Soil Microbial Community Varied with Vegetation Types on a Small Regional Scale of the Qilian Mountains
Abstract
:1. Introduction
2. Methods and Materials
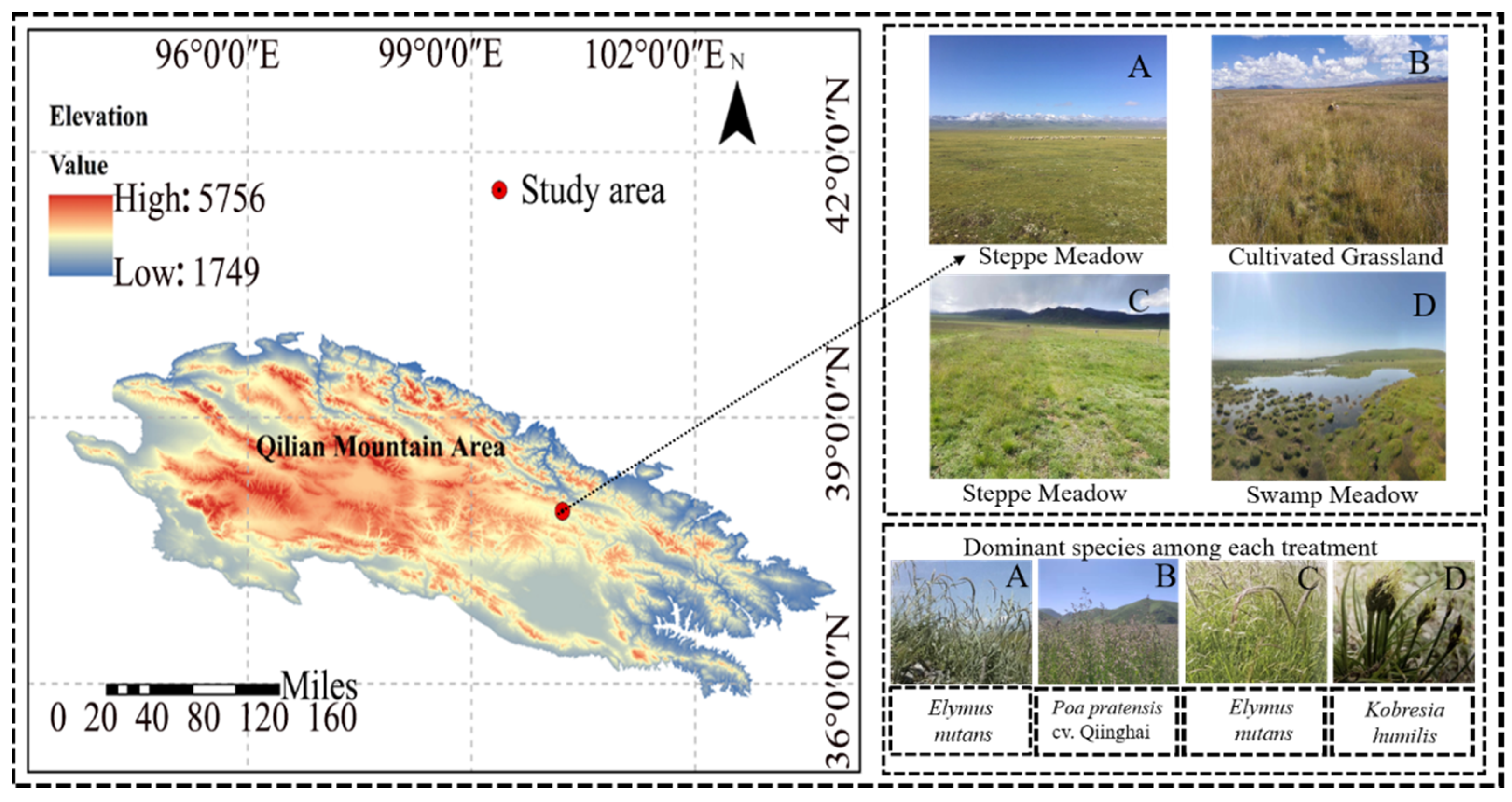
2.1. Study Area
2.2. Experimental Design, Plant Investigation, and Soil Sampling
2.3. Measurement of Soil Physicochemical Properties
2.4. Bioinformatics Analyses
2.5. Statistical Analyses
3. Results
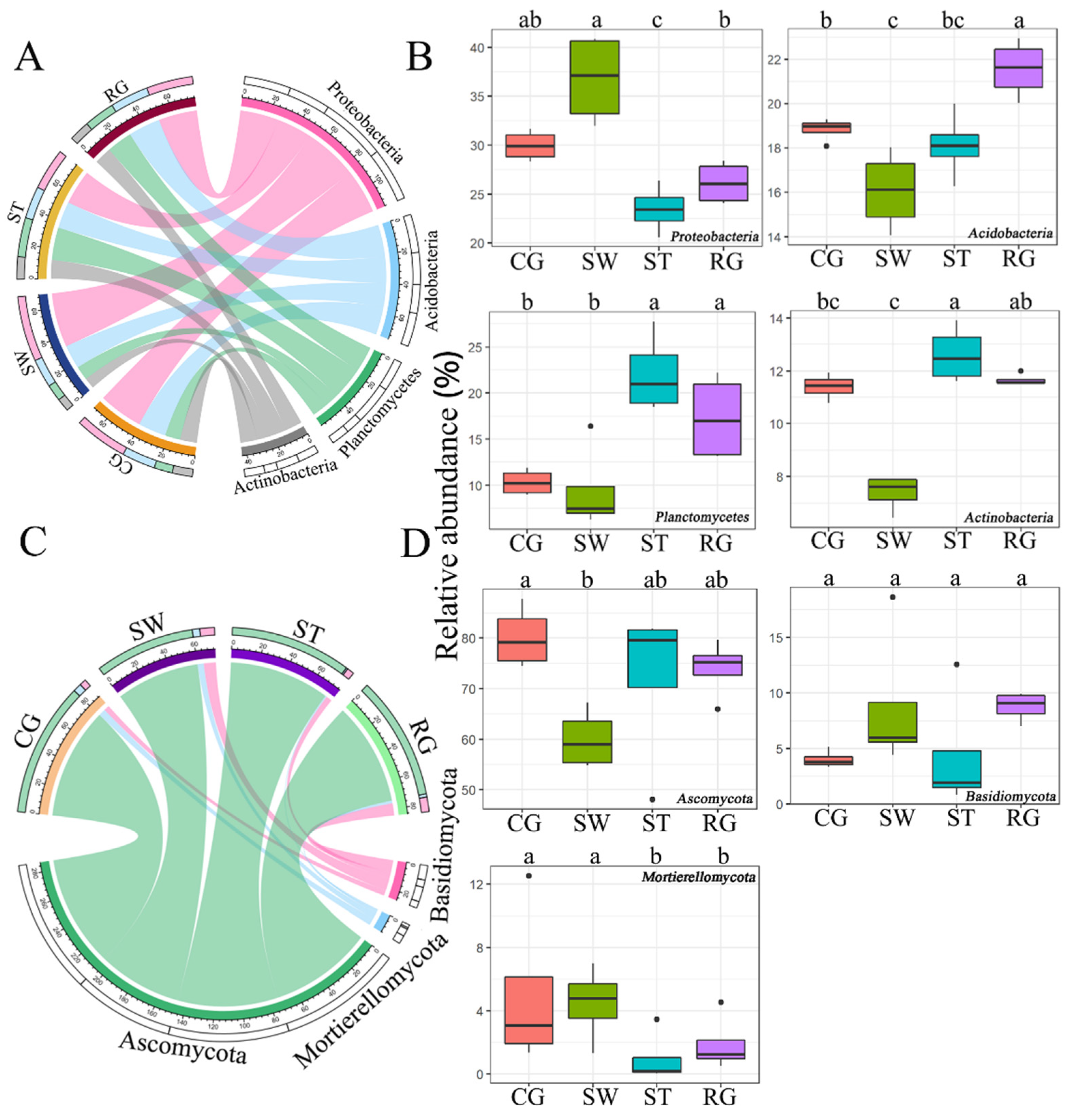
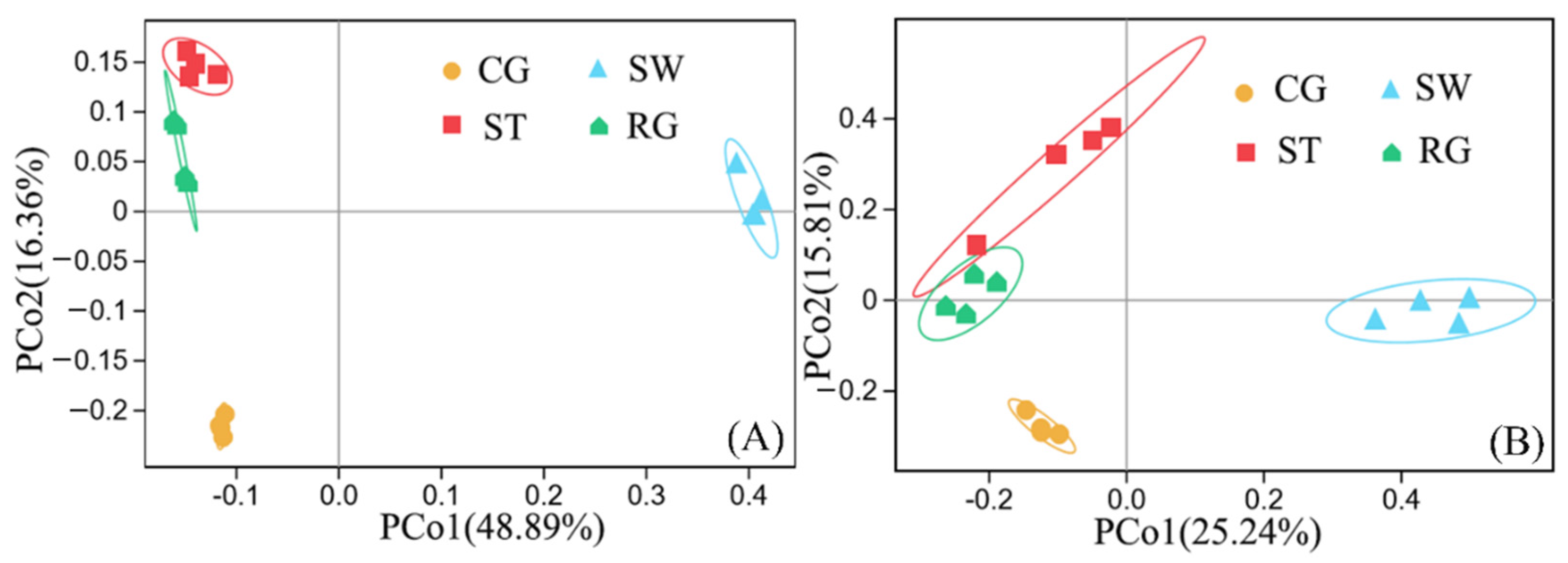
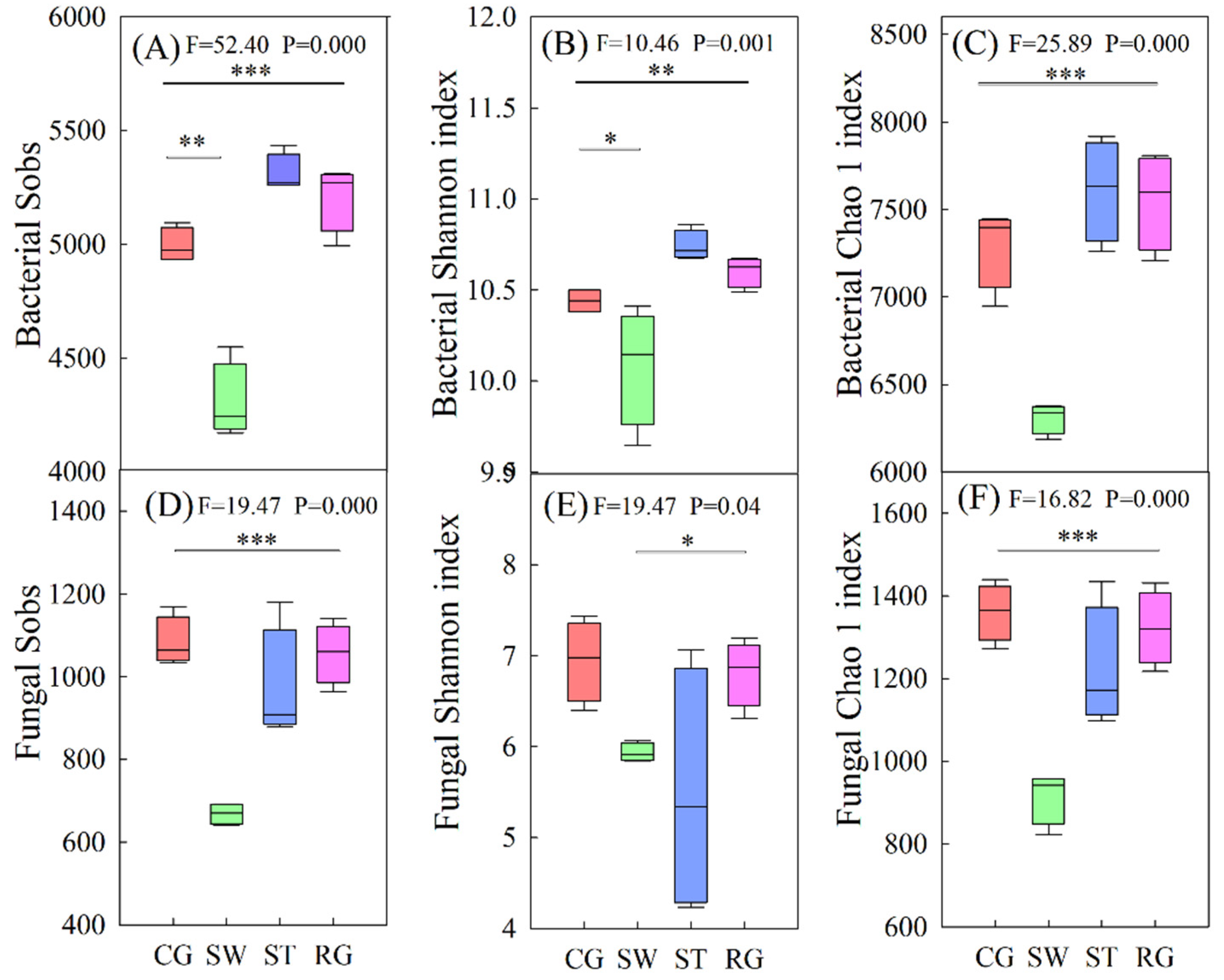
3.1. Microbial Community Composition and Diversity
3.2. Differences in Soil Physicochemical Characteristics among Different Vegetation Types
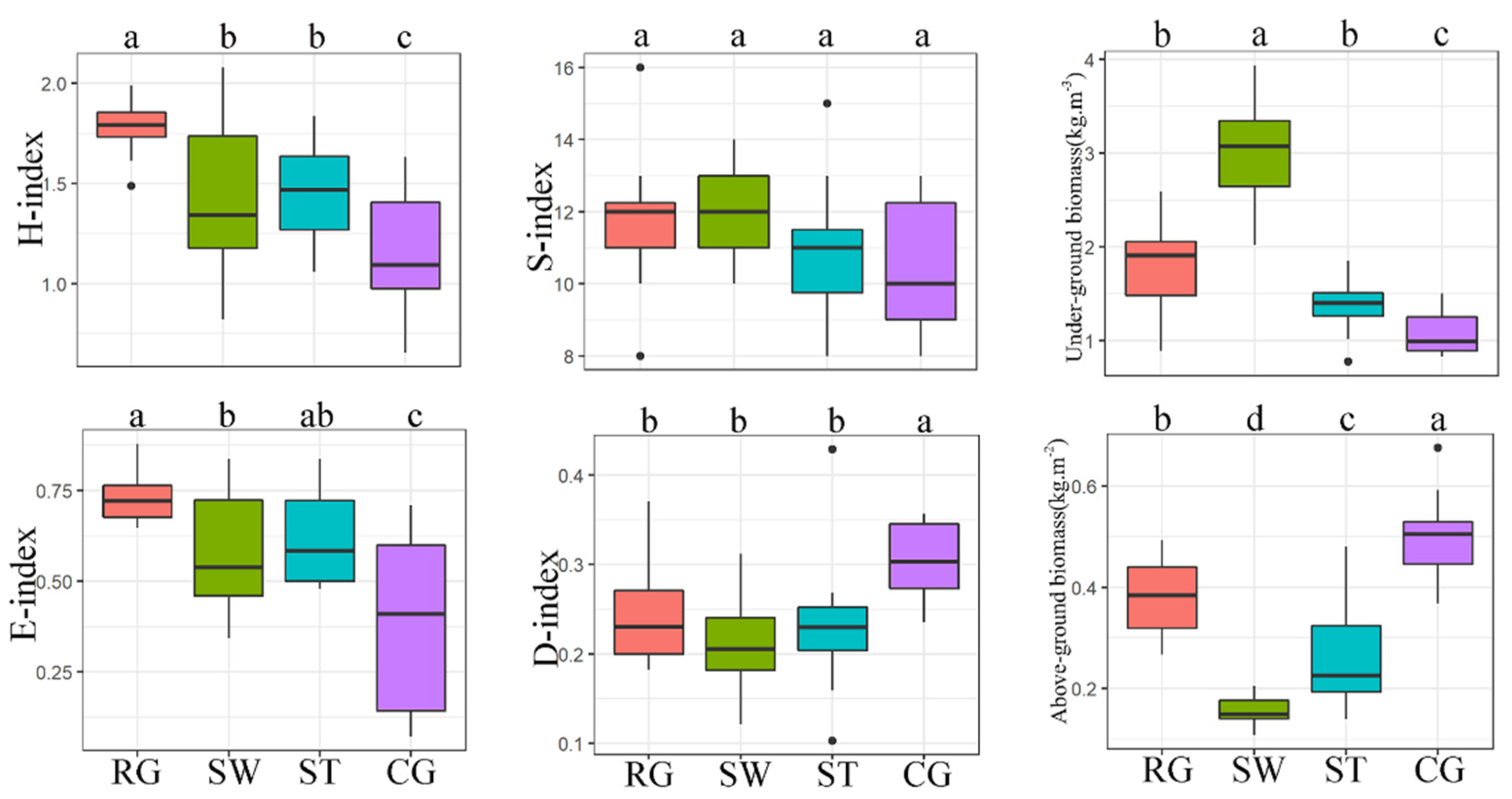
3.3. Differences in the Plant Community among Different Vegetation Types
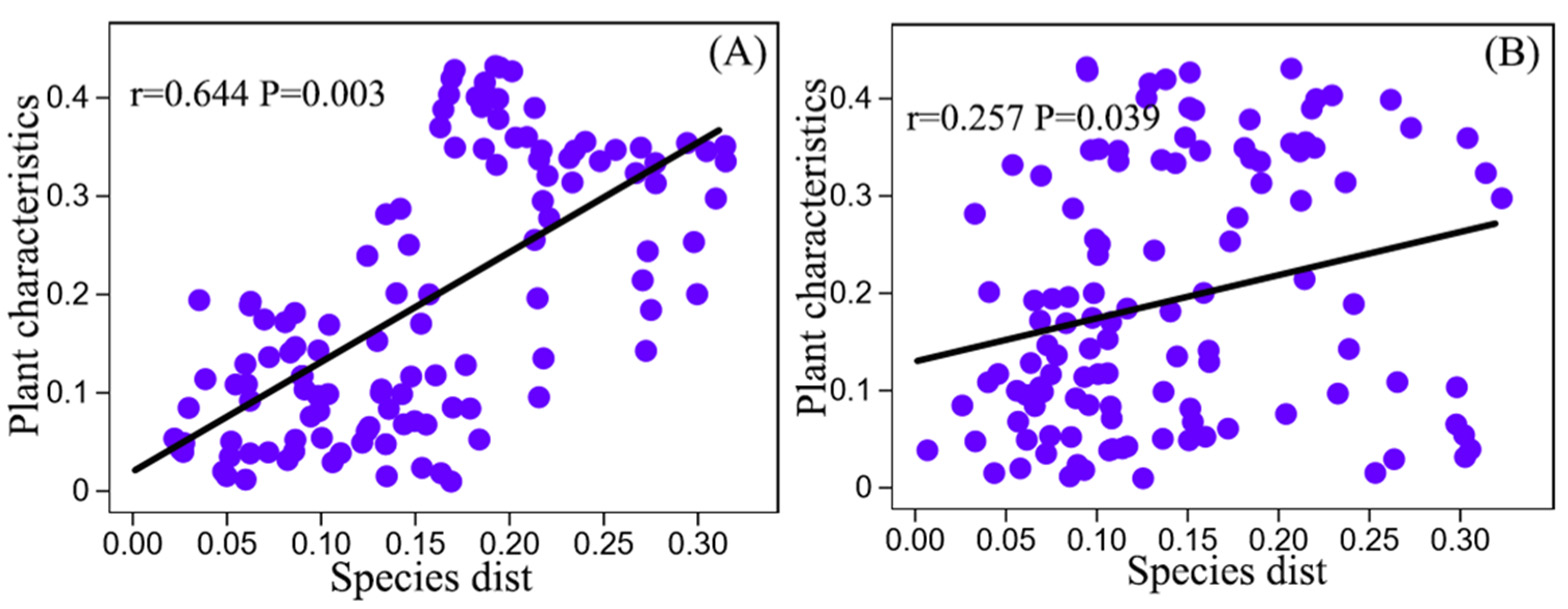
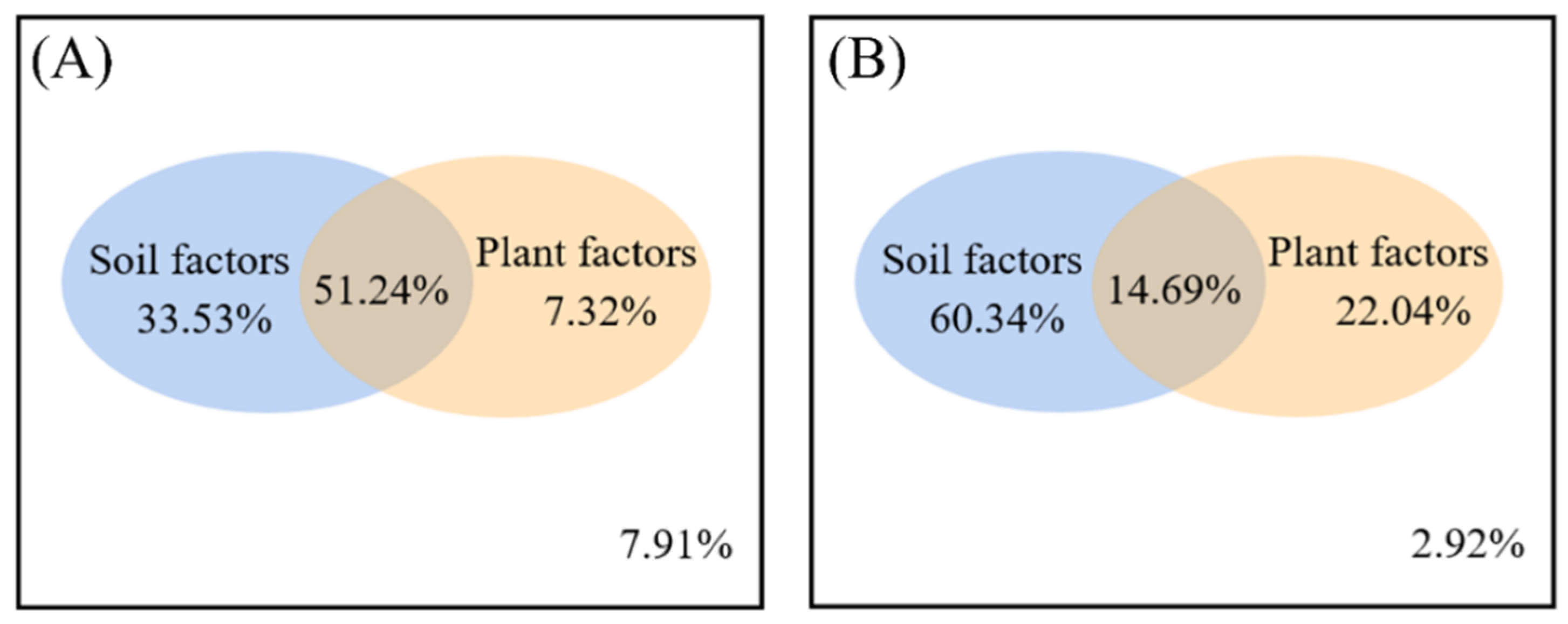
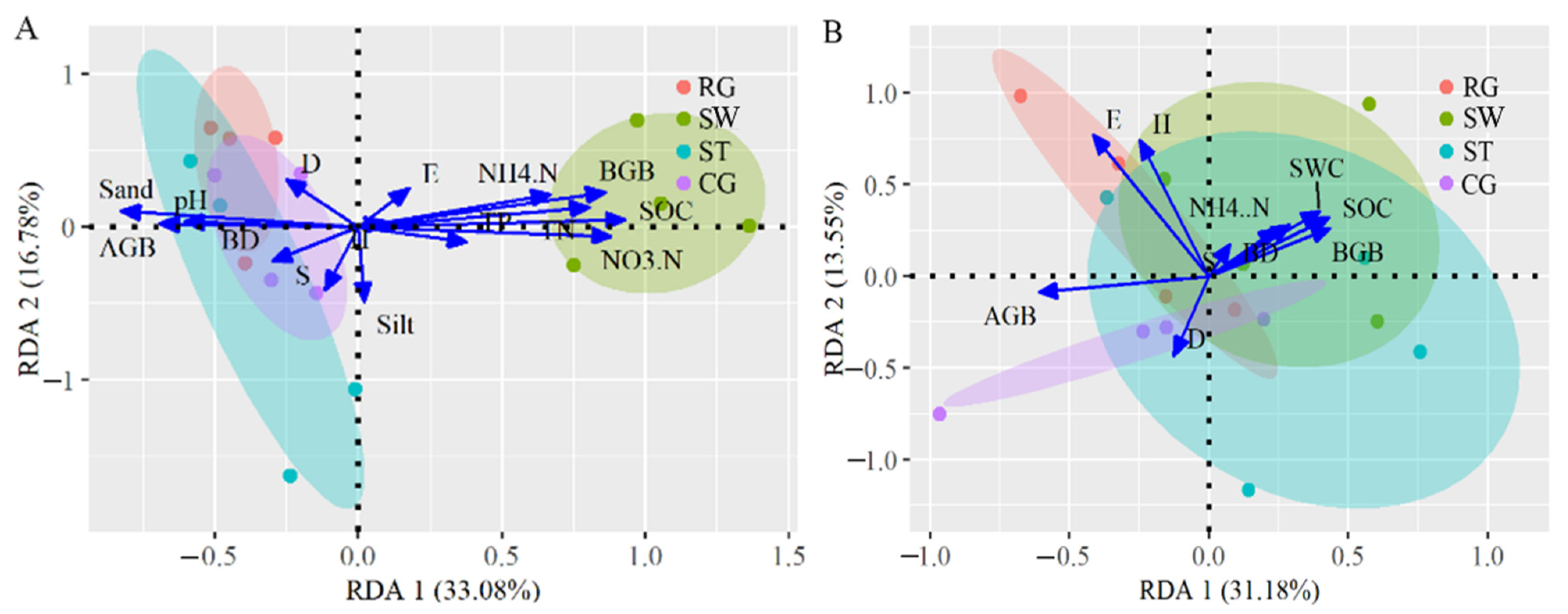
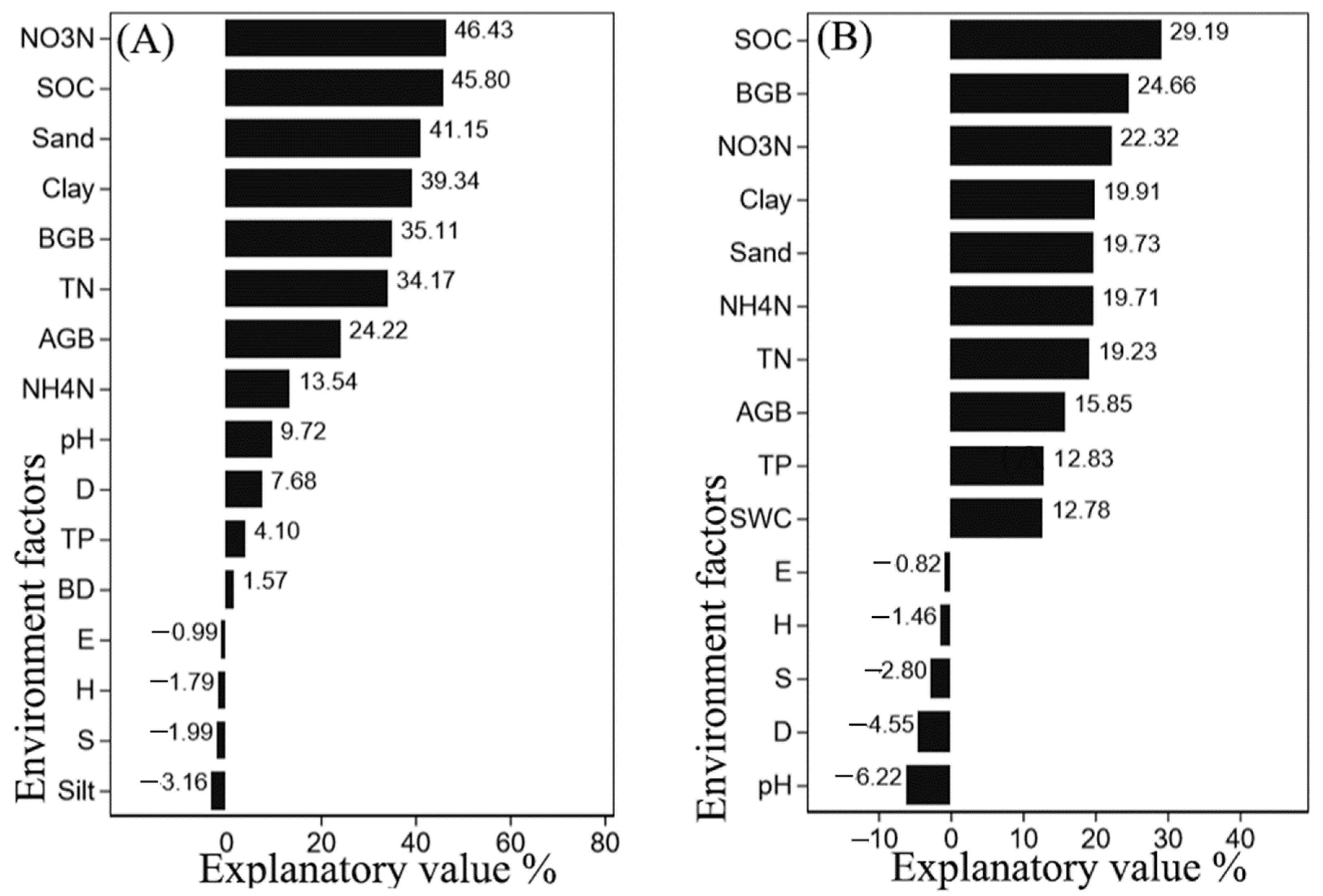
3.4. Correlation of Soil Physicochemical Properties and Plant Factors with Microbial Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, J.; Wang, H.; Holden, N.M.; Adamowski, J.F.; Biswas, A.; Zhang, X.; Feng, Q. Soil properties and microbiome of annual and perennial cultivated grasslands on the Qinghai–Tibetan Plateau. Land Degrad. Dev. 2021, 32, 5306–5321. [Google Scholar] [CrossRef]
- Franklin, R.B.; Mills, A.L. Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol. Ecol. 2003, 44, 335–346. [Google Scholar] [CrossRef] [Green Version]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; van Bodegom, P.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Maestre, F.T.; Delgado-Baquerizo, M.; Jeffries, T.C.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Quero, J.L.; García-Gómez, M.; Gallardo, A.; Ulrich, W.; et al. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. USA 2015, 112, 15684–15689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, M.; Tripathi, B.M.; Shi, Y.; Adams, J.M.; Zhu, Y.; Chu, H. Interpreting distance-decay pattern of soil bacteria via quantifying the assembly processes at multiple spatial scales. MicrobiologyOpen 2019, 8, e00851. [Google Scholar] [CrossRef] [Green Version]
- Franklin, R.B.; Mills, A.L. Importance of spatially structured environmental heterogeneity in controlling microbial community composition at small spatial scales in an agricultural field. Soil Biol. Biochem. 2009, 41, 1833–1840. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Liu, J.; Liu, T.; Cheng, J.; Wei, G.; Lin, Y. Temporal and spatial succession and dynamics of soil fungal communities in restored grassland on the Loess Plateau in China. Land Degrad. Dev. 2019, 30, 1273–1287. [Google Scholar] [CrossRef]
- Juyal, A.; Otten, W.; Falconer, R.; Hapca, S.; Schmidt, H.; Baveye, P.; Eickhorst, T. Combination of techniques to quantify the distribution of bacteria in their soil microhabitats at different spatial scales. Geoderma 2019, 334, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Wang, Z.; Niu, J.; Dang, K.; Zhang, S.; Wang, S.; Wang, Z. Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American ginseng). Plant Soil 2021, 463, 427–446. [Google Scholar] [CrossRef]
- Cruz-Paredes, C.; Tájmel, D.; Rousk, J. Can moisture affect temperature dependences of microbial growth and respiration? Soil Biol. Biochem. 2021, 156, 108223. [Google Scholar] [CrossRef]
- Xun, W.; Yan, R.; Ren, Y.; Jin, D.; Xiong, W.; Zhang, G.; Cui, Z.; Xin, X.; Zhang, R. Grazing-induced microbiome alterations drive soil organic carbon turnover and productivity in meadow steppe. Microbiome 2018, 6, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prober, S.M.; Leff, J.W.; Bates, S.T.; Borer, E.T.; Firn, J.; Harpole, W.S.; Lind, E.M.; Seabloom, E.W.; Adler, P.B.; Bakker, J.D.; et al. Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 2015, 18, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Grogan, P. Soil microbial biomass, nutrient availability and nitrogen mineralization potential among vegetation-types in a low arctic tundra landscape. Plant Soil 2009, 329, 411–420. [Google Scholar] [CrossRef]
- Dang, Z.Q.; Huang, Z.; Tian, F.P.; Liu, Y.; López-Vicente, M.; Wu, G.L. Five-year soil moisture response of typical cultivated grasslands in a semiarid area: Implications for vegetation restoration. Land Degrad. Dev. 2020, 31, 1078–1085. [Google Scholar] [CrossRef]
- Yokobe, T.; Hyodo, F.; Tateno, R.; Tokuchi, N. Linkage of fine and coarse litter traits to soil microbial characteristics and nitrogen mineralization across topographic positions in a temperate natural forest. Plant Soil 2020, 459, 261–276. [Google Scholar] [CrossRef]
- Jangid, K.; Williams, M.A.; Franzluebbers, A.J.; Schmidt, T.M.; Coleman, D.; Whitman, W.B. Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Boil. Biochem. 2011, 43, 2184–2193. [Google Scholar] [CrossRef]
- Fierer, N.; Bradfora, M.A.; Jackson, R.B. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, X.; Wu, Y.; Zhang, L.; Cheng, J.; Wei, G.; Lin, Y. Natural revegetation of a semiarid habitat alters taxonomic and functional diversity of soil microbial communities. Sci. Total Environ. 2018, 635, 598–606. [Google Scholar] [CrossRef]
- Nottingham, A.T.; Turner, B.L.; Whitaker, J.; Ostle, N.J.; McNamara, N.P.; Bardgett, R.D.; Salinas, N.; Meir, P. Soil microbial nutrient constraints along a tropical forest elevation gradient: A belowground test of a biogeochemical paradigm. Biogeosciences 2015, 12, 6071–6083. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Wang, D.; Zhang, J.; Zhou, S.; Cui, X.; Xue, K.; Wang, F.; Tang, L.; Hu, R.; Che, R.; et al. Soil microbial communities in alpine grasslands on the Tibet Plateau and their influencing factors. Chin. Sci. Bull. 2019, 64, 2915–2927. [Google Scholar] [CrossRef] [Green Version]
- Che, R.; Deng, Y.; Wang, F.; Wang, W.; Xu, Z.; Hao, Y.; Xue, K.; Zhang, B.; Tang, L.; Zhou, H.; et al. Autotrophic and symbiotic diazotrophs dominate nitrogen-fixing communities in Tibetan grassland soils. Sci. Total Environ. 2018, 639, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Holtkamp, R.; van der Wal, A.; Kardol, P.; van der Putten, W.H.; de Ruiter, P.; Dekker, S.C. Modelling C and N mineralisation in soil food webs during secondary succession on ex-arable land. Soil Biol. Biochem. 2011, 43, 251–260. [Google Scholar] [CrossRef]
- Bahram, M.; Netherway, T.; Frioux, C.; Ferretti, P.; Coelho, L.P.; Geisen, S.; Bork, P.; Hildebrand, F. Metagenomic assessment of the global diversity and distribution of bacteria and fungi. Environ. Microbiol. 2021, 23, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-G.; Wang, L.-D.; Yao, T.; Li, H.-Y.; Gao, Y.-M.; Yang, X.-M.; Li, C.-N.; Li, Q.; Feng, Y.; Hu, Y.-T. Plant community structure and species diversity differences in alpine grassland in the Qilian Mountains with different levels of degradation. Acta Prataculturae Sin. 2019, 28, 15–25. [Google Scholar]
- Yang, T.; Adams, J.; Shi, Y.; He, J.-S.; Jing, X.; Chen, L.; Tedersoo, L.; Chu, H. Soil fungal diversity in natural grasslands of the Tibetan Plateau: Associations with plant diversity and productivity. New Phytol. 2017, 215, 756–765. [Google Scholar] [CrossRef] [Green Version]
- Zhen, W.; Huiwen, Z.; Shiyin, L. Dynamic process simulation of a glacier on Qilian Mountain based on a thermo-mechanically coupled model. Sci. Total Environ. 2021, 781, 147027. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, Y.-F.; Cui, Z.; Liu, Y.; Wang, D.; Tian, F.-P.; Wu, G.-L. Natural grasslands maintain soil water sustainability better than planted grasslands in arid areas. Agric. Ecosyst. Environ. 2019, 286, 106683. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, D.; Jiang, Z.; Sun, P.; Xiao, H.; Yuxin, W.; Chen, J. Changes in the soil microbial communities of alpine steppe at Qinghai-Tibetan Plateau under different degradation levels. Sci. Total Environ. 2019, 651 Pt 2, 2281–2291. [Google Scholar] [CrossRef]
- Cao, J.; Li, G.; Adamowski, J.F.; Holden, N.M.; Deo, R.C.; Hu, Z.; Zhu, G.; Xu, X.; Feng, Q. Suitable exclosure duration for the restoration of degraded alpine grasslands on the Qinghai-Tibetan Plateau. Land Use Policy 2019, 86, 261–267. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Li, S.; Shen, F.; Jia, M.; Zhang, J.; Xu, X.; Lei, J. Soil aggregation formation in relation to planting time, water salinity, and species in the Taklimakan Desert Highway shelterbelt. J. Soils Sediments 2017, 18, 1466–1477. [Google Scholar] [CrossRef]
- Cai, Y.; Tang, Z.; Xiong, G.; Xie, Z.; Liu, Z.; Feng, X. Different composition and distribution patterns of mineral-protected versus hydrolyzable lipids in shrubland soils. J. Geophys. Res. Biogeosci. 2017, 122, 2206–2218. [Google Scholar] [CrossRef]
- Guo, M.; Wu, F.; Hao, G.; Qi, Q.; Li, R.; Li, N.; Wei, L.; Chai, T. Bacillus subtilis Improves Immunity and Disease Resistance in Rabbits. Front. Immunol. 2017, 8, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Jia, X.; Yan, W.; Zhong, Y.; Shangguan, Z. Changes in soil microbial community structure during long-term secondary succession. Land Degrad. Dev. 2020, 31, 1151–1166. [Google Scholar] [CrossRef]
- Jiang, S.; Xing, Y.; Liu, G.; Hu, C.; Wang, X.; Yan, G.; Wang, Q. Changes in soil bacterial and fungal community composition and functional groups during the succession of boreal forests. Soil Biol. Biochem. 2021, 161, 108393. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, Z.; Wang, Y.; Ma, P.; Niu, D.; Fu, H.; Elser, J.J. Soil bacterial communities vary with grassland degradation in the Qinghai Lake watershed. Plant Soil 2021, 460, 541–557. [Google Scholar] [CrossRef]
- Noah Fierer, M.A. Toward an Ecological Classification of Soil Bacteria. Ecol. Soc. Am. 2007, 88, 1354–1364. [Google Scholar]
- Delgado-Baquerizo, M.; Reith, F.; Dennis, P.G.; Hamonts, K.; Powell, J.R.; Young, A.; Singh, B.K.; Bissett, A. Ecological drivers of soil microbial diversity and soil biological networks in the Southern Hemisphere. Ecology 2018, 99, 583–596. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Rui Huang, D.Z. pH affects bacterial community composition in soils across the Huashan Watershed, China. Can. J. Microbiol. 2016, 62, 726–734. [Google Scholar] [CrossRef]
- Timonen, S.; Bomberg, M. Archaea in dry soil environments. Phytochem. Rev. 2009, 8, 505–518. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, J.; Luo, S.; Zhou, X.; Shi, S.; Tian, C. Similar soil microbial community structure across different environments after long-term succession: Evidence from volcanoes of different ages. J. Basic Microbiol. 2018, 58, 704–711. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Van Nostrand, J.D.; Deng, Y.; Lü, X.; Wang, C.; Zhou, J.; Han, X. Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China’s grasslands. FEMS Microbiol. Ecol. 2015, 91, fiv133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, M.; Niklaus, P.; Zimmermann, S.; Schmutz, S.; Kremer, J.; Abarenkov, K.; Lüscher, P.; Widmer, F.; Frey, B. Resistance and resilience of the forest soil microbiome to logging-associated compaction. ISME J. 2014, 8, 226–244. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Yan, W.; Wang, R.; Wang, W.; Shangguan, Z. Decreased occurrence of carbon cycle functions in microbial communities along with long-term secondary succession. Soil Biol. Biochem. 2018, 123, 207–217. [Google Scholar] [CrossRef]
- Connell, J.H. Diversity in Tropical Rain Forests and Coral Reefs. Science 1978, 199, 1302–1310. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.-L.; Liu, Z.-H.; Zhang, L.; Hu, T.-M.; Chen, J.-M. Effects of artificial grassland establishment on soil nutrients and carbon properties in a black-soil-type degraded grassland. Plant Soil 2010, 333, 469–479. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Boil. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Ren, C.; Zhang, W.; Zhong, Z.; Han, X.; Yang, G.; Feng, Y.; Ren, G. Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Sci. Total Environ. 2018, 610–611, 750–758. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Delgado-Baquerizo, M.; Wang, J.-T.; Hu, H.-W.; Cai, Z.-J.; Zhu, Y.-N.; Singh, B.K. Fungal richness contributes to multifunctionality in boreal forest soil. Soil Biol. Biochem. 2019, 136, 107526. [Google Scholar] [CrossRef]
- Buyer, J.S.; Roberts, D.P.; Russek-Cohen, E. Soil and plant effects on microbial community structure. Can. J. Microbiol. 2002, 48, 955–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Wang, C.; Jiang, L.; Luo, Y. Trends in soil microbial communities during secondary succession. Soil Biol. Biochem. 2017, 115, 92–99. [Google Scholar] [CrossRef]
- You, Y.; Wang, J.; Huang, X.; Tang, Z.; Liu, S.; Sun, O.J. Relating microbial community structure to functioning in forest soil organic carbon transformation and turnover. Ecol. Evol. 2014, 4, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Lajtha, K.; Bowden, R.D.; Crow, S.; Fekete, I.; Kotroczo, Z.; Plante, A.; Simpson, M.; Nadelhoffer, K. The Detrital Input and Removal Treatment (DIRT) Network. Sci. Total Environ. 2018, 640, 1112–1120. [Google Scholar] [CrossRef]
- Castle, S.C.; Nemergut, D.R.; Grandy, A.S.; Leff, J.W.; Graham, E.B.; Hood, E.; Schmidt, S.K.; Wickings, K.; Cleveland, C.C. Biogeochemical drivers of microbial community convergence across actively retreating glaciers. Soil Biol. Biochem. 2016, 101, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Collavino, M.M.; Tripp, H.J.; Frank, I.E.; Vidoz, M.L.; Calderoli, P.A.; Donato, M.; Zehr, J.P.; Aguilar, O.M. nifH pyrosequencing reveals the potential for location-specific soil chemistry to influence N2 -fixing community dynamics. Env. Microbiol. 2014, 16, 3211–3223. [Google Scholar] [CrossRef] [Green Version]
- Mooshammer, M.; Wanek, W.; Zechmeister-Boltenstern, S.; Richter, A.A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: Mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 2014, 5, 22. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Chen, Q.; Liu, L.; Wen, X.; Liao, Y. Responses of soil fungi to 5-year conservation tillage treatments in the drylands of northern China. Appl. Soil Ecol. 2016, 101, 132–140. [Google Scholar] [CrossRef]
| Grassland Types | RG | SW | ST | CG | Bacteria | Fungi | ||
|---|---|---|---|---|---|---|---|---|
| r | p | r | p | |||||
| NH4+-N (mg/kg−1) a | 1.56 ± 0.37 c | 3.06 ± 1.37 a | 1.95 ± 1.34 b | 1.45 ± 0.57 c | 0.4578 | 0.019 | 0.5089 | 0.009 |
| NO3−-N (mg/kg−1) a | 1.94 ± 0.36 b | 5.79 ± 1.95 a | 2.21 ± 0.52 b | 1.74 ± 0.31 b | 0.8641 | 0.001 | 0.6676 | 0.003 |
| SOC (g/kg−1) a | 19.45 ± 1.24 c | 172.68 ± 19.84 a | 41.80 ± 5.26 b | 28.51 ± 6.22 bc | 0.9426 | 0.001 | 0.6808 | 0.001 |
| TN (g/kg−1) a | 1.52 ± 0.49 b | 6.68 ± 1.76 a | 2.62 ± 0.70 b | 2.36 ± 0.50 b | 0.7461 | 0.001 | 0.8318 | 0.001 |
| TP (g/kg−1) a | 0.61 ± 0.02 c | 1.09 ± 0.06 a | 1.05 ± 0.03 a | 0.91 ± 0.04 b | 0.4985 | 0.019 | 0.5507 | 0.007 |
| pH | 7.96 ± 0.37 b | 7.21 ± 0.23 b | 7.31 ± 0.14 a | 7.93 ± 0.14 a | 0.5772 | 0.004 | 0.4863 | 0.010 |
| BD (g/cm3) a | 1.48 ± 0.13 a | 1.05 ± 0.26 b | 1.39 ± 0.12 a | 0.88 ± 0.06 b | 0.0388 | 0.772 | 0.3394 | 0.067 |
| SWC (%) | 13.89 ± 1.69 c | 49.19 ± 2.28 a | 18.34 ± 0.85 b | 16.20 ± 1.69 bc | 0.5582 | 0.005 | 0.2484 | 0.151 |
| Sand (%) a | 70.56 ± 11.15 ab | 43.66 ± 7.48 c | 64.93 ± 3.51 b | 79.91 ± 6.85 a | 0.7721 | 0.001 | 0.5185 | 0.010 |
| Silt (%) a | 1.26 ± 0.59 ab | 3.35 ± 1.37 c | 6.29 ± 1.84 b | 2.71 ± 0.61 a | 0.2189 | 0.214 | 0.334 | 0.069 |
| Clay (%) a | 27.42 ± 9.86 b | 52.99 ± 8.84 a | 30.89 ± 3.33 b | 13.80 ± 1.01c | 0.6240 | 0.003 | 0.5694 | 0.007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Yin, Y.; Li, S.; Liu, J.; Dong, Y.; Su, S. Soil Microbial Community Varied with Vegetation Types on a Small Regional Scale of the Qilian Mountains. Sustainability 2022, 14, 7910. https://doi.org/10.3390/su14137910
Zhao W, Yin Y, Li S, Liu J, Dong Y, Su S. Soil Microbial Community Varied with Vegetation Types on a Small Regional Scale of the Qilian Mountains. Sustainability. 2022; 14(13):7910. https://doi.org/10.3390/su14137910
Chicago/Turabian StyleZhao, Wen, Yali Yin, Shixiong Li, Jingjing Liu, Yiling Dong, and Shifeng Su. 2022. "Soil Microbial Community Varied with Vegetation Types on a Small Regional Scale of the Qilian Mountains" Sustainability 14, no. 13: 7910. https://doi.org/10.3390/su14137910





