | ||||
| leaf-off: 95.7% | ||||
| Norway spruce (Picea abies, 688), European beech (Fagus sylvatica, 812), fir (Abies alba, 70), Sycamore maple (Acer pseudoplatanus, 71), Norway maple (Acer platanoides, 21), lime trees (Tilia Europaea, 2) [Mountainous, mixed forest, SE Germany] | Normalized cut segmentation; Expectation-Maximization Algorithm & Maximum Likelihood Classification [1/5 for model training] | ΔDtrunk dist,horiz., EAWobj,mean, EWmean_single, EWmean_first, ERsingle/multiple | c: | [53] |
| leaf-on: 95/97% | ||||
| leaf-off: 94% | ||||
| Douglas-fir (Pseudotsuga menziesii (Mirb.) Franco), western redcedar (Thuja plicata Donn ex D.Don), black cottonwood (Populus balsamifera L. ssp. trichocarpa (Torr. & A. Gray ex Hook.) Brayshaw), bigleaf maple (Acer macrophyllum Pursh), red alder (Alnus rubra Bong.); (22 to 29 trees per species) [Arboretum, Seattle, USA, leaf-on] | Voxel-based segmentation; Support Vector Machine with five-fold cross-validation | Vstats, ΔRij, EAWmean, Pstats,perc, TNostats, PCwave,Fourier | s: 85.4% | [55] |
| Norway spruce (Picea abies, 688), European beech (Fagus sylvatica, 812), fir (Abies alba, 70), Sycamore maple (Acer pseudoplatanus, 71), Norway maple (Acer platanoides, 21), lime trees (Tilia Europaea, 2) [Mountainous, mixed forest, SE Germany] | Watershed segmentation & Normalized cut segmentation; Expectation Maximization Algorithm & Maximum Likelihood | Pstats,perc, Astats, EAWstats, EWstats, filtered by the index of return | c: | [66] |
| leaf-on: 95% | ||||
| leaf-off: 94% | ||||
| lower layers: | ||||
| leaf-on: 86% | ||||
| leaf-off: 95% | ||||
| upper layers: | ||||
| leaf-on: 97% | ||||
| leaf-off: 94% | ||||
| Jack pine (Pinus banksiana Lamb., 158), eastern white pine (Pinus strobus L., 106), sugar maple (Acer saccharum Marsh., 105), trembling aspen (Populus tremuloides Michx., 182) [Forest, Ontario, Canada, leaf-on] | Individual tree crown delineation from optical imagery; Linear Discriminant Analysis with 10-fold cross validation [1/2 for model training] | Pgrid,VTMR, Pdens,L-func, PTIN,Edge | c: 82.2% | [67] |
| s: 77.5% | ||||
| Norway spruce (Picea abies (L.)), Scots pine (Pinus sylvestris, (L.)), birches (Betula pendula (L.) and Betula pubescens (L.)); (in total, 789 trees) [Forest, SW Sweden, leaf-on] | 3D ellipsoid clustering; Linear Discriminant Analysis with leave-one-out cross validation | Astats,obj, EAWstats,obj, EWstats,obj, TNostats, Zobj_ellip, PSheight, PSradius | s: 71% | [73] |
| Scots pine (Pinus sylvestris), Norway spruce (Picea abies), birch (Betula sp.); (in total, 3695) [Forest, South Finland, leaf-on] | Minimum curvature-based region detector; Random Forest with cross validation | ΔR1st/last, RWE, TNo, A1st/2nd/rd/th, Hm,energy | s: 71.5% | [76] |
| Norway spruce (Picea abies (L.), 209), birch (Betula spp., 203), aspen (Poplus tremula L.) [Forest, SE Norway, leaf-on] | Mean crown radius; Linear Discriminant Analysis & ANCOVA with cross-validation | VH_kurtosis, VH_skewness, VH_coeff.var., Aobj,mean, Aobj,max, Pdens,bin | s: 88% | [97] |
Supplementary Files
Supplementary File 1References
- Wagner, W. Radiometric calibration of small-footprint full-waveform airborne laser scanner measurements: Basic physical concepts. ISPRS J. Photogramm. Remote Sens. 2010, 65, 505–513. [Google Scholar] [CrossRef]
- Anderson, K.; Hancock, S.; Disney, M.; Gaston, K.J. Is waveform worth it? A comparison of LiDAR approaches for vegetation and landscape characterization. Remote Sens. Ecol. Conserv. 2015, 2, 5–15. [Google Scholar] [CrossRef]
- Lin, Y.; Hyyppä, J. A comprehensive but efficient framework of proposing and validating feature parameters from airborne LiDAR data for tree species classification. Int. J. Appl. Earth Observ. Geoinf. 2016, 46, 45–55. [Google Scholar] [CrossRef]
- Hug, C.; Ullrich, A.; Grimm, A. Litemapper-5600—A waveform-digitizing LiDAR terrain and vegetation mapping system. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2004, 36, 24–29. [Google Scholar]
- Mallet, C.; Bretar, F. Full-waveform topographic LiDAR: State-of-the-art. ISPRS J. Photogramm. Remote Sens. 2009, 64, 1–16. [Google Scholar] [CrossRef]
- Balenović, I.; Alberti, G.; Marjanović, H. Airborne laser scanning—The status and perspectives for the application in the South-East European forestry. South-East Eur. For. 2013, 4, 59–79. [Google Scholar] [CrossRef]
- Hyyppä, J.; Yu, X.; Hyyppä, H.; Vastaranta, M.; Holopainen, M.; Kukko, A.; Kaartinen, H.; Jaakkola, A.; Vaaja, M.; Koskinen, J.; et al. Advances in forest inventory using airborne laser scanning. Remote Sens. 2012, 4, 1190–1207. [Google Scholar] [CrossRef]
- Næsset, E. Airborne laser scanning as a method in operational forest inventory: Status of accuracy assessments accomplished in Scandinavia. Scand. J. For. Res. 2007, 22, 433–442. [Google Scholar] [CrossRef]
- Pirotti, F.; Laurin, G.V.; Vettore, A.; Masiero, A.; Valentini, R. Small footprint full-waveform metrics contribution to the prediction of biomass in tropical forests. Remote Sens. 2014, 6, 9576–9599. [Google Scholar] [CrossRef]
- Woods, M.; Pitt, D.; Penner, M.; Lim, K.; Nesbitt, D.; Etheridge, D.; Treitz, P. Operational implementation of a LiDAR inventory in Boreal Ontario. For. Chron. 2011, 87, 512–528. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Næsset, E.; Ørka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef]
- Tilly, N.; Hoffmeister, D.; Cao, Q.; Huang, S.; Lenz-Wiedemann, V.; Miao, Y.; Bareth, G. Multitemporal crop surface models: Accurate plant height measurement and biomass estimation with terrestrial laser scanning in paddy rice. J. Appl. Remote Sens. 2014, 8, 083671. [Google Scholar] [CrossRef]
- Höfle, B.; Hollaus, M.; Hagenauer, J. Urban vegetation detection using radiometrically calibrated small-footprint full-waveform airborne LiDAR data. ISPRS J. Photogramm. Remote Sens. 2012, 67, 134–147. [Google Scholar] [CrossRef]
- Borre, J.V.; Paelinckx, D.; Mücher, C.A.; Kooistra, L.; Haest, B.; De Blust, G.; Schmidt, A.M. Integrating remote sensing in Natura 2000 habitat monitoring: Prospects on the way forward. J. Nat. Conserv. 2011, 19, 116–125. [Google Scholar] [CrossRef]
- Cao, L.; Coops, N.C.; Innes, J.L.; Dai, J.; Ruan, H.; She, G. Tree species classification in subtropical forests using small-footprint full-waveform LiDAR data. Int. J. Appl. Earth Observ. Geoinf. 2016, 49, 39–51. [Google Scholar] [CrossRef]
- Latifi, H.; Fassnacht, F.E.; Müller, J.; Tharani, A.; Dech, S.; Heurich, M. Forest inventories by LiDAR data: A comparison of single tree segmentation and metric-based methods for inventories of a heterogeneous temperate forest. Int. J. Appl. Earth Observ. Geoinf. 2015, 42, 162–174. [Google Scholar] [CrossRef]
- Leiterer, R.; Furrer, R.; Schaepman, M.E.; Morsdorf, F. Forest canopy-structure characterization: A data-driven approach. For. Ecol. Manag. 2015, 358, 48–61. [Google Scholar] [CrossRef]
- Lindberg, E.; Olofsson, K.; Holmgren, J.; Olsson, H. Estimation of 3D vegetation structure from waveform and discrete return airborne laser scanning data. Remote Sens. Environ. 2012, 118, 151–161. [Google Scholar] [CrossRef]
- Fieber, K.D.; Davenport, I.J.; Tanase, M.A.; Ferryman, J.M.; Gurney, R.J.; Becerra, V.M.; Walker, J.P.; Hacker, J.M. Validation of Canopy Height Profile methodology for small-footprint full-waveform airborne LiDAR data in a discontinuous canopy environment. ISPRS J. Photogramm. Remote Sens. 2015, 104, 144–157. [Google Scholar] [CrossRef] [Green Version]
- Alonzo, M.; Bookhagen, B.; McFadden, J.P.; Sun, A.; Roberts, D.A. Mapping urban forest leaf area index with airborne LiDAR using penetration metrics and allometry. Remote Sens. Environ. 2015, 162, 141–153. [Google Scholar] [CrossRef]
- Fieber, K.D.; Davenport, I.J.; Tanase, M.A.; Ferryman, J.M.; Gurney, R.J.; Walker, J.P.; Hacker, J.M. Effective LAI and CHP of a single tree from small-footprint full-waveform LiDAR. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1634–1638. [Google Scholar] [CrossRef]
- Allouis, T.; Durrieu, S.; Véga, C.; Couteron, P. Stem volume and above-ground biomass estimation of individual pine trees from LiDAR data: Contribution of full-waveform signals. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2013, 6, 924–934. [Google Scholar] [CrossRef]
- Cao, L.; Coops, N.; Hermosilla, T.; Dai, J. Estimation of forest structural variables using small-footprint full-waveform LiDAR in a subtropical forest, China. In Proceedings of the 2014 3rd International Workshop on Earth Observation and Remote Sensing Applications (EORSA), Changsha, China, 11–14 June 2014.
- Xu, G.; Pang, Y.; Li, Z.; Zhao, D.; Liu, L. Individual trees species classification using relative calibrated full-waveform LiDAR data. In Proceedings of the SilviLaser 2012: 12th International Conference on LiDAR Applications for Assessing Forest Ecosystems, Vancouver, BC, Canada, 16–19 September 2012; Coops, N., Wulder, M., Eds.; pp. 163–174.
- Heinzel, J.; Koch, B. Exploring full-waveform LiDAR parameters for tree species classification. Int. J. Appl. Earth Observ. Geoinf. 2011, 13, 152–160. [Google Scholar] [CrossRef]
- Hollaus, M.; Mücke, W.; Höfle, B.; Dorigo, W.; Pfeifer, N.; Wagner, W.; Bauerhansl, C.; Regner, B. Tree species classification based on full-waveform airborne laser scanning data. In Proceedings of Silvilaser 2009: 9th International Conference on LiDAR Applications for Assessing Forest Ecosystems, College Station, TX, USA, 14–16 October 2009; pp. 54–62.
- Hovi, A.; Korhonen, L.; Vauhkonen, J.; Korpela, I. LiDAR waveform features for tree species classification and their sensitivity to tree- and acquisition related parameters. Remote Sens. Environ. 2016, 173, 224–237. [Google Scholar] [CrossRef]
- Mücke, W.; Hermann, A. Estimation of biodiversity relevant quantities from airborne laser scanning data. Österreichische Z. Vermess. Geoinfor. 2010, 4, 201–210. [Google Scholar]
- Müller, J.; Vierling, K. Assessing Biodiversity by Airborne Laser Scanning. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2014; pp. 357–374. [Google Scholar]
- Hyde, P.; Dubayah, R.; Peterson, B.; Blair, J.B.; Hofton, M.; Hunsaker, C.; Knox, R.; Walker, W. Mapping forest structure for wildlife habitat analysis using waveform LiDAR: Validation of montane ecosystems. Remote Sens. Environ. 2005, 96, 427–437. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Kennedy-Bowdoin, T.; Jones, M.O.; Martin, R.E.; Boardman, J.; Hughes, R.F. Invasive species detection in Hawaiian rainforests using airborne imaging spectroscopy and LiDAR. Remote Sens. Environ. 2008, 112, 1942–1955. [Google Scholar] [CrossRef]
- Dwyer, J.F.; McPherson, E.G.; Schroeder, H.W.; Rowntree, R.A. Assessing the benefits and costs of the urban forest. J. Arboric. 1992, 18, 227–234. [Google Scholar]
- Van Leeuwen, M.; Coops, N.C.; Hilker, T.; Wulder, M.A.; Newnham, G.J.; Culvenor, D.S. Automated reconstruction of tree and canopy structure for modeling the internal canopy radiation regime. Remote Sens. Environ. 2013, 136, 286–300. [Google Scholar] [CrossRef]
- Alexander, C.; Tansey, K.; Kaduk, J.; Holland, D.; Tate, N.J. An approach to classification of airborne laser scanning point cloud data in an urban environment. Int. J. Remote Sens. 2011, 32, 9151–9169. [Google Scholar] [CrossRef]
- Janhäll, S. Review on urban vegetation and particle air pollution—Deposition and dispersion. Atmos. Environ. 2015, 105, 130–137. [Google Scholar] [CrossRef]
- Koma, Z.; Koenig, K.; Höfle, B. Urban Tree Classification Using Full-Waveform Airborne Laser Scanning. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, III-3, 185–192. [Google Scholar] [CrossRef]
- Hyyppä, J.; Hyyppä, H.; Leckie, D.; Gougeon, F.; Yu, X.; Maltamo, M. Review of methods of small-footprint airborne laser scanning for extracting forest inventory data in boreal forests. Int. J. Remote Sens. 2008, 29, 1339–1366. [Google Scholar] [CrossRef]
- Lim, K.; Treitz, P.; Wulder, M.; St-Onge, B.; Flood, M. LiDAR remote sensing of forest structure. Prog. Phys. Geogr. 2003, 27, 88–106. [Google Scholar] [CrossRef]
- Maltamo, M.; Næsset, E.; Vauhkonen, J. Forestry Applications of Airborne Laser Scanning—Concepts and Case Studies; Managing Forest Ecosystems; Springer Netherlands: Dordrecht, The Netherlands, 2014. [Google Scholar]
- Pirotti, F. Analysis of full-waveform LiDAR data for forestry applications: A review of investigations and methods. iForest—Biogeosci. For. 2011, 4, 100–106. [Google Scholar] [CrossRef]
- Yan, W.Y.; Shaker, A.; El-Ashmawy, N. Urban land cover classification using airborne LiDAR data: A review. Remote Sens. Environ. 2015, 158, 295–310. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Ørka, H.O.; Holmgren, J.; Dalponte, M.; Heinzel, J.; Koch, B. Tree species recognition based on airborne laser scanning and complementary data sources. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer Science + Business Media: Dordrecht, The Netherlands, 2014; pp. 135–156. [Google Scholar]
- Wagner, W.; Ullrich, A.; Ducic, V.; Melzer, T.; Studnicka, N. Gaussian decomposition and calibration of a novel small-footprint full-waveform digitising airborne laser scanner. ISPRS J. Photogramm. Remote Sens. 2006, 60, 100–112. [Google Scholar] [CrossRef]
- Wagner, W.; Ullrich, A.; Melzer, T.; Briese, C.; Kraus, K. From single-pulse to full-waveform airborne laser scanners: Potential and practical challenge. Int. Arch. Photogramm. Remote Sens. 2004, 35, 201–206. [Google Scholar]
- Roncat, A.; Morsdorf, F.; Briese, C.; Wagner, W.; Pfeifer, N. Laser pulse interaction with forest canopy: Geometric and radiometric issues. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2014; pp. 19–41. [Google Scholar]
- Eysn, L.; Hollaus, M.; Lindberg, E.; Berger, F.; Monnet, J.M.; Dalponte, M.; Kobal, M.; Pellegrini, M.; Lingua, E.; Mongus, D.; et al. A benchmark of LiDAR-based single tree detection methods using heterogeneous forest data from the Alpine space. Forests 2015, 6, 1721–1747. [Google Scholar] [CrossRef] [Green Version]
- Höfle, B.; Hollaus, M.; Lehner, H.; Pfeifer, N.; Wagner, W. Area-based parameterization of forest structure using full-waveform airborne laser scanning data. In Proceedings of the SilviLaser 2008: 8th International Conference on LiDAR Applications for Assessing Forest Ecosystems, Edinburgh, UK, 17–19 September 2008; pp. 227–236.
- Koch, B.; Kattenborn, T.; Straub, C.; Vauhkonen, J. Segmentation of forest to tree objects. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2014; pp. 89–112. [Google Scholar]
- Gupta, S.; Weinacker, H.; Koch, B. Comparative analysis of clustering-based approaches for 3-D single tree detection using airborne fullwave LiDAR data. Remote Sens. 2010, 2, 968–989. [Google Scholar] [CrossRef]
- Strîmbu, V.F.; Strîmbu, B.M. A graph-based segmentation algorithm for tree crown extraction using airborne LiDAR data. ISPRS J. Photogramm. Remote Sens. 2015, 104, 30–43. [Google Scholar] [CrossRef]
- Zhang, J.; Sohn, G.; Brédif, M. A hybrid framework for single tree detection from airborne laser scanning data: A case study in temperate mature coniferous forests in Ontario, Canada. ISPRS J. Photogramm. Remote Sens. 2014, 98, 44–57. [Google Scholar] [CrossRef]
- Reitberger, J.; Krzystek, P.; Stilla, U. Analysis of full waveform LIDAR data for the classification of deciduous and coniferous trees. Int. J. Remote Sens. 2008, 29, 1407–1431. [Google Scholar] [CrossRef]
- Reitberger, J.; Krzystek, P.; Stilla, U. Benefit of airborne full waveform LiDAR for 3D segmentation and classification of single trees. In Proceedings of the ASPRS 2009 Annual Conference, Baltimore, MD, USA, 9–13 March 2009; pp. 1–9.
- Rutzinger, M.; Höfle, B.; Hollaus, M.; Pfeifer, N. Object-based point cloud analysis of full-waveform airborne laser scanning data for urban vegetation classification. Sensors 2008, 8, 4505–4528. [Google Scholar] [CrossRef]
- Vaughn, N.R.; Moskal, L.M.; Turnblom, E.C. Tree species detection accuracies using discrete point LiDAR and airborne waveform LiDAR. Remote Sens. 2012, 4, 377–403. [Google Scholar] [CrossRef]
- Alexander, C.; Tansey, K.; Kaduk, J.; Holland, D.; Tate, N.J. Backscatter coefficient as an attribute for the classification of full-waveform airborne laser scanning data in urban areas. ISPRS J. Photogramm. Remote Sens. 2010, 65, 423–432. [Google Scholar] [CrossRef]
- Ducic, V.; Hollaus, M.; Ullrich, A.; Wagner, W.; Melzer, T. 3D Vegetation mapping and classification using full-waveform laser scanning. In Proceedings of the EARSeL/ISPRS 2006, Vienna, Austria, 14–15 February 2006.
- Fieber, K.D.; Davenport, I.J.; Ferryman, J.M.; Gurney, R.J.; Walker, J.P.; Hacker, J.M. Analysis of full-waveform LiDAR data for classification of an orange orchard scene. ISPRS J. Photogramm. Remote Sens. 2013, 82, 63–82. [Google Scholar] [CrossRef]
- Hollaus, M.; Wagner, W.; Molnar, G.; Mandlburger, G.; Nothegger, C.; Otepka, J. Delineation of vegetation and building polygons from full-waveform airborne LIDAR data using OPALS software. In Proceedings of the geospatial data and geovisualization: Environment, security, and society, special joint symposium of ISPRS technical commission IV and AutoCarto 2010 in conjunction with ASPRS/GaGIS speciality conference, Orlando, FL, USA, 15–19 November 2010.
- Lin, Y.-C. Normalization of echo features derived from full-waveform airborne laser scanning data. Remote Sens. 2015, 7, 2731–2751. [Google Scholar] [CrossRef]
- Mallet, C.; Bretar, F.; Soergel, U. Analysis of full-waveform LiDAR data for classification of urban areas. Photogramm. Fernerkund. GeoInf. 2008, 5, 337–349. [Google Scholar]
- Stilla, U.; Jutzi, B. Waveform analysis for small-footprint pulsed laser systems. In Topographic Laser Ranging and Scanning: Principles and Processing; CRC Press: Boca Raton, FL, USA, 2008; pp. 215–234. [Google Scholar]
- Tran, G.; Nguyen, D.; Milenkovic, M.; Pfeifer, N. Potential of full waveform airborne laser scanning data for urban area classification—Transfer if classification approaches between missions. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, 40, 1317–1323. [Google Scholar] [CrossRef]
- Tseng, Y.H.; Wang, C.K.; Chu, H.J.; Hung, Y.C. Waveform-based point cloud classification in land-cover identification. Int. J. Appl. Earth Observ. Geoinf. 2015, 34, 78–88. [Google Scholar] [CrossRef]
- Wagner, W.; Hollaus, M.; Briese, C.; Ducic, V. 3D vegetation mapping using small-footprint full-waveform airborne laser scanners. Int. J. Remote Sens. 2008, 29, 1433–1452. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree species classification and estimation of stem volume and DBH based on single tree extraction by exploiting airborne full-waveform LiDAR data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
- Li, J.; Hu, B.; Noland, T.L. Classification of tree species based on structural features derived from high density LiDAR data. Agric. For. Meteorol. 2013, 171, 104–114. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural Features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Höfle, B.; Hollaus, M. Urban vegetation detection using high density full-waveform airborne LiDAR data-combination of object-based image and point cloud analysis. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2010, 38, 281–286. [Google Scholar]
- Hollaus, M.; Mücke, W.; Roncat, A.; Pfeifer, N.; Briese, C. Full-waveform airborne laser scanning systems and their possibilities in forest applications. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2014; pp. 43–61. [Google Scholar]
- Guo, B.; Huang, X.; Zhang, F.; Sohn, G. Classification of airborne laser scanning data using JointBoost. ISPRS J. Photogramm. Remote Sens. 2015, 100, 71–83. [Google Scholar] [CrossRef]
- Gross, H.; Jutzi, B.; Thoennessen, U. Segmentation of tree regions using data of a full-waveform laser. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2007, 36, 57–62. [Google Scholar]
- Lindberg, E.; Eysn, L.; Hollaus, M.; Holmgren, J.; Pfeifer, N. Delineation of tree crowns and tree species classification from full-waveform airborne laser scanning data using 3-D ellipsoidal clustering. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2014, 7, 3174–3181. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Korpela, I.; Maltamo, M.; Tokola, T. Imputation of single-tree attributes using airborne laser scanning-based height, intensity, and alpha shape metrics. Remote Sens. Environ. 2010, 114, 1263–1276. [Google Scholar] [CrossRef]
- Ko, C.; Sohn, G.; Remmel, T.K.; Miller, J. Hybrid ensemble classification of tree genera using airborne LiDAR data. Remote Sens. 2014, 6, 11225–11243. [Google Scholar] [CrossRef]
- Yu, X.; Litkey, P.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Assessment of low density full-waveform airborne laser scanning for individual tree detection and tree species classification. Forests 2014, 5, 1011–1031. [Google Scholar] [CrossRef]
- Englhart, S.; Jubanski, J.; Siegert, F. Quantifying dynamics in tropical peat swamp forest biomass with multi-temporal LiDAR datasets. Remote Sens. 2013, 5, 2368–2388. [Google Scholar] [CrossRef] [Green Version]
- Stereńczak, K. Factors influencing individual tree crowns detection based on airborne laser scanning data. For. Res. Pap. 2013, 74, 323–333. [Google Scholar] [CrossRef]
- Han, A.R.; Lee, S.K.; Suh, G.U.; Park, Y.; Park, P.S. Wind and topography influence the crown growth of Picea jezoensis in a subalpine forest on Mt. Deogyu, Korea. Agric. For. Meteorol. 2012, 166, 207–214. [Google Scholar] [CrossRef]
- Seidel, D.; Hoffmann, N.; Ehbrecht, M.; Juchheim, J.; Ammer, C. How neighborhood affects tree diameter increment—New insights from terrestrial laser scanning and some methodical considerations. For. Ecol. Manag. 2015, 336, 119–128. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Wang, T.; Isenburg, M.; Khoshelham, K. Effect of slope on treetop detection using a LiDAR Canopy Height Model. ISPRS J. Photogramm. Remote Sens. 2015, 104, 44–52. [Google Scholar] [CrossRef]
- Romanczyk, P.; van Aardt, J.; Cawse-Nicholson, K.; Kelbe, D.; McGlinchy, J.; Krause, K. Assessing the impact of broadleaf tree structure on airborne full-waveform small-footprint LiDAR signals through simulation. Can. J. Remote Sens. 2013, 39, S60–S72. [Google Scholar] [CrossRef]
- Duan, Z.; Zhao, D.; Zeng, Y.; Zhao, Y.; Wu, B.; Zhu, J. Assessing and correcting topographic effects on forest canopy height retrieval using airborne LiDAR data. Sensors 2015, 15, 12133–12155. [Google Scholar] [CrossRef] [PubMed]
- Vega, C.; Hamrouni, A.; El Mokhtari, S.; Morel, J.; Bock, J.; Renaud, J.P.; Bouvier, M.; Durrieu, S. PTrees: A point-based approach to forest tree extraction from LiDAR data. Int. J. Appl. Earth Observ. Geoinf. 2014, 33, 98–108. [Google Scholar] [CrossRef]
- Molnar, B.; Laky, S.; Toth, C. Using full waveform data in urban areas. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2013, 38, 203–208. [Google Scholar] [CrossRef]
- Hopkinson, C.; Chasmer, L.E.; Sass, G.; Creed, I.F.; Sitar, M.; Kalbfleisch, W.; Treitz, P. Vegetation class dependent errors in LiDAR ground elevation and canopy height estimates in a boreal wetland environment. Can. J. Remote Sens. 2005, 31, 191–206. [Google Scholar] [CrossRef]
- Hermosilla, T.; Coops, N.C.; Ruiz, L.A.; Moskal, L.M. Deriving pseudo-vertical waveforms from small-footprint full-waveform LiDAR data. Remote Sens. Lett. 2014, 5, 332–341. [Google Scholar] [CrossRef]
- Buddenbaum, H.; Seeling, S.; Hill, J. Fusion of full-waveform LiDAR and imaging spectroscopy remote sensing data for the characterization of forest stands. Int. J. Remote Sens. 2013, 34, 4511–4524. [Google Scholar] [CrossRef]
- Hermosilla, T.; Ruiz, L.A.; Kazakova, A.N.; Coops, N.C.; Moskal, L.M. Estimation of forest structure and canopy fuel parameters from small-footprint full-waveform LiDAR data. Int. J. Wildland Fire 2014, 23, 224–233. [Google Scholar] [CrossRef]
- Chauve, A.; Vega, C.; Durrieu, S.; Bretar, F.; Allouis, T.; Pierrot Deseilligny, M.; Puech, W. Advanced full-waveform LiDAR data echo detection: Assessing quality of derived terrain and tree height models in an alpine coniferous forest. Int. J. Remote Sens. 2009, 30, 5211–5228. [Google Scholar] [CrossRef]
- Lukeš, P.; Stenberg, P.; Rautiainen, M.; Mõttus, M.; Vanhatalo, K.M. Optical properties of leaves and needles for boreal tree species in Europe. Remote Sens. Lett. 2013, 4, 667–676. [Google Scholar] [CrossRef]
- Woolley, J.T. Reflectance and Transmittance of Light by Leaves. Plant Physiol. 1971, 47, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Briese, C.; Pfennigbauer, M.; Lehner, H.; Ullrich, A.; Wagner, W.; Pfeifer, N. Radiometric calibration of multi-wavelength airborne laser scanning data. ISPRS Ann. Photogramm. Remote Sens. Spatial Inf. Sci. 2012, 1, 335–340. [Google Scholar] [CrossRef]
- Reitberger, J.; Schnörr, C.; Krzystek, P.; Stilla, U. 3D segmentation of single trees exploiting full waveform LIDAR data. ISPRS J. Photogramm. Remote Sens. 2009, 64, 561–574. [Google Scholar] [CrossRef]
- Ko, C.; Sohn, G.; Remmel, T.K. The impact of LiDAR point density on classifying tree genus: Using geometric features and vertical profile features. In Proceedings of the SilviLaser 2012: 12th International Conference on LiDAR Applications for Assessing Forest, Vancouver, BC, Canada, 16–19 September 2012; Coops, N., Wulder, M., Eds.;
- Ko, C.; Sohn, G.; Remmel, T.K. A comparitive study using geometric and vertical profile features derived from airborne LIDAR for classifying tree genera. ISPRS Ann. Photogramm. Remote Sens. Spatial Inf. Sci. 2012, 1, 129–134. [Google Scholar] [CrossRef]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Classifying species of individual trees by intensity and structure features derived from airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1163–1174. [Google Scholar] [CrossRef]
| Type | References (Applied at c: Class Level; s: Species Level) |
|---|---|
| Geometric | |
| Point-assigned: Elevation difference ΔHij between echo i and reference echo j, Elevation variance of all echoes ΔHvar, Elevation difference between highest and lowest elevation value; Point density Pdens, Penetration Index PI, Echo Ratio ER3D/2D (slope adaptive) and ERME (echo index) | c: [23,54,56,60,61,71,72] |
| Planarity: Plane residuals Ɽ, Deviation of local normal vector ηZ, Structure tensor planarity TP and omnivariance TO | c: [54,60,61,72] |
| Height layer/bin/percentiles: Average echo number Navg,bin, Maximum echo number deviation from the average echo number Nnb,bin, Point density Pdens,bin, Graph features of connected height layers (e.g., top distance TD), Variance-to-mean-ratio of number (VTMR) of echoes of gridded height layer Pgrid,VTMR, Mean trunk distance ΔDtrunk dist, horiz., Statistics *1 of TIN edges PTIN,Edge, filtered *2 statistics *1 of height Hperc,stats,filter, L-function features of echo number Pdens,L-func * | c: [50,71] |
| s: [52,55,66,67,73] | |
| Raster Statistics *1 (and Additional Filters *2): Number of echoes TNorast,stats,filter | s: [25,66] |
| Voxel: Echo number voxel column ratio TNovoxel,column, echo number voxel area ratio ΔAreavoxel,ch, Haralick’s texture features of echo number voxels HTi | s: [55,67] |
| Radiometric | |
| Point-assigned: Echo width EW, Fuzzy small membership of echo width FEWCV, Product of echo amplitude and width EAW | c: [60] |
| s: [25,66] | |
| Raster Statistics *1 (and Additional Filters *2): Amplitude Arast,stats,filter, | s: [25,66] |
| Height layer/bin/percentiles: filtered *2 statistics *1 of amplitude Aperc,stats,filter | s: [66] |
| Type | References (Applied at c: Class Level; s: Species Level) |
|---|---|
| Geometric | |
| Statistics *1 (and Additional Filters *2): Number of echoes TNostats,obj by index ERsingle/multiple,obj, normalized height values Znorm,stats,obj, Voxel neighborhood statistics Voxneighbor,stats | c: [64] |
| s: [24,26,53,55,66,73,76] | |
| Point distribution: Echo Ratio ER3D/2D (slope adaptive) and ERME (echo index), Height of center of gravity Hgravity, Percentage of laser echoes above step-off count TNoPerc,Hmin,obj, Sphere-based minimum projection area Areaproj, Crown related distributions (e.g., CRpdens,derivatives) | c: [13,59] |
| Vertical profile: Height values VH,stats | s: [47] |
| Shape: Ellipsoid features (e.g., height Zobj_ellip), Parabolic features (e.g., radius PSradius), Crown features (e.g., crown length—tree height—ratio CRlt) | s: [28,66,73] |
| Waveform | |
| Statistics *1: Sum of waveform amplitude Asum,obj, Returned waveform energy RWE and the height value Hm,energy of e.g., 50% of the total energy, Number of echoes per waveform TNowave,stats and between peak indices ΔNoij, distance between peak indices in range ΔRij and time ΔTij, distance between waveform metrics in range ΔWRij and time ΔWTij, Overlap width of first and second echo ΔOW1st/2nd,stats, Front slope angle from waveform beginning to first peak FS, Product of echo amplitude and width EAWstats,obj, Echo width EWstats,obj, Shape parameter of Gaussian decomposition αdecomp,wave (e.g., skewness) | c: [15,64] |
| s: [15,55,66,73,76] | |
| Vertical Profile *1: Echo width VEW | c: [13] |
| s: [47] | |
| Radiometric | |
| Statistics *1: Amplitude Astats,obj, Backscatter cross-section σstats,obj, Backscatter coefficient γstats,obj, | c: [13,47,54,64] |
| s: [24,26,52,53,55,58,66,73] | |
| Vertical Profile *1: Amplitude VA, backscatter cross-section Vγ | c: [13] |
| s: [47] |
| Taxonomy (Tree Class and Species) | Feature Characteristic |
|---|---|
| Coniferous trees (vs. deciduous trees) | |
| Larch |
|
| Pine (vs. deciduous trees) |
|
| Spruce |
|
| Deciduous trees |
|
| Aspen |
|
| Beech | |
| Birch | |
| Maple | |
| Oak | |
| Poplar |
|
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koenig, K.; Höfle, B. Full-Waveform Airborne Laser Scanning in Vegetation Studies—A Review of Point Cloud and Waveform Features for Tree Species Classification. Forests 2016, 7, 198. https://doi.org/10.3390/f7090198
Koenig K, Höfle B. Full-Waveform Airborne Laser Scanning in Vegetation Studies—A Review of Point Cloud and Waveform Features for Tree Species Classification. Forests. 2016; 7(9):198. https://doi.org/10.3390/f7090198
Chicago/Turabian StyleKoenig, Kristina, and Bernhard Höfle. 2016. "Full-Waveform Airborne Laser Scanning in Vegetation Studies—A Review of Point Cloud and Waveform Features for Tree Species Classification" Forests 7, no. 9: 198. https://doi.org/10.3390/f7090198
APA StyleKoenig, K., & Höfle, B. (2016). Full-Waveform Airborne Laser Scanning in Vegetation Studies—A Review of Point Cloud and Waveform Features for Tree Species Classification. Forests, 7(9), 198. https://doi.org/10.3390/f7090198