Assessing Monkeypox Virus Prevalence in Small Mammals at the Human–Animal Interface in the Democratic Republic of the Congo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Animal Capture and Sample Collection
2.3. Laboratory Diagnostics
2.4. DNA Sequencing
2.5. Habitat Analysis
3. Results
3.1. Animal Capture
3.2. Laboratory Diagnostics
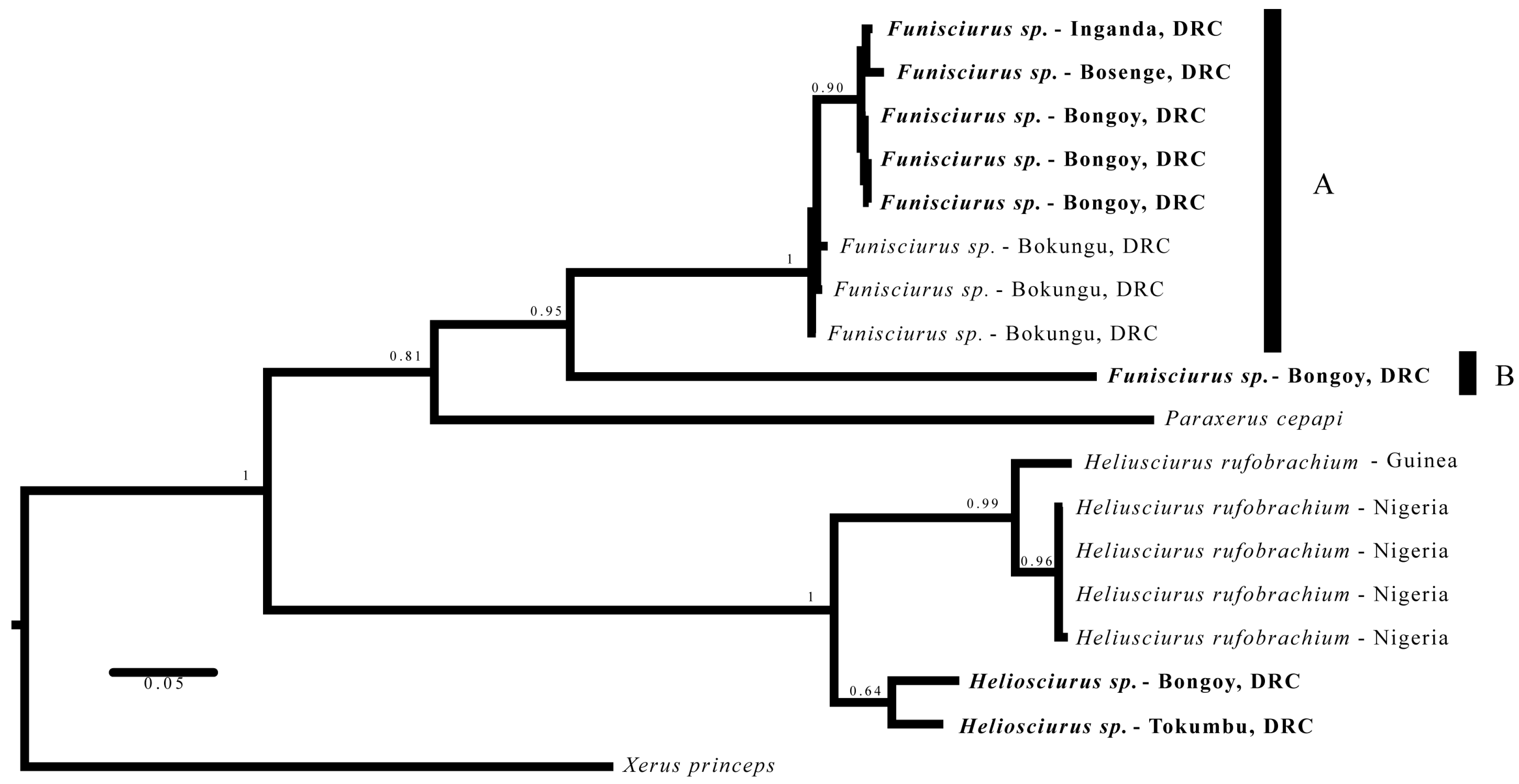
3.3. Phylogenetics
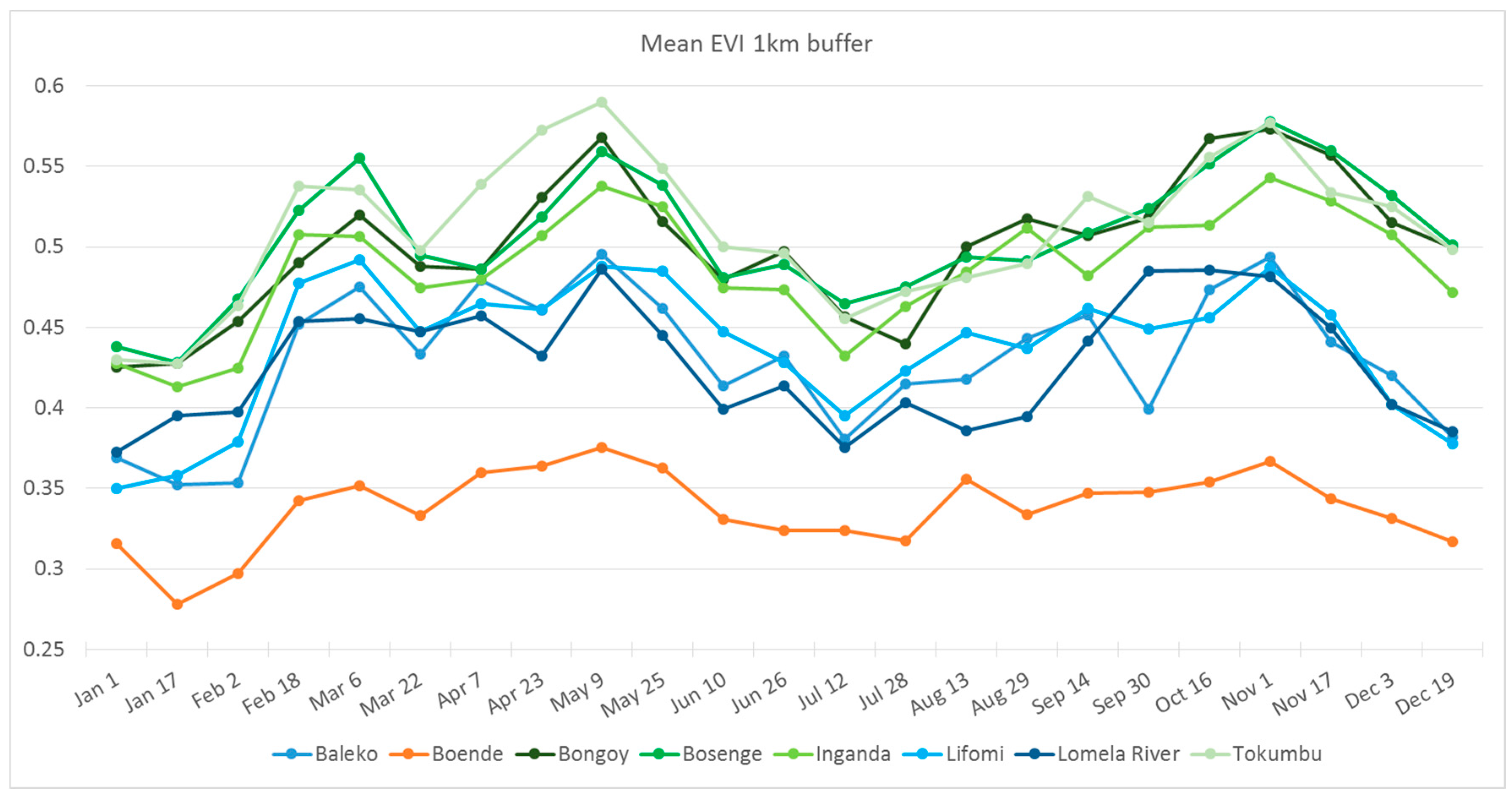
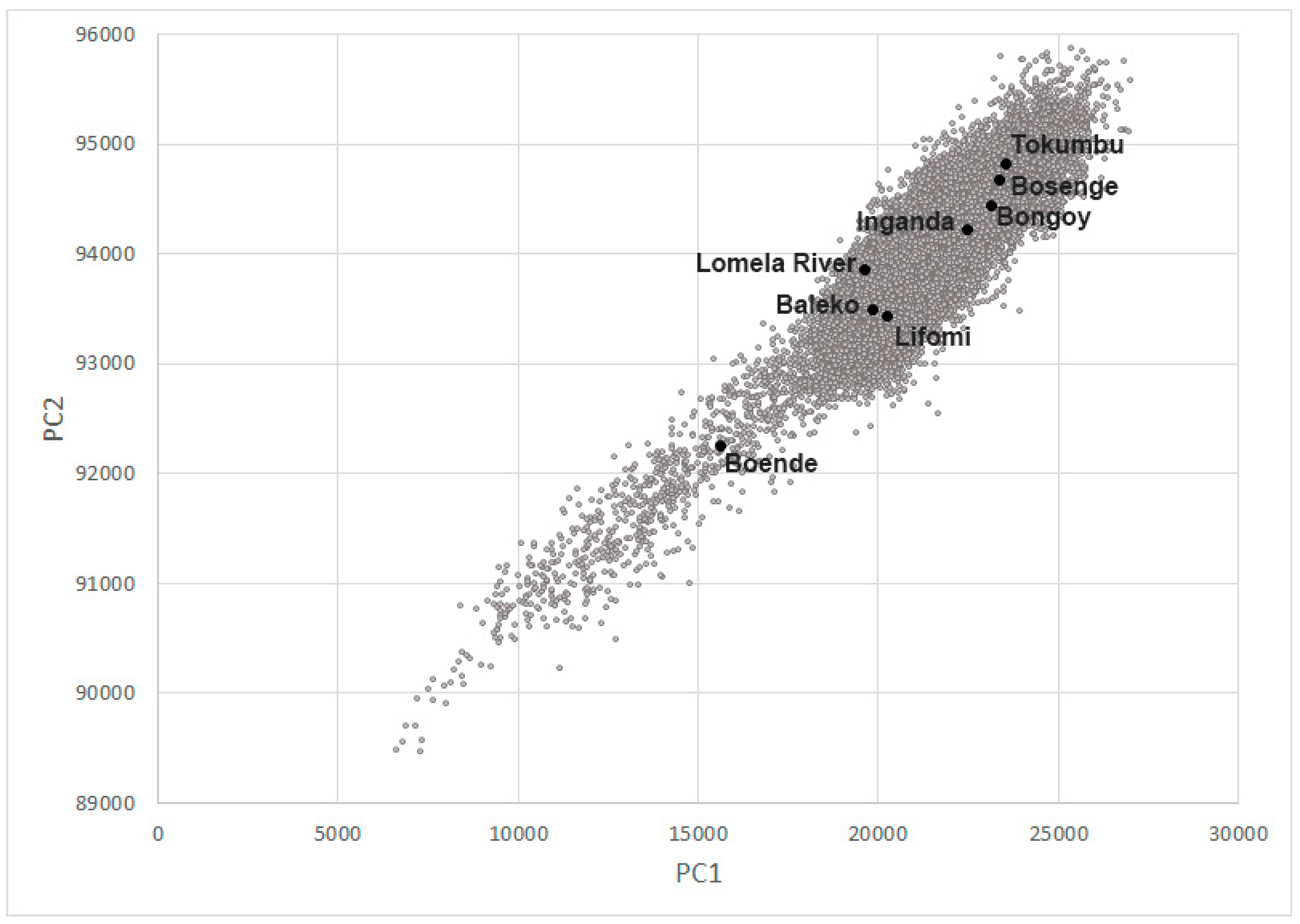
3.4. Habitat Analysis
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
| Order | Genus | No. Sampled | No. IgG Positive |
|---|---|---|---|
| Rodentia | Praomys | 102 | 0 |
| Rodentia | Graphiurus | 13 | 1, 7.7% |
| Rodentia | Oenomys | 14 | 1, 7.1% |
| Rodentia | Cricetomys | 10 | 1, 10% |
| Rodentia | Funisciurus | 6 | 2, 33.3% |
| Rodentia | Heliosciurus | 3 | 1, 33.3% |
| Rodentia | Lemniscomys | 6 | 0 |
| Rodentia | Unknown squirrel | 1 | 0 |
| Rodentia | Rattus | 29 | 0 |
| Rodentia | Mus | 21 | 0 |
| Rodentia | Dendromus | 1 | 0 |
| Rodentia | Deomys | 4 | 0 |
| Rodentia | Lophuromys | 23 | 0 |
| Rodentia | Hylomyscus | 5 | 0 |
| Rodentia | Hybomys | 16 | 0 |
| Rodentia | Malacomys | 4 | 0 |
| Rodentia | Atherurus | 2 | 0 |
| Rodentia | Anomalurus | 1 | 0 |
| Rodentia | Stochomys | 1 | 0 |
| Soricomorpha | Crocidura | 51 | 0 |
| Soricomorpha | Sylvisorex | 13 | 0 |
| Soricomorpha | Scutisorex | 3 | 0 |
| Macroscelidea | Petrodromus | 16 | 1, 6.3% |
| Pholidota | Manis | 6 | 0 |
| Insectivora | Potamogale | 1 | 0 |
| Hydracoidea | Dendrohyrax | 1 | 0 |
| Total | 353 | 7, 2% |
References
- Magnus, P.V.; Andersen, E.K.; Petersen, K.B.; Birch-Andersen, A. A pox-like disease in cynomolgus monkeys. Acta Pathol. Microbiol. Scand. 1959, 46, 156–176. [Google Scholar] [CrossRef]
- Arita, I.; Henderson, D.A. Smallpox and monkeypox in non-human primates. Bull. World Health Org. 1968, 39, 277–283. [Google Scholar] [PubMed]
- Likos, A.M.; Sammons, S.A.; Olson, V.A.; Frace, A.M.; Li, A.M.; Olsen-Rasmussen, M.; Davidson, W.; Galloway, R.; Khistova, M.L.; Reynolds, M.G.; et al. A tale of two clades: Monkeypox viruses. J. Gen. Virol. 2005, 86, 2661–2672. [Google Scholar] [CrossRef] [PubMed]
- Breman, J.G.; Bernadou, J.; Nakano, J.H. Poxvirus in West African nonhuman primates: Serological survey results. Bull. World Health Org. 1977, 55, 605–612. [Google Scholar] [PubMed]
- Khodakevich, L.; Ježek, Z.; Messinger, D. Monkeypox virus: Ecology and public health significance. Bull. World Health Org. 1988, 66, 747–752. [Google Scholar] [PubMed]
- Hutin, Y.J.; Williams, R.J.; Malfait, P.; Pebody, R.; Loparev, V.N.; Ropp, S.L.; Rodriguez, M.; Knight, J.C.; Tshioko, F.K.; Khan, A.S.; et al. Outbreak of human monkeypox, Democratic Republic of Congo, 1996 to 1997. Emerg. Infect. Dis. 2001, 7, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Nolen, L.D.; Osadebe, L.; Katomba, J.; Likofata, J.; Mukadi, D.; Monroe, B.; Doty, J.B.; Kalemba, L.; Malekani, J.; Kabamba, J.; et al. Introduction of monkeypox into a community and household: Risk factors and zoonotic reservoirs in the Democratic Republic of Congo. Am. J. Trop. Med. Hyg. 2015, 93, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.G.; Carroll, D.S.; Olson, V.A.; Hughes, C.; Galley, J.; Likos, A.; Montgomery, J.M.; Suu-Ire, R.; Kwasi, M.O.; Root, J.J.; et al. A silent enzootic of an orthopoxvirus in Ghana, West Africa: Evidence for multi-species involvement in the absence of widespread human disease. Am. J. Trop. Med. Hyg. 2010, 82, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Khodakevich, L.; Ježek, Z.; Kinzanzka, K. Isolation of monkeypox virus from wild squirrel infected in nature. Lancet 1986, 1, 98–99. [Google Scholar] [CrossRef]
- Radonic, A.; Metzger, S.; Dabrowski, P.W.; Couacy-Hymann, E.; Schuenadel, L.; Kurth, A.; Matz-Rensing, K.; Boesch, C.; Leendertz, F.H.; Nitsche, A. Fatal monkeypox in wild-living sooty mangabey, Cote d’Ivoire, 2012. Emerg. Infect. Dis. 2014, 20, 1009–1011. [Google Scholar] [CrossRef] [PubMed]
- Rimoin, A.W.; Mulembakani, P.M.; Johnston, S.C.; Lloyd Smith, J.O.; Kisalu, N.K.; Kinkela, T.L.; Blumberg, S.; Thomassen, H.A.; Pike, B.L.; Fair, J.N.; et al. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. USA 2010, 107, 16262–16267. [Google Scholar] [CrossRef] [PubMed]
- Quiner, C.A.; Moses, C.; Monroe, B.P.; Nakazawa, Y.J.; Doty, J.B.; Hughes, C.M.; McCollum, A.M.; Ibata, S.; Malekani, J.; Okitolonda, E.; et al. Presumptive risk factors for monkeypox in rural communities in the Democratic Republic of the Congo. PLoS ONE 2017, 12, e0168664. [Google Scholar] [CrossRef] [PubMed]
- Fa, J.E.; Ryan, S.F.; Bell, D.J. Hunting vulnerability, ecological characteristics and harvest rates of bushmeat species in Afrotropical forests. Biol. Conserv. 2005, 121, 167–176. [Google Scholar] [CrossRef]
- Wilkie, D.S.; Carpenter, J.F. Bushmeat hunting in the Congo Basin: An assessment of impacts and options for mitigation. Biodivers. Conserv. 1999, 8, 927–955. [Google Scholar] [CrossRef]
- Gaubert, P.; Njiokou, F.; Olayemi, A.; Pagani, P.; Dufour, S.; Danquah, E.; Nutsuakor, M.E.K.; Ngua, G.; Missoup, A.D.; Tedesco, P.A.; et al. Bushmeat genetics: Setting up a reference framework for the DNA typing of African forest bushmeat. Mol. Ecol. Resour. 2015, 15, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Monroe, B.P.; Doty, J.B.; Moses, C.; Ibata, S.; Reynolds, M.; Carroll, D.S. Collection and utilization of animal carcasses associated with zoonotic disease in Tshuapa District, DRC, 2012. J. Wildl. Dis. 2015, 5, 734–738. [Google Scholar] [CrossRef] [PubMed]
- Karem, K.L.; Reynolds, M.; Braden, Z.; Lou, G.; Bernard, N.; Patton, J.; Damon, I.K. Characterization of acute-phase humoral immunity to monkeypox: Use of immunoglobulin M enzyme-linked immunosorbent assay for detection of monkeypox infection during the 2003 North American outbreak. Clin. Diagn. Lab. Immunol. 2005, 12, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Hutson, C.L.; Olson, V.A.; Carroll, D.S.; Abel, J.A.; Hughes, C.M.; Braden, Z.H.; Weiss, S.; Self, J.; Osorio, J.E.; Hudson, P.N.; et al. A prairie dog model of systemic orthopoxvirus disease using West African and Congo Basin strains of monkeypox virus. J. Gen. Virol. 2009, 90, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Olson, V.A.; Laue, T.; Laker, M.T.; Damon, I.K. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. 2006, 36, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.W.; Bradley, R.D. Molecular systematics and historical phylobiogeography of the Neotoma mexicana species group. J. Mammal. 2002, 83, 20–30. [Google Scholar] [CrossRef]
- Bickham, J.W.; Wood, C.C.; Patton, J.C. Biogeographic implications of cytochrome b sequences and allozymes in sockeye (Oncorhynchus nerka). J. Hered. 1995, 86, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Bickham, J.W.; Patton, J.C.; Schlitter, D.A.; Rautenbach, I.L.; Honeycutt, R.L. Molecular phylogenetics, karyotypic diversity, and partition of the genus Myotis (Chiroptera: Vespertilionidae). Mol. Phylogenet. Evol. 2004, 33, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.F.; Patton, J.L. The diversification of South American murid rodents: Evidence from mitochondrial DNA sequence data for the akodontine tribe. Biol. J. Linnean Soc. 1993, 50, 149–177. [Google Scholar] [CrossRef]
- Peppers, L.L.; Bradley, R.D. Cryptic species in Sigmodon hispidus: Evidence from DNA sequences. J. Mammal. 2000, 81, 332–343. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Mercer, J.M.; Roth, V.L. The effects of Cenozoic global change on squirrel phylogeny. Science 2003, 299, 1568–1572. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.T.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Khodakevich, L.; Szsceniowski, M.; Disu, N.M.; Ježek, Z.; Marennikova, S.; Nakano, J.; Meier, F. Monkeypox virus in relation to the ecological features surrounding human settlements in Bumba Zone, Zaire. Trop. Geogr. Med. 1987, 39, 56–63. [Google Scholar] [PubMed]
- Khodakevich, L.; Szsceniowski, M.; Disu, N.M.; Jezek, Z.; Marennikova, S.; Nakano, J.; Messinger, D. The role of squirrels in sustaining monkeypox virus transmission. Trop. Geogr. Med. 1987, 39, 115–122. [Google Scholar] [PubMed]
- Malekani, J.M.; University of Kinshasa, Kinshasa, DRC. Personal communication, 2015.
- Kingdon, J. The Kingdon Field Guide to African Mammals; A&C Black Publishers Ltd.: London, UK, 1997; p. 476. [Google Scholar]
- Bradley, R.D.; Baker, R.J. A test of genetic species concept: Cytochrome-b sequences and mammals. J. Mammal. 2010, 82, 960–973. [Google Scholar] [CrossRef]
- Verhegghen, A.; Mayaux, P.; de Wasseige, C.; Defourny, P. Mapping Congo Basin vegetation types from 300 m and 1 km multi-sensor time series for carbon stocks and forest areas estimation. Biogeosciences 2012, 9, 5061–5079. [Google Scholar] [CrossRef] [Green Version]
- Moss, B. Poxviridae: The viruses and their replication. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 2, pp. 2905–2946. [Google Scholar]


| Baleko | Boende | Bongoy | Bosenge | Inganda | Lifomi | Lomela | Tokumbu | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Rodentia | 86 | 29 | 6 | 1 | 6 | 95 | 14 | 25 | 262 |
| Soricomorpha | 33 | 6 | 26 | 2 | 67 | ||||
| Pholidota | 6 | 6 | |||||||
| Macroscelidea | 12 | 4 | 16 | ||||||
| Hyracoidea | 1 | 1 | |||||||
| Insectivora | 1 | 1 | |||||||
| Total | 120 | 36 | 18 | 1 | 10 | 127 | 16 | 25 | 353 |
| Genus | No. Sampled |
|---|---|
| Rattus | 7 |
| Mus | 6 |
| Crocidura | 3 |
| Total | 16 |
| 2012 Localities | 2013 Localities | 2015 Localities | IgG Positive | |
|---|---|---|---|---|
| Funisciurus spp. | None | Inganda | Bongoy, Bosenge | 2/6, 33.3% |
| Graphiurus lorraineus | None | None | Inganda, Tokumbu | 1/13, 7.7% |
| Cricetomys emini | None | None | Boende, Tokumbu | 1/9, 11.1% |
| Heliosciurus rufobrachium | None | None | Bongoy, Tokumbu | 1/3, 33.3% |
| Oenomys hypoxanthus | Baleko | Lifomi | Inganda, Tokumbu | 1/22, 4.5% |
| Petrodromus tetradactylus | None | Inganda | Bongoy | 1/17, 5.9% |
| FuniA | FuniB | HelioSamples | HelioRefs | Paraxerus | |
|---|---|---|---|---|---|
| FuniB | 24.23% | ||||
| HelioSamples | 30.22% | 33.65% | |||
| HelioRefs | 25.62% | 28.04% | 11.39% | ||
| Paraxerus | 29.05% | 30.75% | 36.38% | 30.05% | |
| Xerus | 29.88% | 33.43% | 33.61% | 32.46% | 34.15% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doty, J.B.; Malekani, J.M.; Kalemba, L.N.; Stanley, W.T.; Monroe, B.P.; Nakazawa, Y.U.; Mauldin, M.R.; Bakambana, T.L.; Liyandja Dja Liyandja, T.; Braden, Z.H.; et al. Assessing Monkeypox Virus Prevalence in Small Mammals at the Human–Animal Interface in the Democratic Republic of the Congo. Viruses 2017, 9, 283. https://doi.org/10.3390/v9100283
Doty JB, Malekani JM, Kalemba LN, Stanley WT, Monroe BP, Nakazawa YU, Mauldin MR, Bakambana TL, Liyandja Dja Liyandja T, Braden ZH, et al. Assessing Monkeypox Virus Prevalence in Small Mammals at the Human–Animal Interface in the Democratic Republic of the Congo. Viruses. 2017; 9(10):283. https://doi.org/10.3390/v9100283
Chicago/Turabian StyleDoty, Jeffrey B., Jean M. Malekani, Lem’s N. Kalemba, William T. Stanley, Benjamin P. Monroe, Yoshinori U. Nakazawa, Matthew R. Mauldin, Trésor L. Bakambana, Tobit Liyandja Dja Liyandja, Zachary H. Braden, and et al. 2017. "Assessing Monkeypox Virus Prevalence in Small Mammals at the Human–Animal Interface in the Democratic Republic of the Congo" Viruses 9, no. 10: 283. https://doi.org/10.3390/v9100283





