Increased Presence of Circulating Cell-Free, Fragmented, Host DNA in Pigs Infected with Virulent African Swine Fever Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples from ASFV-Infected Pigs
2.1.1. Experiment A
2.1.2. Experiment B
2.2. Samples from CSFV-Infected Pigs
2.3. Laboratory Analyses
2.3.1. Viral Genome Detection
2.3.2. Host Genomic and Mitochondrial DNA Detection
2.3.3. Genome Copy Number Determination
2.3.4. Lactate Dehydrogenase (LDH) Assays
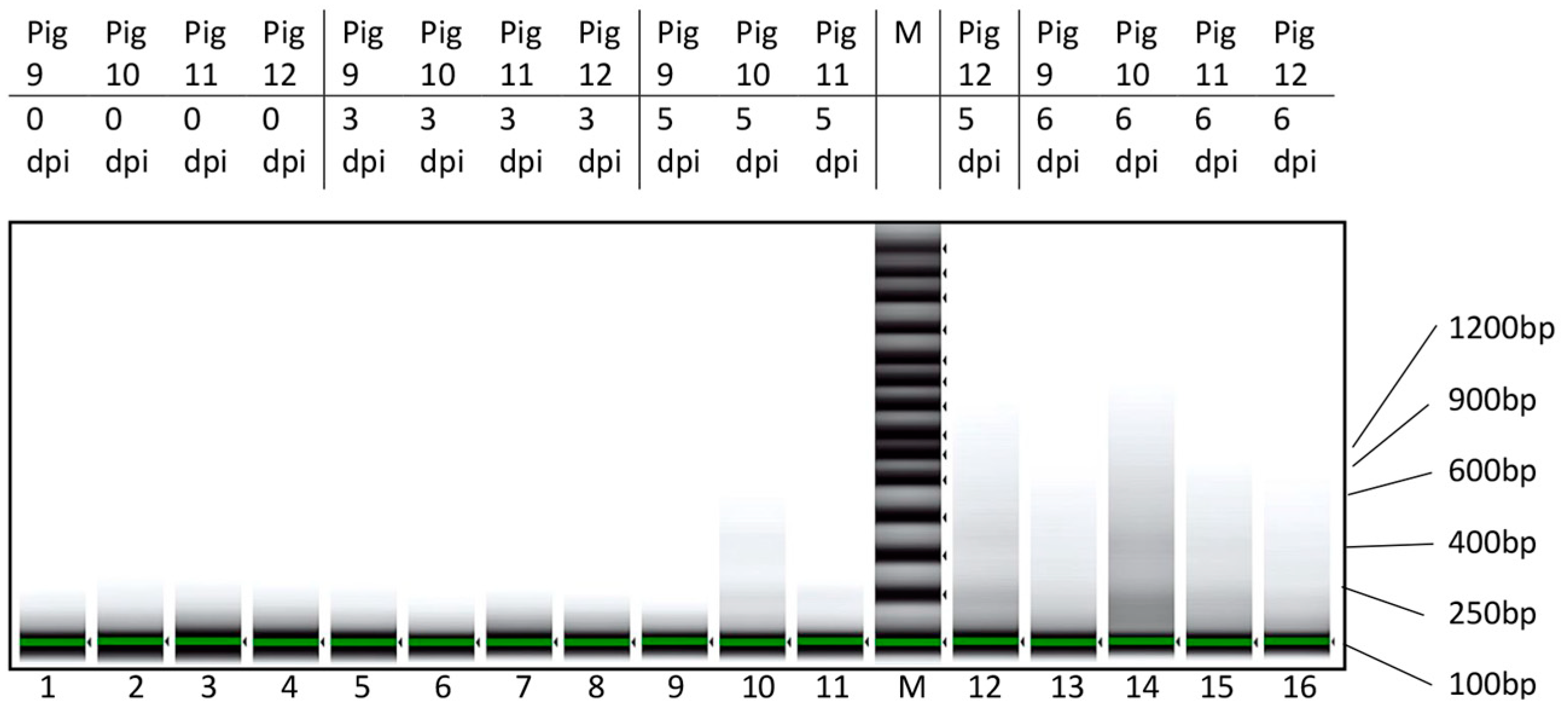
2.3.5. DNA Fragment Size Determination
2.3.6. Data Presentation
3. Results
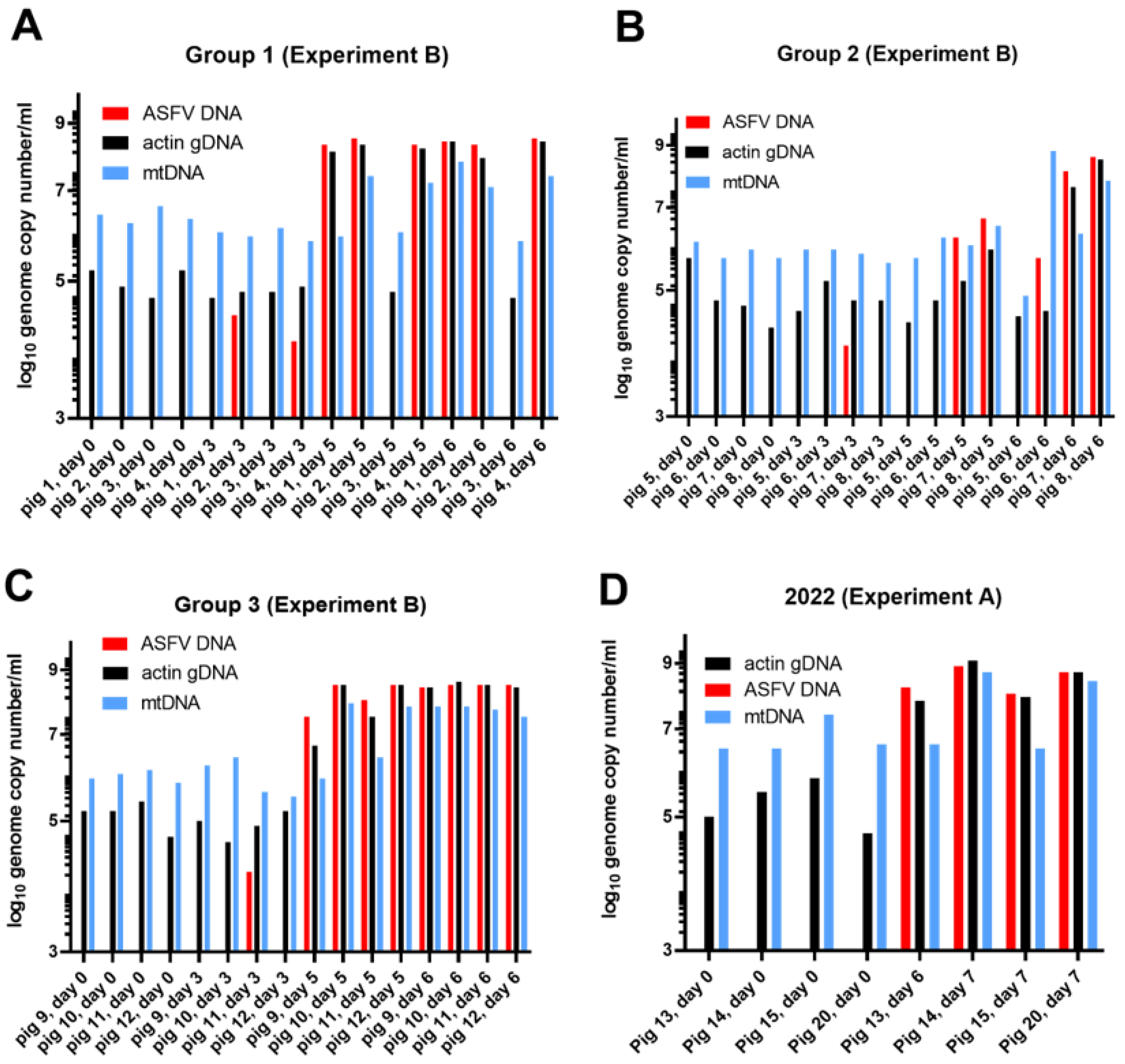
3.1. Characterization of ASFV Infections in Pigs
3.1.1. Experiment A
Assessment of Body Temperature, Clinical Signs, and Virus Replication
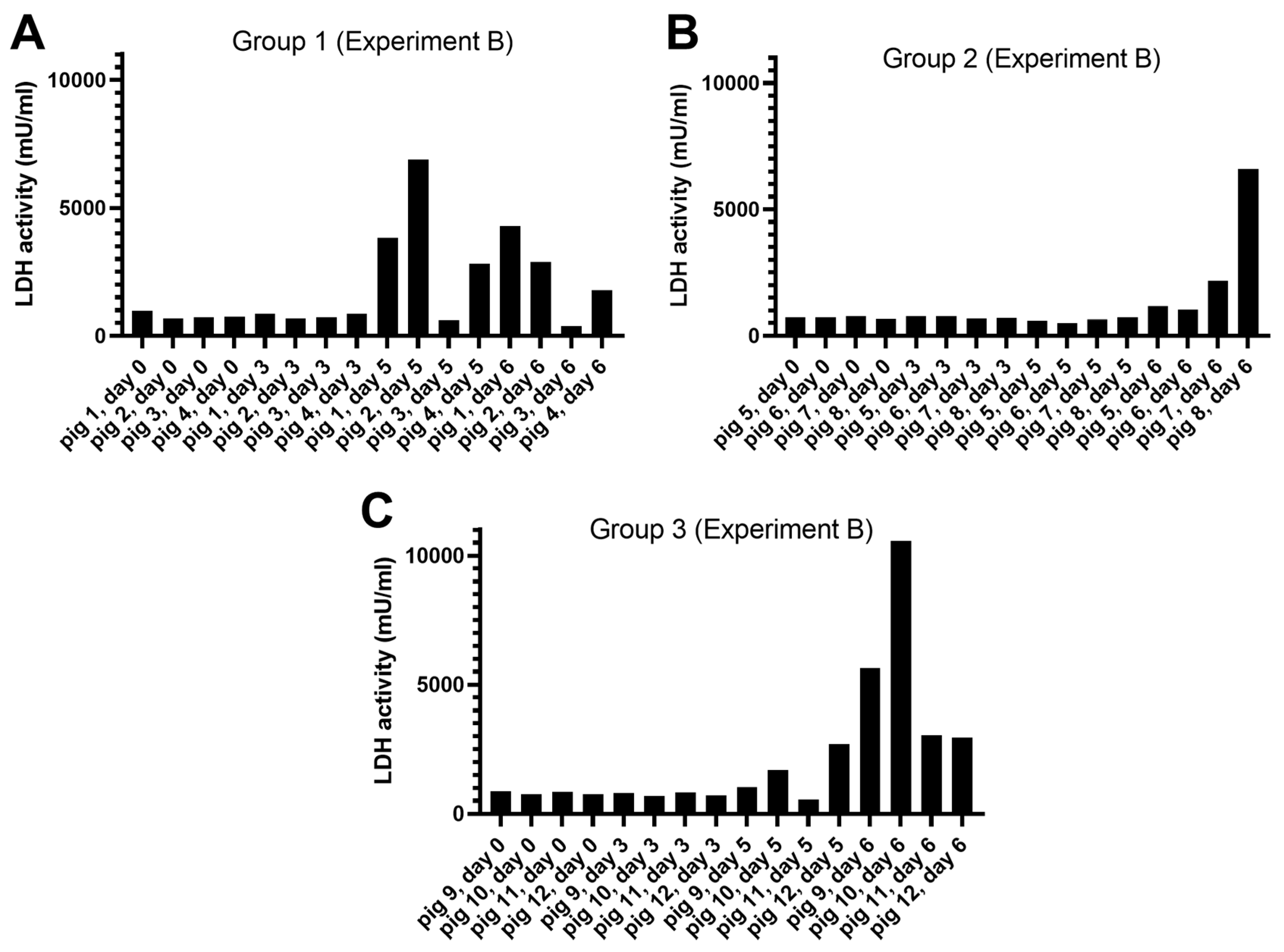
3.1.2. Experiment B
Release of LDH Activity
3.2. Characterization of cfDNA in ASFV-Infected Pig Sera
3.3. CSFV Infection Studies
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dixon, L.; Sun, H.; Roberts, H. African swine fever. Antivir. Res. 2019, 165, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Sauter-Louis, C.; Conraths, F.J.; Probst, C.; Blohm, U.; Schulz, K.; Sehl, J.; Fischer, M.; Forth, J.H.; Zani, L.; Depner, K.; et al. African Swine Fever in Wild Boar in Europe—A Review. Viruses 2021, 13, 1717. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Cackett, G.; Matelska, D.; Sýkora, M.; Portugal, R.; Malecki, M.; Bähler, J.; Dixon, L.; Werner, F. The African Swine Fever Virus Transcriptome. J. Virol. 2020, 94, e00119-20. [Google Scholar] [CrossRef] [PubMed]
- Bastos, A.D.S.; Penrith, M.-L.; Crucière, C.; Edrich, J.L.; Hutchings, G.; Roger, F.; Couacy-Hymann, E.R.; Thomson, G.R. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch. Virol. 2003, 48, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Stahl, K.; Jori, F.; Vial, L.; Pfeiffer, D.U. African swine fever epidemiology and control. Annu. Rev. Anim. Biosci. 2020, 8, 221–246. [Google Scholar] [CrossRef]
- Ruiz-Saenz, J.; Diaz, A.; Bonilla-Aldana, D.K.; Rodríguez-Morales, A.J.; Martinez-Gutierrez, M.; Aguilar, P.V. African swine fever virus: A re-emerging threat to the swine industry and food security in the Americas. Front. Microbiol. 2022, 13, 1011891. [Google Scholar] [CrossRef]
- EFSA; Ståhl, K.; Boklund, A.; Podgórski, T.; Vergne, T.; Abrahantes, J.C.; Papanikolaou, A.; Zancanaro, G.; Mur, L. Epidemiological analysis of Afrcian swine fever in the European Union during 2022. EFSA J. 2023, 21, e08016. [Google Scholar] [CrossRef]
- Mighell, E.; Ward, M.P. African Swine Fever spread across Asia, 2018–2019. Transbound Emerg Dis. 2021, 68, 2722–2732. [Google Scholar] [CrossRef]
- Blome, S.; Franzke, K.; Beer, M. African swine fever—A review of current knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef]
- Salguero, F.J. Comparative Pathology and Pathogenesis of African Swine Fever Infection in Swine. Front. Vet. Sci. 2020, 7, 282. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Soler, A.; Nieto, R.; Cano, C.; Pelayo, V.; Sánchez, M.A.; Pridotkas, G.; Fernandez-Pinero, J.; Briones, V.; Arias, M. Experimental Infection of Domestic Pigs with African Swine Fever Virus Lithuania 2014 Genotype II Field Isolate. Transbound Emerg. Dis. 2017, 64, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Nurmoja, I.; Mõtus, K.; Kristian, M.; Niine, T.; Schulz, K.; Depner, K.; Viltrop, A. Epidemiological analysis of the 2015–2017 African swine fever outbreaks in Estonia. Prev. Vet. Med. 2020, 181, 1044556. [Google Scholar] [CrossRef] [PubMed]
- Olesen, A.S.; Lohse, L.; Boklund, A.; Halasa, T.; Gallardo, C.; Pejsak, Z.; Belsham, G.J.; Rasmussen, T.B.; Bøtner, A. Transmission of African swine fever virus from infected pigs by direct contact and aerosol routes. Vet. Microbiol. 2017, 211, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Guinat, C.; Reis, A.L.; Netherton, C.L.; Goatley, L.; Pfeiffer, D.U.; Dixon, L. Dynamics of African swine fever virus shedding and excretion in domestic pigs infected by intramuscular inoculation and contact transmission. Vet. Res. 2014, 45, 93. [Google Scholar] [CrossRef]
- Blome, S.; Gabriel, C.; Beer, M. Pathogenesis of African swine fever in domestic pigs and European wild boar. Virus Res. 2013, 173, 122–130. [Google Scholar] [CrossRef]
- Pikalo, J.; Zani, L.; Hühr, J.; Beer, M.; Blome, S. Pathogenesis of African swine fever in domestic pigs and European wild boar—Lessons learned from recent animal trials. Virus Res. 2019, 271, 197614. [Google Scholar] [CrossRef]
- Oura, C.A.; Powell, P.P.; Parkhouse, R.M. African swine fever: A disease characterized by apoptosis. J. Gen. Virol. 1998, 79, 1427–1438. [Google Scholar] [CrossRef]
- Ramiro-Ibanez, F.; Ortega, A.; Brun, A.; Escribano, J.M.; Alonso, C. Apoptosis: A mechanism of cell killing and lymphoid organ impairment during acute African swine fever virus infection. J. Gen. Virol. 1996, 77, 2209–2219. [Google Scholar] [CrossRef]
- Ramiro-Ibanez, F.; Ortega, A.; Ruiz-Gonzalvo, F.; Escribano, J.M.; Alonso, C. Modulation of immune cell populations and activation markers in the pathogenesis of African swine fever virus infection. Virus Res. 1997, 47, 31–40. [Google Scholar] [CrossRef]
- Celec, P.; Vlková, B.; Lauková, L.; Bábíčková, J.; Boor, P. Cell-free DNA: The role in pathophysiology and as a biomarker in kidney diseases. Expert. Rev. Mol. Med. 2018, 20, e1. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.T.N.; Huy, N.T.; Murao, L.A.; Lan, N.T.P.; Thuy, T.T.; Tuan, H.M.; Nga, C.T.P.; Van Tuong, V.; Van Dat, T.; Kikuchi, M.; et al. Elevated Levels of Cell-Free Circulating DNA in Patients with Acute Dengue Virus Infection. PLoS ONE 2011, 6, e25969. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Zhang, Y.; Zhang, Y.; Ma, Y.; Zhang, C.; Li, Q.; Liu, B.; Liu, Z.; Liu, J.; Zhang, X.; et al. Increased Plasma Cell-Free DNA Level during HTNV Infection: Correlation with Disease Severity and Virus Load. Viruses 2014, 6, 2723–2734. [Google Scholar] [CrossRef] [PubMed]
- Hoeter, K.; Neuberger, E.; Fischer, S.; Herbst, M.; Juškevičiūtė, E.; Rossmann, H.; Sprinzl, M.F.; Bodenstein, M.; Schäfer, K.E. Evidence for the utility of cfDNA plasma concentrations to predict disease severity in COVID-19: A retrospective pilot study. Peer J. 2023, 11, e16072. [Google Scholar] [CrossRef]
- Kakarla, R.; Hur, J.; Kim, Y.J.; Kim, J.; Chwae, Y.J. Apoptotic cell-derived exosomes: Messages from dying cells. Exp. Mol. Med. 2020, 52, 1–6. [Google Scholar] [CrossRef]
- Olesen, A.S.; Kodama, M.; Lohse, L.; Accensi, F.; Rasmussen, T.B.; Lazov, C.M.; Limborg, M.T.; Gilbert, M.T.P.; Bøtner, A.; Belsham, G.J. Identification of African Swine Fever Virus Transcription within Peripheral Blood Mononuclear Cells of Acutely Infected Pigs. Viruses 2021, 13, 2333. [Google Scholar] [CrossRef]
- Olesen, A.S.; Lazov, C.M.; Lecocq, A.; Accensi, F.; Jensen, A.B.; Lohse, L.; Rasmussen, T.B.; Belsham, G.J.; Bøtner, A. Uptake and Survival of African Swine Fever Virus in Mealworm (Tenebrio molitor) and Black Soldier Fly (Hermetia illucens) Larvae. Pathogens 2023, 12, 47. [Google Scholar] [CrossRef]
- Olesen, A.S.; Lohse, L.; Accensi, F.; Goldswain, H.; Belsham, G.J.; Bøtner, A.; Netherton, C.L.; Dixon, L.K.; Portugal, R. Inefficient transmission of African swine fever virus to sentinel pigs from environmental contamination under experimental conditions. bioRxiv 2023. [Google Scholar]
- Lohse, L.; Nielsen, J.; Uttenthal, Å. Early pathogenesis of classical swine fever virus (CSFV) strains in Danish pigs. Vet. Microbiol. 2012, 159, 327–336. [Google Scholar] [CrossRef]
- Tignon, M.; Gallardo, C.; Iscaro, C.; Hutet, E.; Van der Stede, Y.; Kolbasov, D.; De Mia, G.M.; Le Potier, M.-F.; Bishop, R.P.; Arias, M.; et al. Development and inter-laboratory validation study of an improved new real-time PCR assay with internal control for detection and laboratory diagnosis of African swine fever virus. J. Virol. Methods 2011, 178, 161–170. [Google Scholar] [CrossRef]
- Hoffmann, B.; Beer, M.; Schelp, C.; Schirrmeier, H.; Depner, K. Validation of real-time RT-PCR assay for sensitive and specific detection of classical swine fever. J. Virol. Methods 2005, 130, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Forth, J.H. Standardisierung Eines Nicht-Invasiven Beprobungssystems zur Infektionsüberwachung bei Wildschweinen (Sus scrofa); Ernst-Moritz-Arndt-Universität Greifswald, Mathematisch-Naturwissenschaftliche Fakultät: Greifswald, Germany, 2015; Available online: https://www.openagrar.de/receive/openagrar_mods_00012495 (accessed on 13 July 2023).
- Dahlén, A.; Mertens, F.; Mandahl, N.; Panagopoulos, I. ACTB (Actin, beta). Atlas Cytogenet. Oncol. Haematol. 2005, 9, 129–130. [Google Scholar] [CrossRef]
- Olesen, A.S.; Kodama, M.; Skovgaard, K.; Møbjerg, A.; Lohse, L.; Limborg, M.T.; Bøtner, A.; Belsham, G.J. Influence of African Swine Fever Virus on Host Gene Transcription within Peripheral Blood Mononuclear Cells from Infected Pigs. Viruses 2022, 14, 2147. [Google Scholar] [CrossRef] [PubMed]
- Scozzi, D.; Cano, M.; Ma, L.; Zhou, D.; Zhu, J.H.; O’halloran, J.A.; Goss, C.W.; Rauseo, A.M.; Liu, Z.; Sahu, S.K.; et al. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight 2021, 6, e143299. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Avula, K.; Sufi, S.A.; Parwin, N.; Das, S.; Alam, M.F. Defective Mitochondrial Quality Control during Dengue Infection Contributes to Disease Pathogenesis. J. Virol. 2022, 96, e0082822. [Google Scholar] [CrossRef]
- Phung, Q.; Lin, M.J.; Xie, H.; Greninger, A.L. Fragment Size-Based Enrichment of Viral Sequences in Plasma Cell-Free DNA. J. Mol. Diagn. 2022, 24, 476–484. [Google Scholar] [CrossRef]
- Bogenhagen, D.F. Mitochondrial DNA nucleoid structure. BBA-Gene Regul. Mech. 2012, 1819, 914–920. [Google Scholar] [CrossRef]
- Sherwood, K.; Weimer, E.T. Characteristics, properties, and potential applications of circulating cell-free DNA in clinical diagnostics: A focus on transplantation. J. Immunol. Meth. 2018, 463, 27–38. [Google Scholar] [CrossRef]
- Karalyan, Z.; Zakaryan, H.; Arakelova, E.; Aivazyan, V.; Tatoyan, M.; Kotsinyan, A.; Izmailyan, R.; Karalova, E. Evidence of hemolysis in pigs infected with highly virulent African swine fever virus. Vet. World 2016, 9, 1413–1419. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olesen, A.S.; Lohse, L.; Johnston, C.M.; Rasmussen, T.B.; Bøtner, A.; Belsham, G.J. Increased Presence of Circulating Cell-Free, Fragmented, Host DNA in Pigs Infected with Virulent African Swine Fever Virus. Viruses 2023, 15, 2133. https://doi.org/10.3390/v15102133
Olesen AS, Lohse L, Johnston CM, Rasmussen TB, Bøtner A, Belsham GJ. Increased Presence of Circulating Cell-Free, Fragmented, Host DNA in Pigs Infected with Virulent African Swine Fever Virus. Viruses. 2023; 15(10):2133. https://doi.org/10.3390/v15102133
Chicago/Turabian StyleOlesen, Ann Sofie, Louise Lohse, Camille Melissa Johnston, Thomas Bruun Rasmussen, Anette Bøtner, and Graham J. Belsham. 2023. "Increased Presence of Circulating Cell-Free, Fragmented, Host DNA in Pigs Infected with Virulent African Swine Fever Virus" Viruses 15, no. 10: 2133. https://doi.org/10.3390/v15102133




