Nucleolin-Targeting AS1411 Aptamer-Conjugated Nanospheres for Targeted Treatment of Glioblastoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Quantitative Real-Time RT-PCR
2.3. Western Blot Analysis
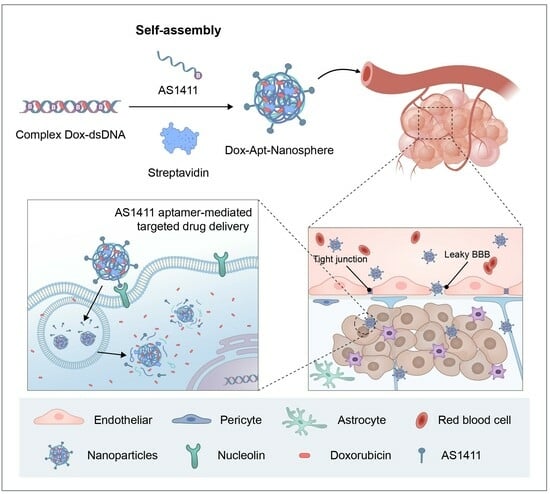
2.4. Synthesis of AS1411 Aptamer-Conjugated Nanospheres
2.4.1. Nanoparticle Synthetic Materials and Instruments
2.4.2. Synthesis of DNAProtein Hybrid Nanospheres Using Self-Assembly
2.4.3. Conjugation of the AS1411 Aptamer to Nanospheres
2.5. Agarose Gel Electrophoresis
2.6. Characterization of Drug Loading and Release
2.7. Cytotoxicity Assay
2.8. Flow Cytometric Analysis
2.9. Confocal Laser-Scanning Microscopy (CLSM)
2.10. Three-Dimensional Tumor Spheroid Formation
2.11. In Vitro BTB Penetration Assays
2.12. In Vivo Anti-Tumor Experiments
2.13. Statistical Analysis
3. Results
3.1. Generation and Characterization of AS1411 Aptamer-Conjugated Nanospheres
3.2. Ability to Selectively Bind Glioblastoma Cells to AS1411 Aptamer-Conjugated Nanospheres
3.3. Transport Efficiency and Cellular Uptake of AS1411 Aptamer-Conjugated Nanospheres with Doxorubicin
3.4. Cytotoxicity of AS1411 Aptamer-Conjugated Nanospheres with Doxorubicin
3.5. Anti-Tumor Effect of AS1411 Aptamer-Conjugated Nanospheres with Doxorubicin
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, O.G.; Brzozowski, J.S.; Skelding, K.A. Glioblastoma Multiforme: An Overview of Emerging Therapeutic Targets. Front. Oncol. 2019, 9, 963. [Google Scholar] [CrossRef] [PubMed]
- van Tellingen, O.; Yetkin-Arik, B.; de Gooijer, M.C.; Wesseling, P.; Wurdinger, T.; de Vries, H.E. Overcoming the blood-brain tumor barrier for effective glioblastoma treatment. Drug Resist. Updat. 2015, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lingineni, K.; Belekar, V.; Tangadpalliwar, S.R.; Garg, P. The role of multidrug resistance protein (MRP-1) as an active efflux transporter on blood-brain barrier (BBB) permeability. Mol. Divers. 2017, 21, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Niksic, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Esteve, J.; et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.M.; Patel, B.M. Crossing the Blood-Brain Barrier: Recent Advances in Drug Delivery to the Brain. CNS Drugs 2017, 31, 109–133. [Google Scholar] [CrossRef] [PubMed]
- Dong, X. Current Strategies for Brain Drug Delivery. Theranostics 2018, 8, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Phoenix, T.N.; Patmore, D.M.; Boop, S.; Boulos, N.; Jacus, M.O.; Patel, Y.T.; Roussel, M.F.; Finkelstein, D.; Goumnerova, L.; Perreault, S.; et al. Medulloblastoma Genotype Dictates Blood Brain Barrier Phenotype. Cancer Cell 2016, 29, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Pardridge, W.M. Blood-brain barrier endogenous transporters as therapeutic targets: A new model for small molecule CNS drug discovery. Expert. Opin. Ther. Targets 2015, 19, 1059–1072. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Porter, M.; Konstantopoulos, A.; Zhang, P.; Cui, H. Preclinical development of drug delivery systems for paclitaxel-based cancer chemotherapy. J. Control. Release 2017, 267, 100–118. [Google Scholar] [CrossRef]
- Pais, V.; Danaila, L.; Pais, E. Ultrastructural patterns of the activated cell death programs in the human brain. Ultrastruct. Pathol. 2013, 37, 110–120. [Google Scholar] [CrossRef]
- Wang, K.; Huang, Q.; Qiu, F.; Sui, M. Non-viral Delivery Systems for the Application in p53 Cancer Gene Therapy. Curr. Med. Chem. 2015, 22, 4118–4136. [Google Scholar] [CrossRef] [PubMed]
- Qu, N.; Sun, Y.; Li, Y.; Hao, F.; Qiu, P.; Teng, L.; Xie, J.; Gao, Y. Docetaxel-loaded human serum albumin (HSA) nanoparticles: Synthesis, characterization, and evaluation. Biomed. Eng. Online 2019, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Meredith, A.M.; Dass, C.R. Increasing role of the cancer chemotherapeutic doxorubicin in cellular metabolism. J. Pharm. Pharmacol. 2016, 68, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Cagel, M.; Grotz, E.; Bernabeu, E.; Moretton, M.A.; Chiappetta, D.A. Doxorubicin: Nanotechnological overviews from bench to bedside. Drug Discov. Today 2017, 22, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xiang, M.H.; Liu, J.W.; Tang, H.; Jiang, J.H. DNA Polymer Nanoparticles Programmed via Supersandwich Hybridization for Imaging and Therapy of Cancer Cells. Anal. Chem. 2018, 90, 12951–12958. [Google Scholar] [CrossRef] [PubMed]
- Ramya, A.N.; Joseph, M.M.; Maniganda, S.; Karunakaran, V.; Sreelekha, T.T.; Maiti, K.K. Emergence of Gold-Mesoporous Silica Hybrid Nanotheranostics: Dox-Encoded, Folate Targeted Chemotherapy with Modulation of SERS Fingerprinting for Apoptosis toward Tumor Eradication. Small 2021, 17, e2007852. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Guo, D.S.; Wang, X.; Liu, Y. Multistimuli responsive supramolecular vesicles based on the recognition of p-Sulfonatocalixarene and its controllable release of doxorubicin. ACS Nano 2011, 5, 2880–2894. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Hu, X.Y.; Li, Y.; Zou, X.; Xiong, S.; Lin, C.; Shen, Y.Z.; Wang, L. Multistimuli-responsive supramolecular vesicles based on water-soluble pillar [6]arene and SAINT complexation for controllable drug release. J. Am. Chem. Soc. 2014, 136, 10762–10769. [Google Scholar] [CrossRef]
- Klaikherd, A.; Nagamani, C.; Thayumanavan, S. Multi-stimuli sensitive amphiphilic block copolymer assemblies. J. Am. Chem. Soc. 2009, 131, 4830–4838. [Google Scholar] [CrossRef]
- Dehghani, S.; Alibolandi, M.; Tehranizadeh, Z.A.; Oskuee, R.K.; Nosrati, R.; Soltani, F.; Ramezani, M. Self-assembly of an aptamer-decorated chimeric peptide nanocarrier for targeted cancer gene delivery. Colloids Surf. B Biointerfaces 2021, 208, 112047. [Google Scholar] [CrossRef] [PubMed]
- Thelu, H.V.P.; Atchimnaidu, S.; Perumal, D.; Harikrishnan, K.S.; Vijayan, S.; Varghese, R. Self-Assembly of an Aptamer-Decorated, DNA-Protein Hybrid Nanogel: A Biocompatible Nanocarrier for Targeted Cancer Therapy. ACS Appl. Bio Mater. 2019, 2, 5227–5234. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Girvan, A.C.; Casson, L.K.; Pierce, W.M., Jr.; Qian, M.; Thomas, S.D.; Bates, P.J. AS1411 alters the localization of a complex containing protein arginine methyltransferase 5 and nucleolin. Cancer Res. 2007, 67, 10491–10500. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Hogberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xia, D.; Klausen, L.H.; Dong, M. The self-assembled behavior of DNA bases on the interface. Int. J. Mol. Sci. 2014, 15, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Perbandt, M.; Bruns, O.; Vallazza, M.; Lamla, T.; Betzel, C.; Erdmann, V.A. High resolution structure of streptavidin in complex with a novel high affinity peptide tag mimicking the biotin binding motif. Proteins 2007, 67, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Chilkoti, A.; Tan, P.H.; Stayton, P.S. Site-directed mutagenesis studies of the high-affinity streptavidin-biotin complex: Contributions of tryptophan residues 79, 108, and 120. Proc. Natl. Acad. Sci. USA 1995, 92, 1754–1758. [Google Scholar] [CrossRef] [PubMed]
- Park, T.J.; Lee, K.B.; Lee, S.J.; Park, J.P.; Lee, Z.W.; Choi, S.K.; Jung, H.C.; Pan, J.G.; Lee, S.Y.; Choi, I.S. Micropatterns of spores displaying heterologous proteins. J. Am. Chem. Soc. 2004, 126, 10512–10513. [Google Scholar] [CrossRef]
- Muller, M.K.; Petkau, K.; Brunsveld, L. Protein assembly along a supramolecular wire. Chem. Commun. (Camb) 2011, 47, 310–312. [Google Scholar] [CrossRef]
- Weber, P.C.; Ohlendorf, D.H.; Wendoloski, J.J.; Salemme, F.R. Structural origins of high-affinity biotin binding to streptavidin. Science 1989, 243, 85–88. [Google Scholar] [CrossRef]
- Jain, A.; Cheng, K. The principles and applications of avidin-based nanoparticles in drug delivery and diagnosis. J. Control. Release 2017, 245, 27–40. [Google Scholar] [CrossRef]
- Kincanon, M.; Murphy, C.J. Nanoparticle Size Influences the Self-Assembly of Gold Nanorods Using Flexible Streptavidin-Biotin Linkages. ACS Nano 2023, 17, 24090–24103. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Chen, Q.; Liu, J.; Yang, X.; Huang, J.; Li, L.; Guo, X.; Zhang, J.; Wang, K. Programmable Self-Assembly of DNA-Protein Hybrid Hydrogel for Enzyme Encapsulation with Enhanced Biological Stability. Biomacromolecules 2016, 17, 1543–1550. [Google Scholar] [CrossRef]
- Lee, D.; Baek, S.; Kim, Y.Y.; Bang, Y.; Jung, H.N.; Im, H.J.; Song, Y.K. Self-Assembled DNA-Protein Hybrid Nanospheres: Biocompatible Nano-Drug-Carriers for Targeted Cancer Therapy. ACS Appl. Mater. Interfaces 2022, 14, 37493–37503. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.Y.; Bang, Y.; Lee, A.H.; Song, Y.K. Multivalent Traptavidin-DNA Conjugates for the Programmable Assembly of Nanostructures. ACS Nano 2019, 13, 1183–1194. [Google Scholar] [CrossRef]
- Chen, W.L.; Li, F.; Tang, Y.; Yang, S.D.; Li, J.Z.; Yuan, Z.Q.; Liu, Y.; Zhou, X.F.; Liu, C.; Zhang, X.N. Stepwise pH-responsive nanoparticles for enhanced cellular uptake and on-demand intracellular release of doxorubicin. Int. J. Nanomed. 2017, 12, 4241–4256. [Google Scholar] [CrossRef] [PubMed]
- de Lazaro, I.; Mooney, D.J. A nanoparticle’s pathway into tumours. Nat. Mater. 2020, 19, 486–487. [Google Scholar] [CrossRef]
- Aravind, A.; Yoshida, Y.; Maekawa, T.; Kumar, D.S. Aptamer-conjugated polymeric nanoparticles for targeted cancer therapy. Drug Deliv. Transl. Res. 2012, 2, 418–436. [Google Scholar] [CrossRef]
- Cerchia, L.; de Franciscis, V. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 2010, 28, 517–525. [Google Scholar] [CrossRef]
- Herr, J.K.; Smith, J.E.; Medley, C.D.; Shangguan, D.; Tan, W. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal. Chem. 2006, 78, 2918–2924. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tian, T.; Zhou, R.; Li, S.; Ma, W.; Zhang, Y.; Liu, N.; Shi, S.; Li, Q.; Xie, X.; et al. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat. Protoc. 2020, 15, 2728–2757. [Google Scholar] [CrossRef] [PubMed]
- Vahidfar, N.; Aghanejad, A.; Ahmadzadehfar, H.; Farzanehfar, S.; Eppard, E. Theranostic Advances in Breast Cancer in Nuclear Medicine. Int. J. Mol. Sci. 2021, 22, 4597. [Google Scholar] [CrossRef] [PubMed]
- Saberian, M.; Hamzeiy, H.; Aghanejad, A.; Asgari, D. Aptamer-based Nanosensors: Juglone as an Attached-Redox Molecule for Detection of Small Molecules. Bioimpacts 2011, 1, 31–36. [Google Scholar] [CrossRef]
- Deng, R.; Shen, N.; Yang, Y.; Yu, H.; Xu, S.; Yang, Y.W.; Liu, S.; Meguellati, K.; Yan, F. Targeting epigenetic pathway with gold nanoparticles for acute myeloid leukemia therapy. Biomaterials 2018, 167, 80–90. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; Bambury, R.M.; Van Allen, E.M.; Drabkin, H.A.; Lara, P.N., Jr.; Harzstark, A.L.; Wagle, N.; Figlin, R.A.; Smith, G.W.; Garraway, L.A.; et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Investig. New Drugs 2014, 32, 178–187. [Google Scholar] [CrossRef]
- Bunka, D.H.; Platonova, O.; Stockley, P.G. Development of aptamer therapeutics. Curr. Opin. Pharmacol. 2010, 10, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef]
- Mongelard, F.; Bouvet, P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr. Opin. Mol. Ther. 2010, 12, 107–114. [Google Scholar]
- Esposito, C.L.; Nuzzo, S.; Catuogno, S.; Romano, S.; de Nigris, F.; de Franciscis, V. STAT3 Gene Silencing by Aptamer-siRNA Chimera as Selective Therapeutic for Glioblastoma. Mol. Ther. Nucleic Acids 2018, 10, 398–411. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhao, G.; Zhang, S.; Nigim, F.; Zhou, G.; Yu, Z.; Song, Y.; Chen, Y.; Li, Y. AS1411-Induced Growth Inhibition of Glioma Cells by Up-Regulation of p53 and Down-Regulation of Bcl-2 and Akt1 via Nucleolin. PLoS ONE 2016, 11, e0167094. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef]
- Xu, X.; Hamhouyia, F.; Thomas, S.D.; Burke, T.J.; Girvan, A.C.; McGregor, W.G.; Trent, J.O.; Miller, D.M.; Bates, P.J. Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J. Biol. Chem. 2001, 276, 43221–43230. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Gorospe, M. RNA-binding protein nucleolin in disease. RNA Biol. 2012, 9, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Galzio, R.; Rosati, F.; Benedetti, E.; Cristiano, L.; Aldi, S.; Mei, S.; D’Angelo, B.; Gentile, R.; Laurenti, G.; Cifone, M.G.; et al. Glycosilated nucleolin as marker for human gliomas. J. Cell. Biochem. 2012, 113, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Goldshmit, Y.; Trangle, S.S.; Kloog, Y.; Pinkas-Kramarski, R. Interfering with the interaction between ErbB1, nucleolin and Ras as a potential treatment for glioblastoma. Oncotarget 2014, 5, 8602–8613. [Google Scholar] [CrossRef]
- Luo, Z.; Yan, Z.; Jin, K.; Pang, Q.; Jiang, T.; Lu, H.; Liu, X.; Pang, Z.; Yu, L.; Jiang, X. Precise glioblastoma targeting by AS1411 aptamer-functionalized poly (l-gamma-glutamylglutamine)-paclitaxel nanoconjugates. J. Colloid Interface Sci. 2017, 490, 783–796. [Google Scholar] [CrossRef]
- Huang, P.H.; Xu, A.M.; White, F.M. Oncogenic EGFR signaling networks in glioma. Sci. Signal. 2009, 2, re6. [Google Scholar] [CrossRef]
- Wu, J.; Song, C.; Jiang, C.; Shen, X.; Qiao, Q.; Hu, Y. Nucleolin targeting AS1411 modified protein nanoparticle for antitumor drugs delivery. Mol. Pharm. 2013, 10, 3555–3563. [Google Scholar] [CrossRef]
- Alexis, F.; Pridgen, E.; Molnar, L.K.; Farokhzad, O.C. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 2008, 5, 505–515. [Google Scholar] [CrossRef]
- Gao, H.; Qian, J.; Cao, S.; Yang, Z.; Pang, Z.; Pan, S.; Fan, L.; Xi, Z.; Jiang, X.; Zhang, Q. Precise glioma targeting of and penetration by aptamer and peptide dual-functioned nanoparticles. Biomaterials 2012, 33, 5115–5123. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Gao, X.; Su, L.; Xia, H.; Gu, G.; Pang, Z.; Jiang, X.; Yao, L.; Chen, J.; Chen, H. Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials 2011, 32, 8010–8020. [Google Scholar] [CrossRef] [PubMed]
- Hovanessian, A.G.; Soundaramourty, C.; El Khoury, D.; Nondier, I.; Svab, J.; Krust, B. Surface expressed nucleolin is constantly induced in tumor cells to mediate calcium-dependent ligand internalization. PLoS ONE 2010, 5, e15787. [Google Scholar] [CrossRef] [PubMed]
- McMillan, J.R.; Hayes, O.G.; Remis, J.P.; Mirkin, C.A. Programming Protein Polymerization with DNA. J. Am. Chem. Soc. 2018, 140, 15950–15956. [Google Scholar] [CrossRef] [PubMed]
- Taki, S.; Ardestani, M.S. Novel nanosized AS1411-chitosan-BODIPY conjugate for molecular fluorescent imaging. Int. J. Nanomed. 2019, 14, 3543–3555. [Google Scholar] [CrossRef] [PubMed]
- Cherepanova, A.; Tamkovich, S.; Pyshnyi, D.; Kharkova, M.; Vlassov, V.; Laktionov, P. Immunochemical assay for deoxyribonuclease activity in body fluids. J. Immunol. Methods 2007, 325, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Tamkovich, S.N.; Cherepanova, A.V.; Kolesnikova, E.V.; Rykova, E.Y.; Pyshnyi, D.V.; Vlassov, V.V.; Laktionov, P.P. Circulating DNA and DNase activity in human blood. Ann. N. Y. Acad. Sci. 2006, 1075, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Mendes, B.; Marques, C.; Carvalho, I.; Costa, P.; Martins, S.; Ferreira, D.; Sarmento, B. Influence of glioma cells on a new co-culture in vitro blood-brain barrier model for characterization and validation of permeability. Int. J. Pharm. 2015, 490, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Zhao, H.; Zhu, T.; Yang, Z.; Xu, H.; Fu, Y.; Lin, F.; Pan, X.; Li, L.; et al. Enhanced in Vivo Blood-Brain Barrier Penetration by Circular Tau-Transferrin Receptor Bifunctional Aptamer for Tauopathy Therapy. J. Am. Chem. Soc. 2020, 142, 3862–3872. [Google Scholar] [CrossRef]
- So, Y.H. The effect of limited monomer solubility in heterogeneous step-growth polymerization. Acc. Chem. Res. 2001, 34, 753–758. [Google Scholar] [CrossRef]
- Lu, X.; Fu, H.; Shih, K.C.; Jia, F.; Sun, Y.; Wang, D.; Wang, Y.; Ekatan, S.; Nieh, M.P.; Lin, Y.; et al. DNA-Mediated Step-Growth Polymerization of Bottlebrush Macromonomers. J. Am. Chem. Soc. 2020, 142, 10297–10301. [Google Scholar] [CrossRef]
- Holmberg, A.; Blomstergren, A.; Nord, O.; Lukacs, M.; Lundeberg, J.; Uhlen, M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis 2005, 26, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, C.; Tang, T.H.; Tominaga, J.; Tan, S.C.; Gopinath, S.C. Single-stranded DNA (ssDNA) production in DNA aptamer generation. Analyst 2012, 137, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Perez-Arnaiz, C.; Busto, N.; Leal, J.M.; Garcia, B. New insights into the mechanism of the DNA/doxorubicin interaction. J. Phys. Chem. B 2014, 118, 1288–1295. [Google Scholar] [CrossRef]
- Ali-Boucetta, H.; Al-Jamal, K.T.; McCarthy, D.; Prato, M.; Bianco, A.; Kostarelos, K. Multiwalled carbon nanotube-doxorubicin supramolecular complexes for cancer therapeutics. Chem. Commun. (Camb) 2008, 459–461. [Google Scholar] [CrossRef]
- Gnapareddy, B.; Dugasani, S.R.; Ha, T.; Paulson, B.; Hwang, T.; Kim, T.; Kim, J.H.; Oh, K.; Park, S.H. Chemical and Physical Characteristics of Doxorubicin Hydrochloride Drug-Doped Salmon DNA Thin Films. Sci. Rep. 2015, 5, 12722. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef]
- Lim, W.; Hoang, H.H.; You, D.; Han, J.; Lee, J.E.; Kim, S.; Park, S. Formation of size-controllable tumour spheroids using a microfluidic pillar array (muFPA) device. Analyst 2018, 143, 5841–5848. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, K.M.; MacParland, S.A.; Ma, X.Z.; Spetzler, V.N.; Echeverri, J.; Ouyang, B.; Fadel, S.M.; Sykes, E.A.; Goldaracena, N.; Kaths, J.M.; et al. Mechanism of hard-nanomaterial clearance by the liver. Nat. Mater. 2016, 15, 1212–1221. [Google Scholar] [CrossRef]
- Schlageter, K.E.; Molnar, P.; Lapin, G.D.; Groothuis, D.R. Microvessel organization and structure in experimental brain tumors: Microvessel populations with distinctive structural and functional properties. Microvasc. Res. 1999, 58, 312–328. [Google Scholar] [CrossRef]
- Subhan, M.A.; Yalamarty, S.S.K.; Filipczak, N.; Parveen, F.; Torchilin, V.P. Recent Advances in Tumor Targeting via EPR Effect for Cancer Treatment. J. Pers. Med. 2021, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Ginisty, H.; Sicard, H.; Roger, B.; Bouvet, P. Structure and functions of nucleolin. J. Cell Sci. 1999, 112 Pt 6, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Dam, D.H.; Lee, J.H.; Sisco, P.N.; Co, D.T.; Zhang, M.; Wasielewski, M.R.; Odom, T.W. Direct observation of nanoparticle-cancer cell nucleus interactions. ACS Nano 2012, 6, 3318–3326. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, L.; Wang, L.; Jiang, W. A dual-targeting DNA tetrahedron nanocarrier for breast cancer cell imaging and drug delivery. Talanta 2018, 179, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Huang, Y.; Gao, P.; Chen, T. Nucleus-targeted DNA tetrahedron as a nanocarrier of metal complexes for enhanced glioma therapy. Chem. Commun. 2018, 54, 9394–9397. [Google Scholar] [CrossRef]
- Sun, Q.; Zhou, Z.; Qiu, N.; Shen, Y. Rational Design of Cancer Nanomedicine: Nanoproperty Integration and Synchronization. Adv. Mater. 2017, 29, 1606628. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, K.; Hwang, K.; Nam, K.M.; Kim, M.J.; Song, Y.-K.; Kim, C.-Y. Nucleolin-Targeting AS1411 Aptamer-Conjugated Nanospheres for Targeted Treatment of Glioblastoma. Pharmaceutics 2024, 16, 566. https://doi.org/10.3390/pharmaceutics16040566
Seo K, Hwang K, Nam KM, Kim MJ, Song Y-K, Kim C-Y. Nucleolin-Targeting AS1411 Aptamer-Conjugated Nanospheres for Targeted Treatment of Glioblastoma. Pharmaceutics. 2024; 16(4):566. https://doi.org/10.3390/pharmaceutics16040566
Chicago/Turabian StyleSeo, Kyeongjin, Kihwan Hwang, Kyung Mi Nam, Min Ju Kim, Yoon-Kyu Song, and Chae-Yong Kim. 2024. "Nucleolin-Targeting AS1411 Aptamer-Conjugated Nanospheres for Targeted Treatment of Glioblastoma" Pharmaceutics 16, no. 4: 566. https://doi.org/10.3390/pharmaceutics16040566