Effects of Host Variability on the Spread of Invasive Forest Diseases
Abstract
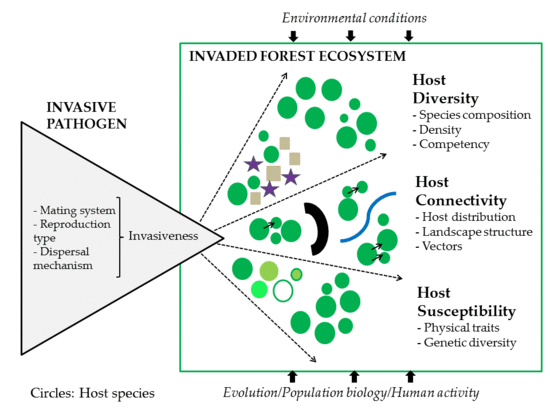
:1. Introduction
1.1. Novel Environments, Novel Hosts
1.2. Pathogen Invasiveness Affected by Species Traits
2. Host Factors
2.1. Diversity
2.2. Connectivity
2.3. Susceptibility
3. Case Studies
3.1. Ash Dieback
3.2. Chestnut Blight
4. “The Way Forward”
4.1. Host Diversity Threshold
4.2. Spread Dynamics
4.3. How to Slow the Spread
4.4. Evolution of Invasive Forest Pathogens
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ricciardi, A. Are Modern Biological Invasions an Unprecedented Form of Global Change? Conserv. Biol. 2007, 21, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Crowl, T.A.; Cris, T.O.; Parmenter, R.R.; Belovsky, G.; Lugo, A.E. The spread of invasive species and infectious disease as drivers of ecosystem change. Front Ecol. Environ. 2008, 6, 238–246. [Google Scholar] [CrossRef]
- Lovett, G.M.; Weiss, M.; Liebhold, A.M.; Holmes, T.P.; Leung, B.; Lambert, K.F.; Orwig, D.A.; Campbell, F.T.; Rosenthal, J.; McCullough, D.G.; et al. Nonnative forest insects and pathogens in the United States: Impacts and policy options. Ecol. Appl. 2016, 26, 1437–1455. [Google Scholar] [CrossRef] [PubMed]
- Aukema, J.E.; McCullough, D.G.; Von Holle, B.; Liebhold, A.J.; Britton, K.O.; Frankel, S.J. Historical accumulation of non-indigenous forest pests in the continental United States. BioScience 2010, 60, 886–897. [Google Scholar] [CrossRef]
- Fisher, M.C.; Henk, D.A.; Briggs, C.; Brownstein, J.S.; Madoff, L.; McCraw, S.L.; Gurr, S. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Santini, A.; Ghelardini, L.; de Pace, C.; Desprez-Loustau, M.L.; Capretti, P.; Chandelier, A.; Cech, T.; Chira, D.; Diamandis, S.; Gaitniekis, T.; et al. Biogeographical, patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013, 197, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Liebhold, A.M.; Brockerhoff, E.G.; Garrett, L.J.; Parke, J.L.; Britton, K.O. Live plant imports: The major pathway for forest insect and pathogen invasions of the US. Front. Ecol. Environ. 2012, 10, 135–143. [Google Scholar] [CrossRef]
- Ghelardini, L.; Pepori, A.L.; Luchi, N.; Capretti, P.; Santini, A. Drivers of emerging fungal diseases of forest trees. For. Ecol. Manag. 2016, 381, 235–246. [Google Scholar] [CrossRef]
- Anagnostakis, S.L. Chestnut blight: The classical problem of an introduced pathogen. Mycologia 1987, 79, 23–37. [Google Scholar] [CrossRef]
- Hansen, E.M.; Kanaskie, A.; Prospero, S.; McWilliams, M.; Goheen, E.M.; Osterbauer, N.; Reeser, P.; Sutton, W. Epidemiology of Phytophthora ramorum in Oregon tanoak forests. Can. J. For. Res. 2008, 38, 1133–1143. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Garbelotto, M. Sudden oak death: Endangering California and Oregon forest ecosystems. Front. Ecol. Environ. 2003, 1, 197–204. [Google Scholar] [CrossRef]
- Hui, C.; Krug, R.M.; Richardson, D.M. Modelling spread in invasion ecology: A synthesis. In Fifty Years of Invasion Ecology: The Legacy of Charles Elton, 1st ed.; Richardson, D.M., Ed.; Wiley-Blackwell: Oxford, UK, 2011; pp. 329–343. [Google Scholar]
- Shigesada, N.; Kawasaki, K.; Takeda, Y. Modelling stratified diffusion in biological invasions. Am. Nat. 1995, 146, 229–251. [Google Scholar] [CrossRef]
- Valéry, L.; Fritz, H.; Lefeuvre, J.C.; Simberloff, D. In search of a real definition of the biological invasion phenomenon itself. Biol. Invasions 2008, 10, 1345–1351. [Google Scholar] [CrossRef]
- Wilson, J.R.U.; Dormontt, E.E.; Prentis, P.J.; Lowe, A.J.; Richardson, D.M. Biogeographic concepts define invasion biology. TRENDS Ecol. Evol. 2009, 24, 586. [Google Scholar] [CrossRef]
- Lockwood, J.L.; Hoopes, M.F.; Marchetti, M.P. Invasion Ecology, 2nd ed.; Wiley Blackwell: West Sussex, UK, 2013. [Google Scholar]
- Smith, C.S.; Lonsdale, W.M.; Fortune, J. When to ignore advice: Invasion predictions and decision theory. Biol. Invasions 1999, 1, 89–96. [Google Scholar] [CrossRef]
- Mack, R.N.; Simberloff, D.; Lonsdale, W.M.; Evans, H.; Clout, M.; Bazzaz, F. Biotic invasions: Causes, epidemiology, global consequences, and control. Ecol. Appl. 2000, 10, 689–710. [Google Scholar] [CrossRef]
- Aylor, D.E. Spread of plant disease on a continental scale: Role of aerial dispersal of pathogens. Ecology 2003, 84, 1989–1997. [Google Scholar] [CrossRef]
- Parker, I.M.; Gilbert, G.S. The Evolutionary Ecology of Novel Plant-Pathogen Interactions. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 675–700. [Google Scholar] [CrossRef]
- Hastings, A.; Cuddington, K.; Davies, K.F.; Dugaw, C.J.; Elmendorf, S.; Freestone, A.; Harrison, S.; Holland, M.; Lambrinos, J.; Malvadkar, U.; et al. The spatial spread of invasions: New developments in theory and evidence. Ecol. Lett. 2005, 8, 91–101. [Google Scholar] [CrossRef]
- Menkis, A.; Östbrant, I.-L.; Wågström, K.; Vasaitis, R. Dutch elm disease on the island of Gotland: Monitoring disease vector and combat measures. Scand. J. For. Res. 2016, 31, 237–241. [Google Scholar] [CrossRef]
- Goss, E.M.; Larsen, M.; Vercauteren, A.; Werres, S.; Heungens, K.; Grünwald, N.J. Phytophthora ramorum in Canada: Evidence for migration within North America and from Europe. Phytopathology 2011, 101, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Goss, E.M.; Larsen, M.M.; Chastagner, G.A.; Givens, D.R.; Grünwald, N.J. Population genetic analysis infers migration pathways of Phytophthora ramorum in US nurseries. PLoS Pathol. 2009, 5, e1000583. [Google Scholar] [CrossRef] [PubMed]
- Jules, E.W.S.; Kauffman, M.J.; Ritts, W.D.; Carrol, A.L. Spread of an invasive pathogen over a variable landscape: A nonnative root rot on Port Orford cedar. Ecology 2002, 83, 3167–3181. [Google Scholar] [CrossRef]
- Kelly, M.; Gueo, Q.; Liu, D.; Shaari, D. Modelling the risk for a new invasive forest disease in the United States: An evaluation of five environmental niche models. Comput. Environ. Urban 2007, 31, 689–710. [Google Scholar] [CrossRef]
- Meentemeyer, R.; Rizzo, D.; Mark, W.; Lotz, E. Mapping the risk of establishment and spread of sudden oak death in California. For. Ecol. Manag. 2004, 200, 195–214. [Google Scholar] [CrossRef]
- Condeso, T.E.; Meentemeyer, R.K. Effects of landscape heterogeneity on the emerging forest disease sudden oak death. J. Ecol. 2007, 95, 364–375. [Google Scholar] [CrossRef]
- Filipe, J.A.N.; Cobb, R.C.; Meentemeyer, R.K.; Lee, C.A.; Valachovic, Y.S.; Cook, A.R.; Rizzo, D.M.; Gilligan, C.A. Landscape Epidemiology and Control of Pathogens with Cryptic and Long-Distance Dispersal: Sudden Oak Death in Northern Californian Forests. PLoS Comput. Biol. 2012, 8, e1002328. [Google Scholar] [CrossRef] [PubMed]
- Cushman, J.H.; Meentemeyer, R.K. Multi-scale patterns of human activity and the incidence of an exotic forest pathogen. J. Ecol. 2008, 96, 766–776. [Google Scholar] [CrossRef]
- Harwood, T.D.; Xu, X.; Pautasso, M.; Jeger, M.J.; Shaw, M.W. Epidemiological risk assessment using linked network and grid based modelling: Phytophthora ramorum and Phytophthora kernoviae in the UK. J. Ecol. Model. 2009, 220, 3353–3361. [Google Scholar] [CrossRef]
- Gilbert, M.; Liebhold, A. Comparing methods for measuring the rate of spread of invading populations. Ecography 2010, 33, 809–817. [Google Scholar] [CrossRef]
- Tisseuil, C.; Gryspeirt, A.; Lancelot, R.; Pioz, M.; Liebhold, A.; Gilbert, M. Evaluating methods to quantify spatial variation in the velocity of biological invasions. Ecography 2016, 39, 409–418. [Google Scholar] [CrossRef]
- Philibert, A.; Desprez-Loustau, M.-L.; Fabre, B.; Frey, P.; Halkett, F.; Husson, C.; Lung-Escarmant, B.; Marçais, B.; Robin, C.; Vacher, C.; et al. Predicting invasion success of forest pathogenic fungi from species traits. J. Appl. Ecol. 2011, 48, 1381–1390. [Google Scholar] [CrossRef]
- Janzen, D.H. On Ecological Fitting. Oikos 1985, 45, 308–310. [Google Scholar] [CrossRef]
- Brooks, D.R.; McLennan, D.A. The Nature of Diversity: An Evolutionary Voyage of Discovery; University of Chicago Press: Chicago, IL, USA, 2002. [Google Scholar]
- Agosta, S.J.; Klemens, J.A. Ecological fitting by phenotypically flexible genotypes: Implications for species associations, community assembly and evolution. Ecol. Lett. 2008, 11, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.H.D.; Burdon, J.J. Mating systems and colonizing success in plants. In Colonization, Succession and Stability; Cary, A.J., Crawley, M.J., Edwards, P.J., Eds.; Blackwell Scientific Publications: Oxford, UK, 1987; pp. 115–131. [Google Scholar]
- Bazin, E.; Mathe-Hubert, H.; Facon, B.; Carlier, J.; Ravigné, V. The effect of mating system on invasiveness: Some genetic load may be advantageous when invading new environments. Biol. Invasions 2014, 16, 875–886. [Google Scholar] [CrossRef] [Green Version]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [PubMed]
- Prospero, S.; Rigling, D. Invasion genetics of the chestnut blight fungus Cryphonectria parasitica in Switzerland. Phytopathology 2012, 102, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Pisani, C.; Ploetz, R.C.; Stover, E.; Ritenour, M.; Scully, B. Laurel Wilt in Avocado: Review of an Emerging Disease. Int. J. Plant Biol. Res. 2015, 3, 1043. [Google Scholar]
- Mehrotra, R.S.; Aggarwal, A. Dispersal of plant pathogens. In Plant Pathology, 2nd ed.; Tata McGraw-Hill Publishing Co. Ltd.: New Delhi, India, 2005; pp. 199–211. [Google Scholar]
- Mehrotra, R.S.; Aggarwal, A. Dispersal of plant pathogens. In Fundamentals of Plant Pathology; Tata McGraw-Hill Publishing Co. Ltd.: New Delhi, India, 2013; Chapter 9. [Google Scholar]
- Hengeveld, R. Dynamics of Biological Invasions; Chapman & Hall: London, UK, 1989. [Google Scholar]
- Gladieux, P.; Feurtey, A.; Hood, M.E.; Snirc, A.; Clavel, J.; Dutech, C.; Roy, M.; Giraud, T. The population biology of fungal invasions. Mol. Ecol. 2015, 24, 1969–1986. [Google Scholar] [CrossRef] [PubMed]
- Pyšek, P.; Jarošik, V.; Hulme, P.E.; Kühn, I.; Wild, J.; Arianoutsou, M.; Bacher, S.; Chiron, F.; Didziulis, V.; Essl, F.; et al. Disentangling the role of environmental and human pressures on biological invasions. Proc. Natl. Acad. Sci. USA 2010, 107, 12157–12162. [Google Scholar]
- Keesing, F.; Belden, L.K.; Daszak, P.; Dobson, A.; Harvell, C.D.; Holt, R.D.; Hudson, P.; Jolles, A.; Jones, K.E.; Mitchell, C.E.; et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 2010, 468, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Ostfeld, R.S.; Keesing, F. Effects of Host Diversity on Infectious Disease. Annu. Rev. Ecol. Evol. Syst. 2012, 43, 157–182. [Google Scholar] [CrossRef]
- Pautasso, M.; Holdenrieder, O.; Stenlid, J. Susceptibility to Fungal Pathogens of Forests Differing in Tree Diversity. In Forest Diversity and Function: Temperate and Boreal Systems, Ecological Studies; Scherer-Lorenzen, M., Körner, C., Schulze, E.-D., Eds.; Springer: New York, NY, USA, 2005; Volume 176, pp. 263–289. [Google Scholar]
- Cleary, M.; Nguyen, D.; Marčiulynienė, D.; Berlin, A.; Vasaitis, R.; Stenlid, J. Friend or foe? Biological and ecological traits of the European ash dieback pathogen Hymenoscyphus fraxineus in its native environment. Sci. Rep. 2016, 6, 21895. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.R.; McKinney, L.V.; Hietala, A.M.; Kjær, E.D. The susceptibility of Asian, European and North American Fraxinus species to the ash dieback pathogen Hymenoscyphus fraxineus reflects their phylogenetic history. Eur. J. Forest. Res. 2016. [Google Scholar] [CrossRef]
- McKinney, L.V.; Neilsen, L.R.; Collinge, D.B.; Thomsen, I.M.; Hansen, J.K.; Kjær, E.J. The ash dieback crisis: Genetic variation in resistance can prove a long-term solution. Plant Pathol. 2014, 63, 485–499. [Google Scholar] [CrossRef]
- Hughes, M.A.; Smith, J.A.; Ploetz, R.C.; Kendra, P.E.; Mayfield, A.E., III; Hanula, J.L.; Hulcr, J.; Stelinski, L.L.; Cameron, S.; Riggins, J.J.; et al. Recovery plan for laurel wilt on redbay and other forest species caused by Raffaelea lauricola and disseminated by Xyleborus glabratus. Plant Health Prog. 2015. [Google Scholar] [CrossRef]
- Mitchell, C.E.; Tilman, D.; Groth, J.V. Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology 2002, 83, 1713–1726. [Google Scholar] [CrossRef]
- Keesing, F.; Holt, R.D.; Ostfel, R.S. Effects of species diversity on disease risk. Ecol. Lett. 2006, 9, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Haas, S.E.; Cushman, J.H.; Dillon, W.W.; Rank, N.E.; Rizzo, D.M.; Meentemeyer, R.K. Effects of individual, community, and landscape drivers on the dynamics of a wildland forest epidemic. Ecology 2016, 97, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, C.; Jolles, A.; Seabloom, E.W.; Power, A.G.; Mitchell, C.E.; Borer, E.T. Non-random biodiversity loss underlies predictable increases in viral disease, prevalence. J. R. Soc. Interface 2014, 11, 20130947. [Google Scholar] [CrossRef] [PubMed]
- Burdon, J.J.; Chilvers, G.A. Host density as a factor in plant disease ecology. Annu. Rev. Phytopathol. 1982, 20, 143–166. [Google Scholar] [CrossRef]
- Wolfe, N.D.; Dunavan, C.P.; Diamond, J. Origins of major human infectious diseases. Nature 2007, 447, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Parker, I.M.; Saunders, S.; Bontrager, M.; Weitz, A.P.; Hendricks, R.; Magarey, R.; Suiter, K.; Gilbert, G.S. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 2015, 520, 542–544. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.S.; Liebhold, A.M.; Tobin, P.C.; Gottschalk, K.W.; Luzader, E. Spread of beech bark disease in the eastern United States and its relationship to regional forest composition. Can. J. For. Res. 2007, 37, 726–736. [Google Scholar] [CrossRef]
- Kasson, M.T.; Livingston, W.H. Relationships among beech bark disease, climate, radial growth response and mortality of American beech in northern Maine, USA. For. Pathol. 2012, 42, 199–212. [Google Scholar] [CrossRef]
- Hatala, J.A.; Dietze, M.C.; Crabtree, R.L.; Kendall, K.; Six, D.; Moorcroft, P.R. An ecosystem-scale model for the spread of a host-specific forest pathogen in the Greater Yellowstone Ecosystem. Ecol. Appl. 2011, 21, 1138–1153. [Google Scholar] [CrossRef] [PubMed]
- Weste, G.; Brown, K.; Kennedy, J.; Walshe, T. Phytophthora cinnamomi infestation—A 24 year study of vegetation change in forests and woodlands of the Grampians, Western Victoria. Aust. J. Bot. 2002, 50, 247–274. [Google Scholar] [CrossRef]
- Davidson, J.M.; Patterson, H.A.; Wickland, A.C.; Fichtner, E.J.; Rizzo, D.M. Forest type influences transmission of Phytophthora ramorum in California oak woodlands. Phytopathology 2001, 101, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Purse, B.V.; Graeser, P.; Searle, K.; Edwards, C.; Harris, C. Challenges in predicting invasive reservoir hosts of emerging pathogens: Mapping Rhododendron ponticum as a foliar host for Phytophthora ramorum and Phytophthora kernoviae in the UK. Biol. Invasions 2013, 15, 529–545. [Google Scholar] [CrossRef]
- Kasson, M.T.; Livingston, W.H. Spatial distribution of Neonectria species associated with beech bark disease in northern Maine. Mycologia 2009, 101, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Park, A.W.; Gubbins, S.; Gilligan, C.A. Extinction times for closed epidemics: The effects of host spatial structure. Ecol. Lett. 2002, 5, 747–755. [Google Scholar] [CrossRef]
- Holdenrieder, O.; Pautasso, M.; Weisberg, P.J.; Lonsdale, D. Tree diseases and landscape processes: The challenge of landscape pathology. TRENDS Ecol. Evol. 2004, 19, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, G. On the spatial spread of insect viruses: Theory and experiment. Ecology 1992, 73, 479–494. [Google Scholar] [CrossRef]
- Burdon, J.J.; Jarosz, A.M.; Kirby, G.C. Pattern and patchiness in plant-pathogen interactions: Causes and consequences. Ann. Rev. Ecol. Syst. 1989, 20, 119–136. [Google Scholar] [CrossRef]
- Caraco, T.; Duryea, M.C.; Glavanakov, S.; Maniatty, W.; Szymanski, B.K. Host Spatial Heterogeneity and the Spread of Vector-Borne Infection. Theor. Popul. Biol. 2001, 59, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Broders, K.; Boraks, A.; Barbison, L.; Brown, J.; Boland, G.J. Recent insights into the pandemic disease butternut canker caused by the invasive pathogen Ophiognomonia clavigignenti-juglandacearum. For. Pathol. 2015, 45, 1–8. [Google Scholar]
- Brar, S.; Tsui, C.K.M.; Dhillon, B.; Bergeron, M.-J.; Joly, D.L.; Zambino, P.J.; El-Kassaby, Y.A.; Hamelin, R.C. Colonization History, Host Distribution, Anthropogenic Influence and Landscape Features Shape Populations of White Pine Blister Rust, an Invasive Alien Tree Pathogen. PLos ONE 2015, 10, e0127916. [Google Scholar] [CrossRef] [PubMed]
- Papaïx, J.; Touzeau, S.; Monod, H.; Lannou, C. Can epidemic control be achieved by altering landscape connectivity in agricultural systems? Ecol. Model. 2014, 284, 35–47. [Google Scholar] [CrossRef]
- Real, L.A.; Biek, R. Spatial dynamics and genetics of infectious diseases on heterogeneous landscapes. J. R. Soc. Interface 2007, 4, 935–948. [Google Scholar] [CrossRef] [PubMed]
- Castello, J.D.; Leopold, D.J.; Smallidge, P.J. Pathogens patterns, and processes in forest ecosystems. Bioscience 1995, 45, 16–24. [Google Scholar] [CrossRef]
- Ellis, A.M.; Váklavík, T.; Meentemeyer, R.K. When is connectivity important? A case study of the spatial pattern of sudden oak death. Oikos 2010, 119, 485–493. [Google Scholar] [CrossRef]
- Garnas, J.R.; Auger-Rozenberg, M.-A.; Roques, A.; Bertelsmeier, C.; Wingfield, M.J.; Saccaggi, D.L.; Roy, H.E.; Slippers, B. Complex patterns of global spread in invasive insects: Eco-evolutionary and management consequences. Biol. Invasions 2016, 18, 935–952. [Google Scholar] [CrossRef]
- Dutech, C.; Fabreguettes, O.; Capdevielle, X.; Robin, C. Multiple introductions of divergent genetic lineages in an invasive fungal pathogen, Cryphonectria parasitica, in France. Heredity 2010, 105, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Möykkynen, T.; Capretti, P.; Pukkala, T. Modelling the potential spread of Fusarium circinatum, the causal agent of pitch canker in Europe. Ann. For. Sci. 2015, 72, 169–181. [Google Scholar] [CrossRef]
- Jules, E.S.; Carroll, A.L.; Garcia, A.M.; Steenbock, C.M.; Kauffman, M.J. Host heterogeneity influences the impact of a non-native disease invasion on populations of a foundation tree species. Ecosphere 2014, 5, 1–17. [Google Scholar] [CrossRef]
- Jarosz, A.M.; Burdon, J.J. The effect of small scale environmental changes on disease incidence and severity in a natural plant pathogen interaction. Oecologia 1988, 75, 78–81. [Google Scholar] [CrossRef]
- Smith, J.P.; Hoffman, J.T. Site and stand characteristics related to white pine blister rust in high-elevation forests of Southern Idaho and Western Wyoming. West. N. Am. Nat. 2001, 61, 409–416. [Google Scholar]
- Marçais, B.; Desprez-Loustau, M.-L. European oak powdery mildew: Impact on trees, effects of environmental factors, and potential effects of climate change. Ann. For. Sci. 2014, 71, 633–642. [Google Scholar] [CrossRef]
- Solla, A.; Martín, J.A.; Ouellette, G.B.; Gil, L. Influence of plant age on symptom development in Ulmus minor following inoculation by Ophiostoma novo-ulmi. Plant Dis. 2005, 89, 1035–1040. [Google Scholar] [CrossRef]
- King, K.C.; Lively, C.M. Does genetic diversity limit disease spread in natural host populations? Heredity 2012, 109, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Burdon, R.D. Genetic diversity and disease resistance: Some considerations for research, breeding, and deployment. Can. J. For. Res. 2001, 31, 596–606. [Google Scholar] [CrossRef]
- Garrett, K.A.; Mundt, C.C. Epidemiology in mixed populations. Phytopathology 1999, 89, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Pilet, F.; Chacon, G.; Forbes, G.A.; Adrivon, D. Protection of susceptible potato cultivars against late blight in mixtures increases with decreasing disease pressure. Phytopathology 2006, 96, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Chen, H.; Fan, J.; Wang, Y.; Li, Y.; Chen, J.; Fan, J.X.; Yang, S.; Hu, L.; Leung, H.; et al. Genetic diversity and disease control in rice. Nature 2000, 406, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Barnes, I.; Wingfield, M.J.; Carbone, I.; Kirisits, T.; Wingfield, B.D. Population structure and diversity of an invasive pine needle pathogen reflects anthropogenic activity. Ecol. Evol. 2014, 4, 3642–3661. [Google Scholar] [CrossRef] [PubMed]
- Andjic, V.; Dell, B.; Barber, P.; Hardy, G.; Wingfield, M.; Burgess, T. Plants for planting; indirect evidence for the movement of a serious forest pathogen, Teratosphaeria destructans, in Asia. Eur. J. Plant Pathol. 2011, 131, 49–58. [Google Scholar] [CrossRef]
- Dwyer, G.; Elkinton, J.S.; Buonaccorsi, J.P. Host Heterogeneity in susceptibility and disease dynamics: Tests of a mathematical model. Am. Nat. 1997, 150, 685–707. [Google Scholar] [CrossRef] [PubMed]
- Spielman, D.; Brook, B.W.; Briscoe, D.A.; Frankham, R. Does inbreeding and loss of genetic diversity decrease disease resistance. Conserv. Genet. 2004, 5, 439–448. [Google Scholar] [CrossRef]
- Aegerter, B.J.; Gordon, T.R. Rates of pitch canker induced seedling mortality among Pinus radiate families varying in levels of genetic resistance to Gibberella circinata (anamorph Fusarium circinatum). For. Ecol. Manag. 2006, 235, 14–17. [Google Scholar] [CrossRef]
- Sniezko, R.A. Resistance breeding against nonnative pathogens in forest trees—Current successes in North America. Can. J. Plant Pathol. 2006, 28, S270–S279. [Google Scholar] [CrossRef]
- Ganz, H.H.; Ebert, D. Benefits of host genetic diversity for resistance to infection depend on parasite diversity. Ecology 2010, 91, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Freer-Smith, P.H.; Webber, J.F. Tree pests and diseases: The threat to biodiversity and the delivery of ecosystem services. Biodivers. Conserv. 2015, 1–15. [Google Scholar] [CrossRef]
- Heuch, J. What lessons need to be learnt from the outbreak of Ash Dieback Disease, Chalara fraxinea in the United Kingdom? Arboric. J. 2014, 36, 32–44. [Google Scholar] [CrossRef]
- Mitchell, R.J.; Beaton, J.K.; Bellamy, P.E.; Broome, A.; Chetcuti, J.; Eaton, S.; Ellis, C.J.; Gimona, A.; Harmer, R.; Hester, A.J.; et al. Ash dieback in the UK: A review of the ecological and conservation implications and potential management options. Biol. Conserv. 2014, 175, 95–109. [Google Scholar] [CrossRef]
- Gross, A.; Holdenrieder, O.; Pautasso, M.; Queloz, V.; Sieber, T.N. Hymenoscyphus pseudoalbidus, the causal agent of European ash dieback. Mol. Plant Pathol. 2014, 15, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Børja, I.; Timmermann, V.; Hietala, A.M.; Tollefsrud, M.M.; Nagy, N.E.; Vivian-Smith, A.; Cross, H.; Sønstebø, H.J.; Myking, T.; Solheim, H. Ash dieback in Norway—Current situation. In Dieback of European Ash (Fraxinus spp.)—Consequences and Guidelines for Sustainable Management; Vasaitis, R., Enderle, R., Eds.; SLU Service/Repro: Uppsala, Sweden, 2017; pp. 166–175. [Google Scholar]
- Cleary, M.; Nguyen, D.; Stener, L.-G.; Stenlid, J.; Skovsgaard, J.-P. Ash and ash dieback in Sweden: A review of disease history, current status, pathogen and host dynamics, host tolerance and management options in forests and landscapes. In Dieback of European Ash (Fraxinus spp.)—Consequences and Guidelines for Sustainable Management; Vasaitis, R., Enderle, R., Eds.; SLU Service/Repro: Uppsala, Sweden, 2017; pp. 195–208. [Google Scholar]
- Roane, M.K.; Griffin, G.J.; Elkins, J.R. Chestnut Blight, Other Endothial Diseases, and the Genus Endothia; American Phytopathological Society: St. Paul, MN, USA, 1986. [Google Scholar]
- Braun, E.L. Deciduous Forests of Eastern North America; Hafner: New York, NY, USA, 1950. [Google Scholar]
- Jacobs, D.F. Toward development of silvical strategies for forest restoration of American chestnut (Castanea dentata) using blight-resistant hybrids. Biol. Conserv. 2007, 137, 497–506. [Google Scholar] [CrossRef]
- Graves, A.H. Relative blight resistance in species and hybrids of Castanea. Phytopathology 1950, 40, 1125–1131. [Google Scholar]
- Dane, F.; Lang, P.; Huang, H.; Fu, Y. Intercontinental genetic divergence of Castanea species in eastern Asia and eastern North America. Heredity 2003, 91, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Dutech, C.; Barres, B.; Bridier, J.; Robin, C.; Milgroom, M.G.; Ravigne, V. The chestnut blight fungus world tour: Successive introduction events from diverse origins in an invasive plant fungal pathogen. Mol. Ecol. 2012, 21, 3931–3946. [Google Scholar] [CrossRef] [PubMed]
- Heiniger, U.; Rigling, D. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 1994, 32, 581–599. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Milgroom, M.G.; Cortesi, P. Biological control of chestnut blight with hypovirulence: A critical analysis. Annu. Rev. Phytopathol. 2004, 42, 311–338. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.S.; Choi, G.H.; Nuss, D.L. Mitotic stability and nuclear inheritance of integrated viral cDNA in engineered hypovirulent strains of the chestnut blight fungus. EMBO J. 1993, 12, 2991–2998. [Google Scholar] [PubMed]
- Zhang, D.X.; Nuss, D.L. Engineering super mycovirus donor strains of chestnut blight fungus by systematic disruption of multilocus vic genes. Proc. Natl. Acad. Sci. USA 2016, 113, 2062–2067. [Google Scholar] [CrossRef] [PubMed]
- Diskin, M.; Steiner, K.C.; Hebard, F.V. Recovery of American chestnut characteristics following hybridization and backcross breeding to restore blight-ravaged Castanea dentata. For. Ecol. Manag. 2006, 223, 439–447. [Google Scholar] [CrossRef]
- Newhouse, A.E.; Polin-McGuigan, L.D.; Baier, K.A.; Valletta, K.E.; Rottmann, W.H.; Tschaplinski, T.J.; Maynard, C.A.; Powell, W.A. Transgenic American chestnuts show enhanced blight resistance and transmit the trait to T1 progeny. Plant Sci. 2014, 228, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, T.; Otani, Y.; Furuya, K. Materials for the fungus flora of Japan. Trans. Mycol. Soc. Jpn. 1993, 34, 429–432. [Google Scholar]
- Drenkhan, R.; Hanson, M. New host species for Chalara fraxinea. New Dis. Rep. 2010, 22, 16. [Google Scholar] [CrossRef]
- Cleary, M.R.; Arhipova, N.; Gaitnieks, T.; Stenlid, J.; Vasaitis, R. Natural infection of Fraxinus excelsior seeds by Chalara fraxinea. For. Pathol. 2013, 43, 83–85. [Google Scholar]
- Luchi, N.; Montecchio, L.; Santini, A. Situation with ash in Italy: Stand characteristics, health condition, ongoing work and research needs. In Interim Report from Chalara fraxinea, FRAXBACK Meeting in Vilnius, 13–14 November 2012; Mainprize, N., Hendry, S., Weir, J., Eds.; Forestry Commission: Bristol, UK, 2012; pp. 25–26. [Google Scholar]
- Zhao, Y.-J.; Hosoya, T.; Baral, H.-O.; Hosaka, K.; Kakishima, M. Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. MycoTaxon 2013, 122, 25–41. [Google Scholar] [CrossRef]
- Chandelier, A.; Gerarts, F.; San Martin, G.; Herman, M.; Delahaye, L. Temporal evolution of collar lesions associated with ash dieback and the occurrence of Armillaria in Belgian forests. For. Pathol. 2016. [Google Scholar] [CrossRef]
- Chandelier, A.; Helson, M.; Dvorak, M.; Gischer, F. Detection and quantification of airborne inoculum of Hymenoscyphus pseudoalbidus using real-time PCR assays. Plant Pathol. 2014, 63, 1296–1305. [Google Scholar] [CrossRef]
- Zheng, H.-D.; Zhuang, W.-Y. Hymenoscyphus albidoides sp. nov. and H. pseudoalbidus from China. Mycol. Prog. 2014, 13, 625–638. [Google Scholar] [CrossRef]
- Han, J.-G.; Shrestha, B.; Hosoya, T.; Lee, K.-H.; Sung, G.-H.; Shin, H.-D. First report of the ash dieback pathogen Hymensocyphus fraxineus in Korea. Mycobiology 2014, 42, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Kirisits, T.; Schwanda, K. First definite report of natural infection of Fraxinus ornus by Hymenoscyphus fraxineus. For. Pathol. 2015, 45, 430–432. [Google Scholar]
- Marçais, B.; Husson, C.; Godart, L.; Caël, O. Influence of site and stand factors on Hymenoscyphus fraxineus-induced basal lesion. Plant Pathol. 2016. [Google Scholar] [CrossRef]
- Park, A.W.; Gubbins, S.; Gilligan, C.A. Invasion persistence of plant parasites in a spatially structured host population. Oikos 2001, 94, 162–174. [Google Scholar] [CrossRef]
- Jeger, M.J.; Pautasso, M.; Holdenrieder, O.; Shaw, M.K. Modelling disease spread and control in networks: Implications for plant sciences. New Phytol. 2007, 114, 279–297. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, J.R.; Preisser, E.L.; Fitzpatrick, M.C. Modeling the spread of invasive species using dynamic network models. Biol. Invasions 2014, 16, 949–960. [Google Scholar] [CrossRef]
- Moslonka-Lefebvre, M.; Finley, A.; Dorigatti, I.; Dehnen-Schmutz, K.; Harwood, T.; Jeger, M.J.; Xu, X.; Holdenrieder, O.; Pautasso, M. Networks in plant epidemiology: From genes to landscapes, countries, and continents. Phytopathology 2011, 101, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Biek, R.; Real, L.A. The landscape genetics of infectious disease emergence and spread. Mol. Ecol. 2010, 19, 3515–3531. [Google Scholar] [CrossRef] [PubMed]
- Desprez-Loustau, M.-L. The alien fungi of Europe. In Handbook of Alien Species in Europe, DAISIE. Invading Nature, Series in Invasion Ecology; Drake, J.A., Ed.; Springer: Berlin, Germany, 2009; Volume 3, pp. 15–28. [Google Scholar]
- Horie, T.; Haight, R.G.; Homans, F.R.; Venette, R.C. Optimal strategies for the surveillance and control of forest pathogens: A case study with oak wilt. Ecol. Econ. 2013, 86, 78–85. [Google Scholar] [CrossRef]
- Liebhold, A.M.; Tobin, P.C. Exploiting the Achilles Heels of Pest Invasions: Allee Effects, Stratified Dispersal and Management of Forest Insect Establishment and Spread. N. Zeal. J. For. Sci. 2010, 40, S25–S33. [Google Scholar]
- Homans, F.; Horie, T. Optimal detection strategies for an established invasive pest. Ecol. Econ. 2011, 70, 1129–1138. [Google Scholar] [CrossRef]
- Hänfling, B.; Kollmann, J. An evolutionary perspective of biological invasions. TRENDS Ecol. Evol. 2002, 17, 545–546. [Google Scholar] [CrossRef]
- Lee, C.E. Evolutionary genetics of invasive species. TRENDS Ecol. Evol. 2002, 17, 386–391. [Google Scholar] [CrossRef]
- Woolhouse, M.E.J.; Haydon, D.T.; Antia, R. Emerging pathogens: The epidemiology and evolution of species jumps TRENDS Ecol. Evol. 2005, 20, 238–244. [Google Scholar]
- Desprez-Loustau, M.-L.; Courtecuisse, R.; Robin, C.; Husson, C.; Moreau, P.-A.; Blancard, D.; Selosse, M.-A.; Lung-Escarmant, B.; Piou, D.; Sache, I. Species diversity and drivers of spread of alien fungi (sensu lato) in Europe with a particular focus on France. Biol. Invasions 2010, 12, 157–172. [Google Scholar] [CrossRef]

| Mechanism | Description |
|---|---|
| Autonomous transmission | This dispersal mechanism is characterized by continuous and persistent growth of hyphal strands that can migrate independently through the soil from plant to plant, quite characteristic of soil fungi (e.g., Armillaria spp.). Dispersal can range from a few cm to several meters per year. |
| Wind | Most fungal pathogens that produce spores or conidia externally on host surfaces are easily carried by wind currents (e.g., downy and powdery mildews, rust fungi). Fungal spores behave as inert particles, with terminal velocities ranging from about 0.05 to 2.5 cm per second, with the larger spores falling more rapidly than smaller ones. Turbulence redistributes spores and affects their progressive dilution with increasing distance from its source [43]. With normal wind and turbulence conditions, spores can travel large distances (from several hundred meters to kilometers) |
| Water | Except in the case of streams or rivers that may carry inoculum, water is usually less effective than wind for long-distance dissemination. Rain splash or splatter during heavy rains can locally distribute inoculum on or around the same or neighboring plants. Similarly, rain or irrigation water that moves either through the soil or on the soil surface can disseminate pathogen propagules. |
| Vectors | Some plant pathogens cannot be directly transferred from one plant to another and require a completely unrelated species to act as a vector. Many insects have piercing and sucking mouthparts that penetrate the plant surface and facilitate the transmission and inoculation of host plants (e.g., Dutch elm disease caused by Ophiostoma ulmi or O. novo-ulmi growing within the egg galleries of Scolytus bark beetles, contaminating emerging adults; Laurel wilt caused by Raffaelea lauricola transmitted by species of ambrosia beetles). Thus, vector-transmitted pathogens are usually transferred to the host with great efficiency and play a major role in the infection lifecycle. Some insects cause wounding of plant tissue through which plant pathogens can enter secondarily. Other vectors of pathogens may include nematodes or mammals that may transmit diseases both externally and internally. Most vectors of forest pathogens are usually, but not always, insects, and are sometimes referred to as alternate hosts or as having ‘hitchhiking’ dispersal. Hitchhiking dispersal is favored by typical fungal features such as inconspicuousness, and the production of numerous small propagules [46]. For vector-dependent fungi, if no alternate host exists, the infection cycle is broken. |
| Human | Plant diseases are often dispersed through human-mediated, extra-range dispersal typically through transportation of infected propagative material (e.g., seed, nursery stock, timber, plant products, or soil), or through mishandling or contamination of healthy plants or plant parts during cultivation practices. Organismal spread may be complicated by multiple introductions (genotypes) from multiple sources to multiple locations [7,47]. |
| Common Name | Ash Dieback 1 | Chestnut Blight 2 |
|---|---|---|
| Causal agent | Hymenoscyphus fraxineus (T. Kowalski) Baral, Queloz, Hosoya, comb. nov. (Family Helotiaceae, Order Helotiales, Class Leotiomycetes) | Cryphonectria parasitica (Murr.) Barr (Family Cryphonetriaceae, Order Diaporthales, Class Sordariomycetes) |
| Host species | Major: genus Fraxinus, in particular European ash (F. excelsior), Narrow-leafed ash (F. angustifolia); Black ash (F. nigra); Green ash (F. pennsylvanica); White ash (F. americana); Tianshan ash (F. sogdiana), and Blue ash (F. quadrangulata) Minor 3: Manna ash (F. ornus); Chinese ash (F. chinensis), Manchurian ash (F. mandschurica), Texas ash (F. albicans), Oregon ash (F. latifolia), Spaeth’s ash (F. platypoda), Pumpkin ash (F. profunda), and Velvet ash (F. veluntina). | Major: genus Castanea, in particular American chestnut (C. dentata), European chestnut (C. sativa), Japanese chestnut (C. crenata), and Chinese chestnut (C. mollissima); Minor (incidental): Quercus spp., Acer spp., Carpinus betulus, Castanea pumila |
| Symptoms | Necrotic lesions on leaflets expanding preferentially along leaf veins, and on leaf rachises; leaf wilting; premature leaf abscission; necrosis of buds, perennial necrotic lesions (cankers) on the bark of twigs, branches, and stem; brown discoloration of xylem; dieback of crown; prolific formation of epicormics shoots; basal stem necrosis | Perennial necrotic lesions (cankers) on the bark of above-ground woody parts (stem, branches) of host plants. The plant part distal to the infection point may wilt. |
| Spread mechanism | Sexual spores (ascospores) for short- (local), medium-, and potentially long-distance spread; Over long distances via movement of latently infected plants or plant material | Over short distances mainly via splash dispersed asexual spores (conidia); Over long distances via sexual ascospores or latently infected plants or plant material. |
| Mating system | Random mating, heterothallic, outcrossing | Mixed with outcrossing and self-fertilization occurring at variable frequencies. |
| Native range | Eastern Asia (China, Korea, Japan, Far East Russia) | Eastern Asia (China, Japan, Korea). |
| Invaded range | Europe | North America, Europe. |
| First detection | Early 1990s (Lithuania, Poland) | 1904 (North America), 1938 (Europe). |
| Introduction pathway | Primary pathway for introduction through nursery stock of latently infected plants (e.g., Manchurian ash F. mandshurica) for planting | Most likely infected plants for planting (probably Castanea crenata). |
| Primary dispersal pathway | Wind-dispersed spores (seasonal—between June and September); movement of infected plants or plant material. | Spores (spontaneous dispersal), infected plant material (e.g., plants for planting, wood with bark). |
| Mean dispersal rates | 30 km per year (Norway) 50–60 km per year (Italy) 75 km per year (from east Poland to Switzerland) | In North America, 37 km per year. In Europe (Italy), 29 km per year. |
| Control | Breeding for resistance through traditional screening and selection of disease tolerant genotypes from wild populations. Chemical and biological treatments have been tried on an experimental basis with varied efficacy. | North America: breeding for resistance by backcrossing blight-resistant Chinese chestnut into the American chestnut genome; Transgenic fungal strains and chestnut trees. Europe: biocontrol by a mycovirus in the family Hypoviridae (Cryphonectria hypovirus 1, CHV-1) which reduces both virulence and sporulation of the infected fungal strain (phenomenon called hypovirulence); Hybrids between C. sativa and C. crenata. |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prospero, S.; Cleary, M. Effects of Host Variability on the Spread of Invasive Forest Diseases. Forests 2017, 8, 80. https://doi.org/10.3390/f8030080
Prospero S, Cleary M. Effects of Host Variability on the Spread of Invasive Forest Diseases. Forests. 2017; 8(3):80. https://doi.org/10.3390/f8030080
Chicago/Turabian StyleProspero, Simone, and Michelle Cleary. 2017. "Effects of Host Variability on the Spread of Invasive Forest Diseases" Forests 8, no. 3: 80. https://doi.org/10.3390/f8030080