Diversity and Enzyme Activity of Ectomycorrhizal Fungal Communities Following Nitrogen Fertilization in an Urban-Adjacent Pine Plantation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Sample Collection
2.3. Enzyme Assays
2.4. Identification of ECM Fungi on Root Tips
2.5. Data Analyses
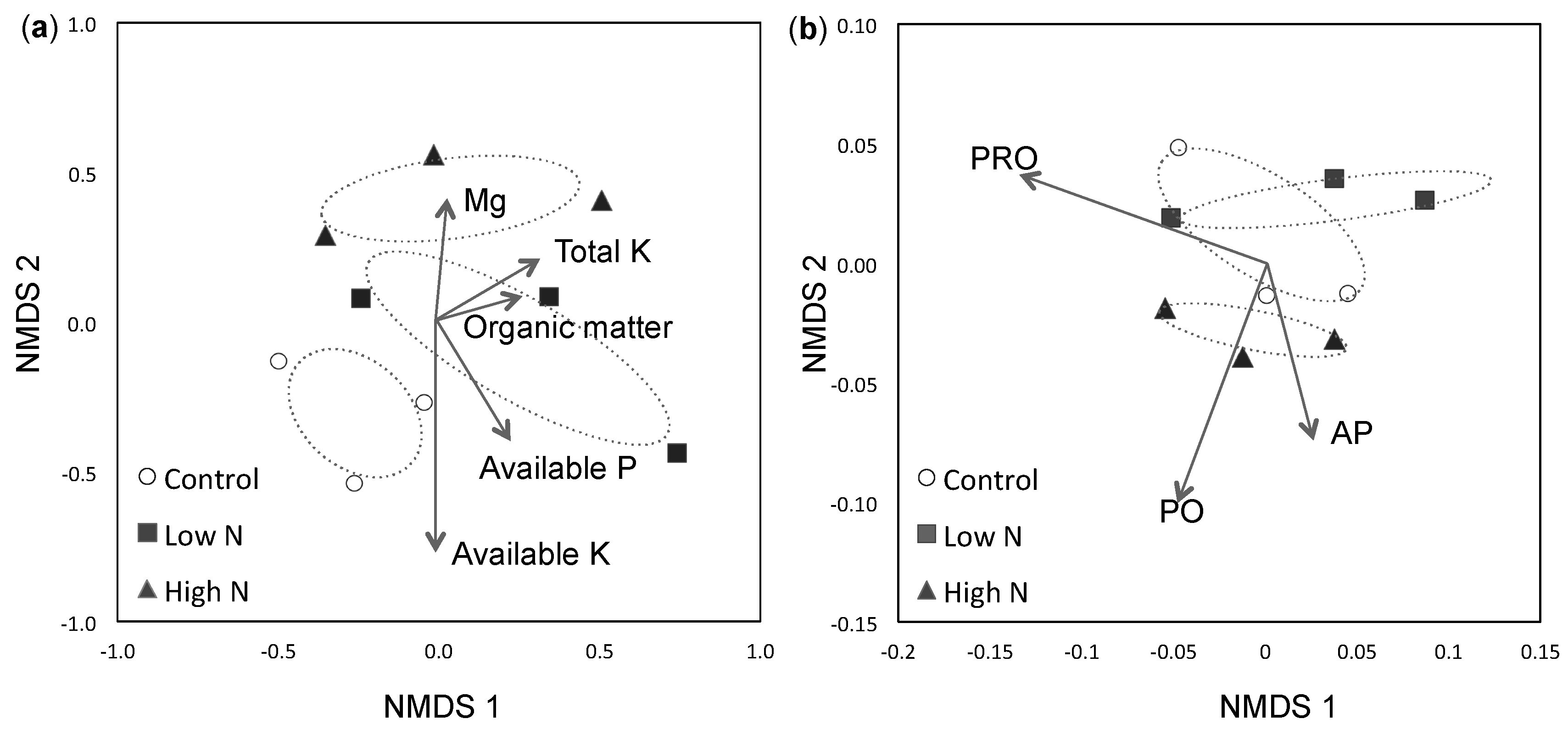
3. Results
3.1. Soil Fertility
3.2. ECM Community Composition and Diversity
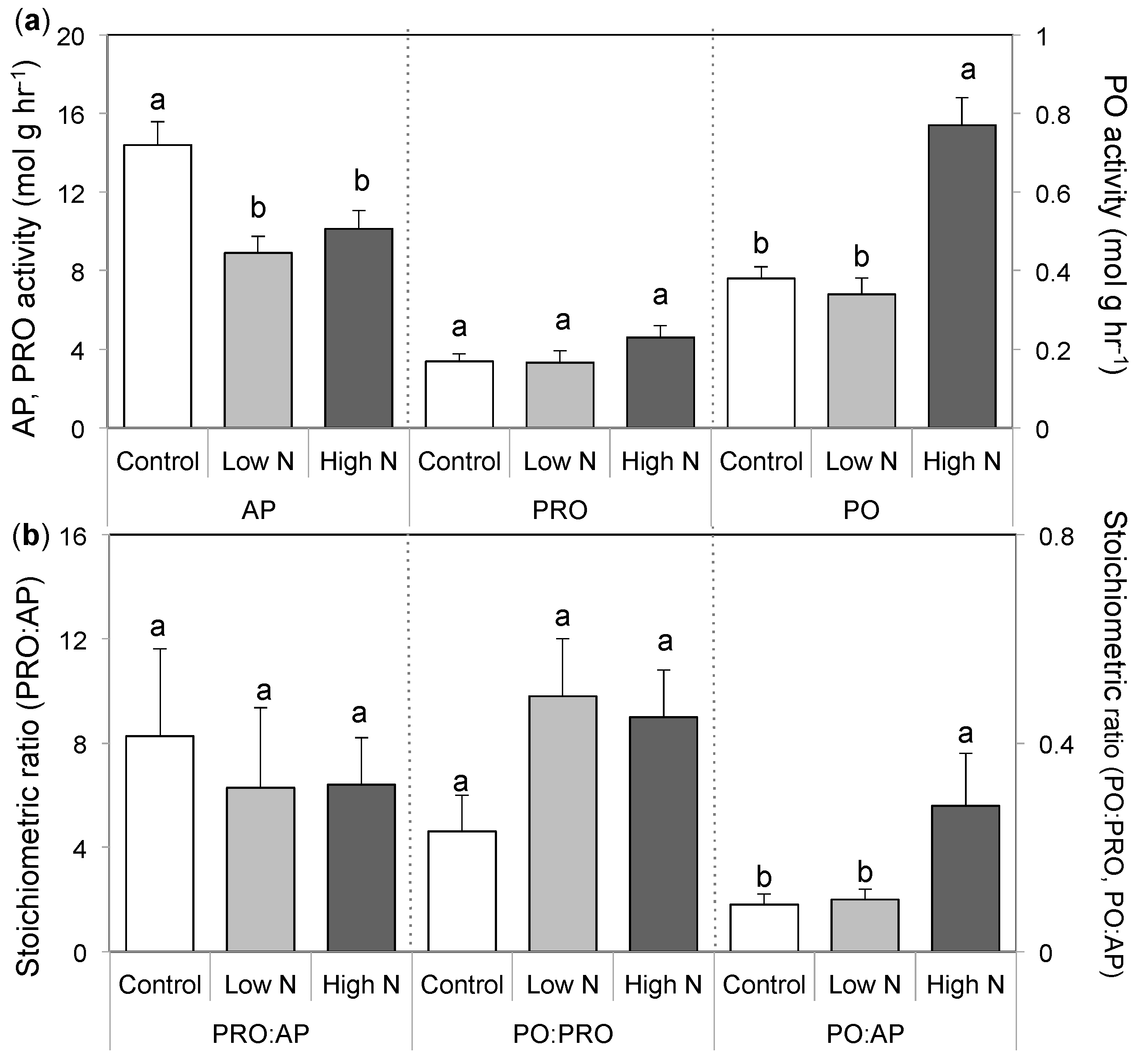
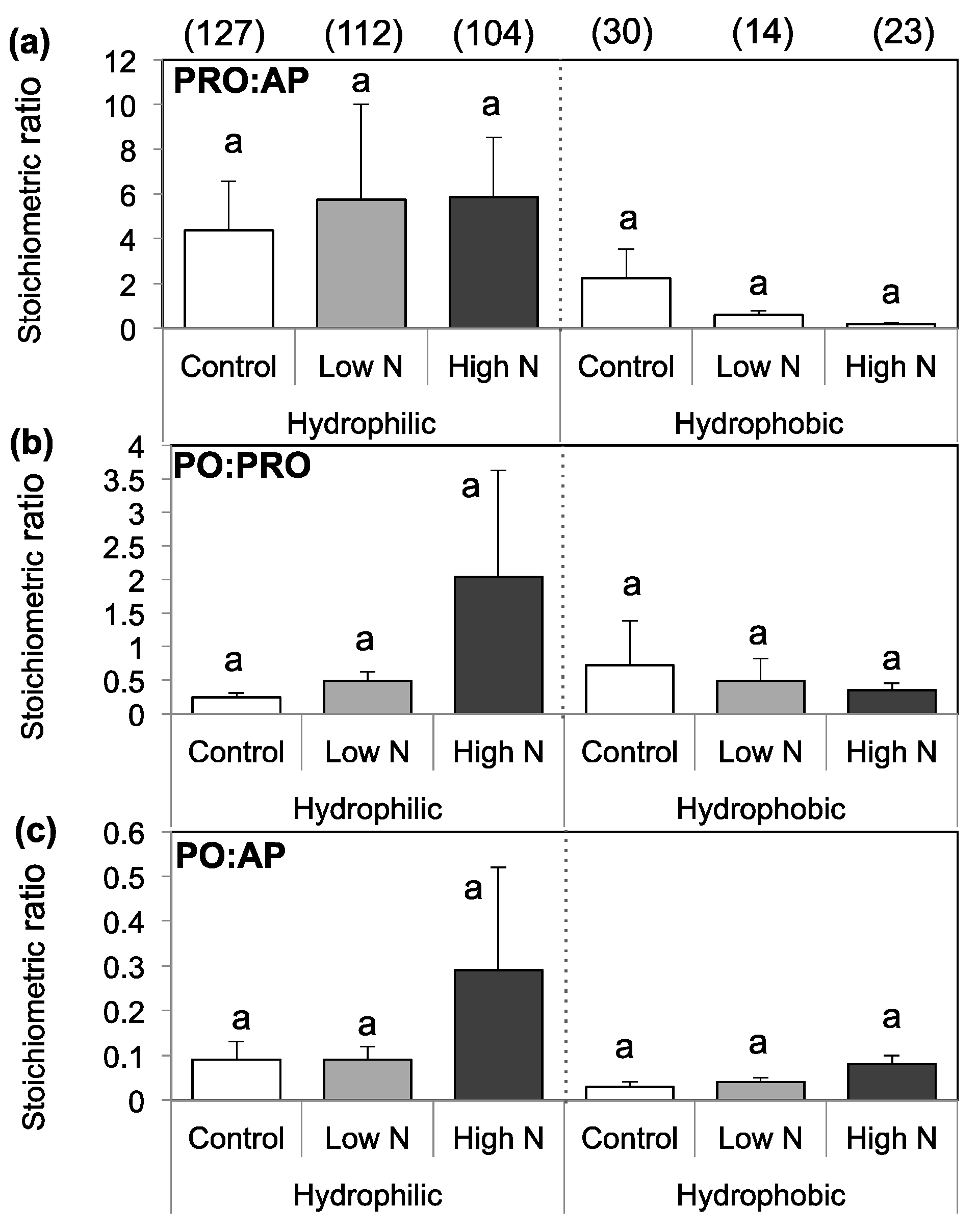
3.3. Extracellular Enzyme Activity
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Galloway, J.N.; Dentener, F.J.; Capone, D.G.; Boyer, R.W.; Howarth, S.P.; Seitzinger, G.P.; Asner, C.C.; Cleveland, P.A.; Green, E.A.; Holland, D.M.; et al. Nitrogen cycles: Past, present, and future. Biogeochemistry 2004, 70, 153–226. [Google Scholar] [CrossRef]
- Carreiro, M.M.; Sinsabaugh, R.L.; Repert, D.A.; Parkhurst, D.F. Microbial enzyme shifts explain litter decay responses to simulated nitrogen deposition. Ecology 2000, 81, 2359–2365. [Google Scholar] [CrossRef]
- Fransson, P.M.; Taylor, A.F.S.; Finlay, R.D. Effects of continuous optimal fertilization on belowground ectomycorrhizal community structure in a Norway spruce forest. Tree Physiol. 2000, 20, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Dupra, C.; Stevens, C.J.; Ranke, T.; Bleeker, A.; Peppler-Lisbach, C.; Gowing, D.J.G.; Dise, N.B.; Dorland, E.; Bobbink, R.; Diekmann, M. Changes in species richness and composition in European acidic grasslands over the past 70 years: the contribution of cumulative atmospheric nitrogen deposition. Glob. Chang. Biol. 2000, 16, 3443–3457. [Google Scholar] [CrossRef]
- Lilleskov, E.A.; Hobbie, E.A.; Horton, T.R. Conservation of ectomycorrhizal fungi: Exploring the linkage between functional and taxonomic responses to anthropogenic N deposition. Fungal Ecol. 2011, 4, 174–183. [Google Scholar] [CrossRef]
- Lilleskov, E.A.; Fahey, T.J.; Horton, T.R.; Lovett, G. Belowground ectomycorrhizal fungal community change over a nitrogen deposition gradient in Alaska. Ecology 2002, 83, 104–115. [Google Scholar] [CrossRef]
- Jones, M.D.; Phillips, L.A.; Treu, R.; Ward, V.; Berch, S.M. Functional responses of ectomycorrhizal fungal communities to long-term fertilization of lodgepole pine (Pinus contorta Dougl. ex Loud. var. latifolia Engelm.) stands in central British Columbia. Appl. Soil. Ecol. 2012, 60, 29–49. [Google Scholar]
- Avis, P.G.; McLaughlin, D.J.; Dentinger, B.C.; Reich, P.B. Long-term increase in nitrogen supply alters above- and below- ground ectomycorrhizal communities and increases the dominance of Russula spp. in a temperate oak savanna. New Phytol. 2003, 160, 239–253. [Google Scholar] [CrossRef]
- Högberg, M.; Blaško, R.; Bach, L.H.; Hasselquist, N.J.; Egnell, G.; Näsholm, T.; Högberg, P. The return of an experimentally N-saturated boreal forest to an N-limited state: observations on the soil microbial community structure, biotic N retention capacity and gross N mineralization. Plan. Soil. 2014, 381, 45–60. [Google Scholar] [CrossRef]
- Parrent, J.L.; Vilgalys, R. Biomass and compositional responses of ectomycorrhizal fungal hyphae to elevated CO2 and nitrogen fertilization. New Phytol. 2007, 176, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Cox, F.; Barsoum, N.; Lilleskov, E.A.; Bidartondo, M.I. Nitrogen availability is a primary determinant of conifer mycorrhizas across complex environmental gradients. Ecol. Lett. 2010, 13, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.W.; Frey, S.D.; Sadowsky, J.J.; Van Diepen, L.T.A.; Thomas, W.K.; Pringle, A. Chronic nitrogen additions fundamentally restructure the soil fungal community in a temperate forest. Fungal Ecol. 2016, 23, 48–57. [Google Scholar] [CrossRef]
- Courty, P.E.; Franc, A.; Garbaye, J. Temporal and functional pattern of secreted enzyme activities in an ectomycorrhizal community. Soil Biol. Biochem. 2010, 42, 2022–2025. [Google Scholar] [CrossRef]
- Courty, P.E.; Labbe, J.; Kohler, A.; Marcais, B.; Bastien, C.; Churin, J.L.; Garbaye, J.; Le Tacon, F. Effect of poplar genotypes on mycorrhizal infection and secreted enzyme activities in mycorrhizal and non-mycorrhizal roots. J. Exp. Bot. 2011, 62, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.D.; Twieg, B.D.; Ward, V.; Barker, J.; Durall, D.M.; Simard, S.W. Functional complementarity of Douglas-fir ectomycorrhizas for extracellular enzyme activity after wildfire or clearcut logging. Funct. Ecol. 2010, 24, 1139–1151. [Google Scholar] [CrossRef]
- Herzo, C.; Peter, M.; Pritsch, K.; Günthardt-Goerg, M.S.; Egli, S. Drought and air warming affects abundance and exoenzyme profiles of Cenococcum geophilum associated with Quercus robur, Q. petraea and Q. pubescens. Plant Biol. 2013, 15, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Courty, P.E.; Bréda, N.; Garbaye, J. Relation between oak tree phenology and the secretion of organic matter degrading enzymes by Lactarius quietus ectomycorrhizas before and during bud break. Soil Biol. Biochem. 2007, 39, 1655–1663. [Google Scholar] [CrossRef]
- Buée, M.; Courty, P.E.; Mignot, D.; Garbaye, J. Soil niche effect on species diversity and catabolic activities in an ectomycorrhizal fungal community. Soil Biol. Biochem. 2007, 39, 1947–1955. [Google Scholar] [CrossRef]
- Courty, P.E.; Pritsch, K.; Schloter, M.; Hartmann, A.; Garbaye, J. Activity profiling of ectomycorrhiza communities in two forest soils using multiple enzymatic tests. New Phytol. 2005, 167, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Pritsch, K.; Garbaye, J. Enzyme secretion by ECM fungi and exploitation of mineral nutrients from soil organic matter. Ann. For. Sci. 2011, 68, 25–32. [Google Scholar] [CrossRef]
- Luis, P.; Kellner, H.; Zimdars, B.; Langer, U.; Martin, F.; Buscot, F. Patchiness and spatial distribution of laccase genes of ectomycorrhizal, saprotrophic, and unknown basidiomycetes in the upper horizons of a mixed forest cambisol. Microb. Ecol. 2005, 50, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.M.; Bruns, T.D.; Smith, D.P.; Branco, S.; Glassman, S.I.; Erlandson, S.; Vilgalys, R.; Peay, K.G. Independent roles of ectomycorrhizal and saprotrophic communities in soil organic matter decomposition. Soil Biol. Biochem. 2013, 57, 282–291. [Google Scholar] [CrossRef]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, E.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saiya-Cork, K.R.; Sinsabaugh, R.L.; Zak, D.R. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 2002, 34, 1309–1315. [Google Scholar] [CrossRef]
- Frey, S.D.; Knorr, M.; Parrent, J.L.; Simpson, R.T. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. For. Ecol. Manag. 2004, 196, 159–171. [Google Scholar] [CrossRef]
- Blackwood, C.B.; Waldrop, M.P.; Zak, D.R.; Sinsabaugh, R.L. Molecular analysis of fungal communities and laccase genes in decomposing litter reveals differences among forest types but no impact of nitrogen deposition. Environ. Microbiol. 2007, 5, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Lucas, R.W.; Casper, B.B. Ectomycorrhizal community and extracellular enzyme activity following simulated atmospheric N deposition. Soil Biol. Biochem. 2008, 40, 1662–1669. [Google Scholar] [CrossRef]
- Eaton, G.K.; Ayres, M.P. Plasticity and constraint in growth and protein mineralization of ectomycorrhizal fungi under simulated nitrogen deposition. Mycologia 2002, 94, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Agerer, R. Fungal relationships and structural identity of their ectomycorrhizae. Mycol. Prog. 2006, 5, 67–107. [Google Scholar] [CrossRef]
- Avis, P.G.; Mueller, G.M.; Lussenhop, J. Ectomycorrhizal fungal communities in two North American oak forests respond to nitrogen addition. New Phytol. 2008, 179, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Lei, P.; Xiang, W.; Yan, W.; Liu, S. Soil microbial biomass carbon and nitrogen in pure and mixed stands of Pinus massoniana and Cinnamomum camphora differing in stand age. For. Ecol. Manag. 2014, 328, 150–158. [Google Scholar] [CrossRef]
- Du, C.Y.; Zeng, G.M.; Zhang, G.; Tang, L.; Li, X.D.; Huang, D.L.; Huang, L.; Jiang, Y.M. Input-output budgets for inorganic nitrogen under acid rain in a subtropical evergreen mixed forest in central-south China. Water Air Soil Pollut. 2008, 190, 171–181. [Google Scholar] [CrossRef]
- Fang, Y.; Gundersen, P.; Vogt, R.D.; Koba, K.; Chen, F.; Chen, X.Y.; Yoh, M. Atmospheric deposition and leaching of nitrogen in Chinese forest ecosystems. J. For. Res. 2011, 16, 341–350. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, X.; Wang, S.; Lu, X.; Ouyang, X. A review of spatial variation of inorganic nitrogen (N) wet deposition in China. PLoS ONE 2016, 11, e0146051. [Google Scholar] [CrossRef] [PubMed]
- Chapela, I.H.; Osher, L.J.; Horton, T.R.; Henn, M.R. Ectomycorrhizal fungi introduced with exotic pine plantations induce soil carbon depletion. Soil. Biol. Biochem. 2001, 32, 1733–1740. [Google Scholar] [CrossRef]
- Bastias, B.A.; Anderson, I.C.; Xu, Z.; Cairney, J.W.G. RNA- and DNA-based profiling of soil fungal communities in a native Australian eucalypt forest and adjacent Pinus elliotti plantation. Soil Biol. Biochem. 2007, 39, 3108–3114. [Google Scholar] [CrossRef]
- Colpaert, J.; Van Laere, A. A comparison of the extracellular enzyme activities of two ectomycorrhizal and a leaf-saprotrophic basidiomycete colonizing beech leaf litter. New Phytol. 1996, 134, 133–141. [Google Scholar] [CrossRef]
- Blankinship, J.C.; Becerra, C.A.; Schaeffer, S.M.; Schinmer, J.P. Separating cellular metabolism from exoenzyme activity in soil organic matter decomposition. Soil. Biol. Biochem. 2014, 71, 68–75. [Google Scholar] [CrossRef]
- Plassard, C.; Louche, J.; Ali, M.A.; Duchemn, M.; Legname, E.; Cloutier-Hurteau, B. Diversity in phosphorus mobilization and uptake in ectomycorrhizal fungi. Ann. For. Sci. 2011, 68, 33–43. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: Above- and below-ground views. Can. J. Bot. 1996, 74, 1572–1583. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myer, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Pengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package Version 2.4-4. Available online: https://github.com/vegandevs/vegan (accessed on 24 August 2017).
- Colwell, R.K. EstimateS: Statistical Estimation of Species Richness and Shared Species from Samples. Version 9. Available online: http://purl.oclc.org/estimates (assessed on 12 August 2016).
- Walker, J.K.M.; Cohen, H.; Higgins, L.M.; Kennedy, P.G. Testing the link between community structure and function for ectomycorrhizal fungi involved in a global tripartite symbiosis. New Phytol. 2014, 202, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Hay, T.N.; Phillips, L.A.; Nicholson, B.A.; Jones, M.D. Ectomycorrhizal community structure and function in interior spruce forests of British Columbia under long-term fertilization. For. Ecol. Manag. 2015, 350, 87–95. [Google Scholar] [CrossRef]
- Berch, S.M.; Brockley, R.P.; Battigelli, J.P.; Haerman, S.; Holl, B. Impacts of repeated fertilization on components of the soil biota under a young lodgepole pine stand in the interior of British Columbia. Can. J. For. Res. 2006, 36, 1415–1426. [Google Scholar] [CrossRef]
- Peter, M.; Ayer, F.; Egli, S. Nitrogen addition in a Norway spruce stand altered macromycete sporocarp production and below-ground ectomycorrhizal species composition. New Phytol. 2001, 149, 311–325. [Google Scholar] [CrossRef]
- Kranabetter, J.M.; Durall, D.; MacKenzie, W. Diversity and species distribution of ectomycorrhizal fungi along productivity gradients of a southern boreal forest. Mycorrhiza 2009, 19, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Hasselquist, N.; Högberg, P. Dosage and duration effects of nitrogen additions on ectomycorrhizal sporocarp production and functioning: An example from two N-limited boreal forests. Ecol. Evol. 2014, 4, 3015–3026. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.; Zimmerman, S.D. The role of mycorrhizal associations in plant potassium nutrition. Front. Plant Sci. 2014, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, B.; Bélanger, G.; Nolin, M.C.; Simard, R. Relationships between soil cations and plant characteristics based on spatial variability in a forage field. Can. J. Plant Sci. 2003, 83, 343–350. [Google Scholar] [CrossRef]
- Wang, L.; Katzensteiner, K.; Schume, H.; van Loo, M.; Godbold, D.L. Potassium fertilization affects the distribution of fine roots but does not change ectomycorrhizal community structure. Ann. For. Sci. 2016, 73, 691–702. [Google Scholar] [CrossRef]
- Finlay, R. Ecological aspects of mycorrhizal symbiosis: With special emphasis on the functional diversity of interactions involving the extraradical mycelium. J. Exp. Bot. 2008, 59, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Olander, L.P.; Vitousek, P.M. Regulation of soil phosphatase and chitinase activity by N and P availability. Biogeochem. 2000, 49, 175–190. [Google Scholar] [CrossRef]
- Phillips, L.A.; Ward, V.; Jones, M.D. Ectomycorrhizal fungi contribute to soil organic matter cycling in sub-boreal forests. ISME J. 2014, 8, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Courty, P.E.; Franc, A.; Pierrat, J.C.; Garbaye, J. Temporal changes in the ectomycorrhizal community in two soil horizons of a temperate oak forest. Appl. Environ. Microbiol. 2008, 74, 5792–5801. [Google Scholar] [CrossRef] [PubMed]
- Rineau, F.; Courty, P. Secreted enzymatic activities of ectomycorrhizal fungi as a case study of functional diversity and functional redundancy. Ann. For. Sci. 2011, 68, 69–80. [Google Scholar] [CrossRef]
- Tedersoo, L.; Kõljalg, U.; Hallenberg, N.; Larsson, K.-H. Fine scale distribution of ectomycorrhizal fungi and roots across substrate layers including coarse woody debris in a mixed forest. New Phytol. 2003, 159, 153–165. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Toots, M.; Diédhiou, A.G.; Henkel, T.W.; Kjøller, R.; Morris, M.H.; Nara, K.; Nouhra, E.; Peay, K.G.; et al. Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol. Ecol. 2012, 21, 4160–4170. [Google Scholar] [CrossRef] [PubMed]
- Courty, P.; Munoz, F.; Selosse, M.-A.; Duchemin, M.; Criquet, S.; Ziarelli, F.; Buée, M.; Plassard, C.; Taudiere, A.; Garbaye, J.; et al. Into the functional ecology of ectomycorrhizal communities: Environmental filtering of enzymatic activities. J. Ecol. 2014, 104, 1585–1598. [Google Scholar]
- Cullings, K.; Ishkhanova, G.; Henson, J. Defoliation effects on enzyme activities of the ectomycorrhizal fungus Suillus granulatus in a Pinus contorta (lodgepole pine) stand in Yellowstone National Park. Oecologia 2008, 158, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, P.G.; Higgins, L.M.; Rogers, R.H.; Weber, M.G. Colonization-competition tradeoffs as a mechanism driving successional dynamics in ectomycorrhizal fungal communities. PLoS ONE 2011, 9, e25126. [Google Scholar] [CrossRef] [PubMed]
- Pickles, B.J.; Genney, D.R.; Anderson, I.C.; Alexander, I.J. Spatial analysis of ectomycorrhizal fungi reveals that root tip communities are structured by competitive interactions. Mol. Ecol. 2012, 21, 5110–5123. [Google Scholar] [CrossRef] [PubMed]
- Clemmensen, K.E.; Finlay, R.D.; Dahlberg, A.; Stenlid, J.; Wardle, D.A.; Lindahl, B.D. Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol. 2015, 205, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Langley, J.A.; Hungate, B.A. Mycorrhizal controls on belowground litter quality. Ecology 2003, 84, 2302–2312. [Google Scholar] [CrossRef]
- Fernandez, C.W.; Koide, R.T. Initial melanin and nitrogen concentrations control the decomposition of ectomycorrhizal fungal litter. Soil Biol. Biochem. 2014, 77, 150–157. [Google Scholar] [CrossRef]
- Schreiner, K.M.; Blair, N.E.; Levinson, W.; Egerton-Warburton, L.M. Contribution of Fungal Macromolecules to Soil Carbon Sequestration. In Progress in Soil Science; Hartemink, A.E., McSweeney, K., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 155–161. [Google Scholar]


| N Fertilization Level | |||
|---|---|---|---|
| Soil nutrient | Control (n = 36) | Low (n = 36) | High (n = 36) |
| Organic matter (g kg−1) | 26 (2) ab | 21 (2) b | 28 (2) a |
| Total N (g kg−1) | 1.34 (0.1) a | 1.26 (0.1) a | 1.48 (0.1) a |
| Organic C:N | 12 (1) a | 10 (1) a | 12 (2) a |
| Available N (µg g−1 soil) | 26 (1.9) a | 25 (1.3) a | 27 (1.7) a |
| Total P (µg g−1 soil) | 110 (4) b | 122 (5) a | 115 (4) b |
| Available P (µg g−1 soil) | 3.5 (0.1) c | 4.6 (0.2) a | 3.0 (0.1) b |
| N:P | 12 (1) a | 11 (1) a | 13 (1) a |
| K (µg g−1 soil) | 69 (6.8) a | 64 (2.7) a | 41 (1.8) b |
| Ca (µg g−1 soil) | 193 (18) a | 257 (15) a | 216 (16) a |
| Mg (µg g−1 soil) | 896 (22) b | 1051 (20) a | 977 (23) a |
| Fe (µg g−1 soil) | 15,986 (266) b | 16,273 (140) b | 17,034 (169) a |
| Mn (µg g−1 soil) | 80 (7) b | 101 (6) a | 114 (6) a |
| CEC (cation exchange capacity) | 20 (1.6) a | 18 (1.2) a | 19 (1.8) a |
| OTU | Accession Number a | Closest Blast Match in Genbank b | Query/Aligned length (bp) (similarity %) c | Closest UNITE Species Match | No. Of root tips/Frequency d | ||
|---|---|---|---|---|---|---|---|
| Control | Low N | High N | |||||
| Tylospora sp. | KP866117 | HM189733 Corticiaceae sp. BB-2010 | 632/637 (99) | SH192265.07FU | 29/2 | 13/1 | 21/3 |
| Atheliaceae sp. | KP866118 | AB839405 Uncultured ECM fungus | 475/512 (93) | SH193510.07FU | 0 | 1/1 | 0 |
| Cenococcum sp.1 | KP866119 | JQ347051 Uncultured Cenococcum | 567/571 (99) | SH214459.07FU | 7/1 | 28/2 | 0 |
| Cenococcum sp.2 | KP866120 | JX456699 Uncultured fungus | 486/497 (98) | SH214466.07FU | 14/3 | 0 | 0 |
| Helotiales sp.1 | KP866121 | KF007259 Uncultured ECM fungus | 608/618 (98) | SH214286.07FU | 18/4 | 24/5 | 40/6 |
| Helotiales sp.2 | KP866122 | AB571492 Uncultured ECM fungus | 637/639 (99) | SH023418.07FU | 16/2 | 7/1 | 15/2 |
| Helotiales sp.3 | KP866123 | AB769894 Uncultured Helotiales | 550/551 (99) | SH201717.07FU | 14/2 | 7/1 | 27/2 |
| Helotiales sp.4 | KP866124 | HM208727 Fungal sp. Phylum141 | 560/562 (99) | SH196495.07FU | 1/1 | 0 | 0 |
| Helotiales sp.5 | - | FN397286 Uncultured fungus | 499/536 (93) | SH211375.07FU | 0 | 0 | 1/1 |
| Lactifluus parvigerardii | KP866125 | JF975641 Lactifluus parvigerardii XHW-2011 | 571/574 (99) | SH012454.07FU | 7/1 | 7/1 | 7/1 |
| Phialocephala fortinii | KP866127 | KF313098 Phialocephala sp. YJM2013 | 562/564 (99) | SH204999.07FU | 1/1 | 0 | 2/1 |
| Russula sp.1 | KP866128 | JX457011 Uncultured fungus | 683/696 (98) | SH017121.07FU | 0 | 7/1 | 7/1 |
| Russula virescens | KP866129 | KM373243 Russula crustosa | 669/716 (93) | SH179774.07FU | 0 | 1/1 | 0 |
| Russula sp.2 | KP866130 | AB597671 Fungal sp. JK-02M | 580/582 (99) | SH017122.07FU | 0 | 7/1 | 0 |
| Scleroderma yunnanense | KP866131 | JQ639046 Scleroderma yunnanense XEX-2012 | 584/588 (100) | SH189277.07FU | 1/1 | 0 | 1/1 |
| Scleroderma citrinum | KP866132 | AB769913 Uncultured Scleroderma citrinum | 561/569 (99) | SH008294.07FU | 0 | 0 | 1/1 |
| Sebacinaceae sp. | KP866133 | KF000673 Uncultured Sebacina clone | 628/657 (96) | SH214656.07FU | 0 | 0 | 1/1 |
| Thelephora terrestris | KP866134 | KJ938034 Uncultured fungus | 677/706 (96) | SH184510.07FU | 7/1 | 14/2 | 0 |
| Tomentella sp.1 | KP866135 | AB769927 Uncultured Thelephoraceae | 663/665 (99) | SH177859.07FU | 14/2 | 7/1 | 0 |
| Tomentella sp.2 | KP866136 | JX456648 Uncultured fungus | 705/706 (99) | SH189353.07FU | 37/5 | 2/1 | 7/1 |
| Ascomycota sp.1 | - | EF619719 Uncultured Orbiliaceae | 525/612 (86) | SH015725.07FU | 2/2 | 0 | 1/1 |
| Ascomycota sp.2 | - | KP323399 Uncultured fungus | 208/226 (92) | SH469383.07FU | 2/2 | 0 | 0 |
| Ascomycota sp.3 | - | KP689247 Ascomycota sp. | 618/625 (99) | SH181934.07FU | 0 | 1/1 | 3/3 |
| Meliniomyces sp. | - | FJ440931 Uncultured ectomycorrhiza | 553/574 (96) | SH181081.07FU | 0 | 2/1 | 0 |
| Sites | Rarified Species Richness | Estimators of Expected Total Species Richness | Diversity Indices | |||
|---|---|---|---|---|---|---|
| Mao Tau | Mao Tau (50 runs mean) | Chao 2 | Jackknife 2 | Shannon’s H’ | Simpson’s 1/D | |
| Control | 5.67 (1.67) | 6.41 (1.58) | 8.54 (1.20) | 8.78 (2.19) | 2.79 (0.96) | 5.89 (1.31) |
| Low N | 4.33 (1.45) | 4.5 (1.08) | 5.1 (1.24) | 6.28 (1.53) | 1.35 (0.25) | 4.06 (0.98) |
| High N | 4.33 (0.67) | 5.1 (0.37) | 5.99 (0.32) | 6.80 (0.12) | 2.84 (1.40) | 4.59 (0.43) |
| p a | 0.729 | 0.508 | 0.119 | 0.52 | 0.519 | 0.444 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ning, C.; Mueller, G.M.; Egerton-Warburton, L.M.; Wilson, A.W.; Yan, W.; Xiang, W. Diversity and Enzyme Activity of Ectomycorrhizal Fungal Communities Following Nitrogen Fertilization in an Urban-Adjacent Pine Plantation. Forests 2018, 9, 99. https://doi.org/10.3390/f9030099
Ning C, Mueller GM, Egerton-Warburton LM, Wilson AW, Yan W, Xiang W. Diversity and Enzyme Activity of Ectomycorrhizal Fungal Communities Following Nitrogen Fertilization in an Urban-Adjacent Pine Plantation. Forests. 2018; 9(3):99. https://doi.org/10.3390/f9030099
Chicago/Turabian StyleNing, Chen, Gregory M. Mueller, Louise M. Egerton-Warburton, Andrew W. Wilson, Wende Yan, and Wenhua Xiang. 2018. "Diversity and Enzyme Activity of Ectomycorrhizal Fungal Communities Following Nitrogen Fertilization in an Urban-Adjacent Pine Plantation" Forests 9, no. 3: 99. https://doi.org/10.3390/f9030099
APA StyleNing, C., Mueller, G. M., Egerton-Warburton, L. M., Wilson, A. W., Yan, W., & Xiang, W. (2018). Diversity and Enzyme Activity of Ectomycorrhizal Fungal Communities Following Nitrogen Fertilization in an Urban-Adjacent Pine Plantation. Forests, 9(3), 99. https://doi.org/10.3390/f9030099





